Methods for Engineering Binders to Multi-Pass Membrane Proteins
Abstract
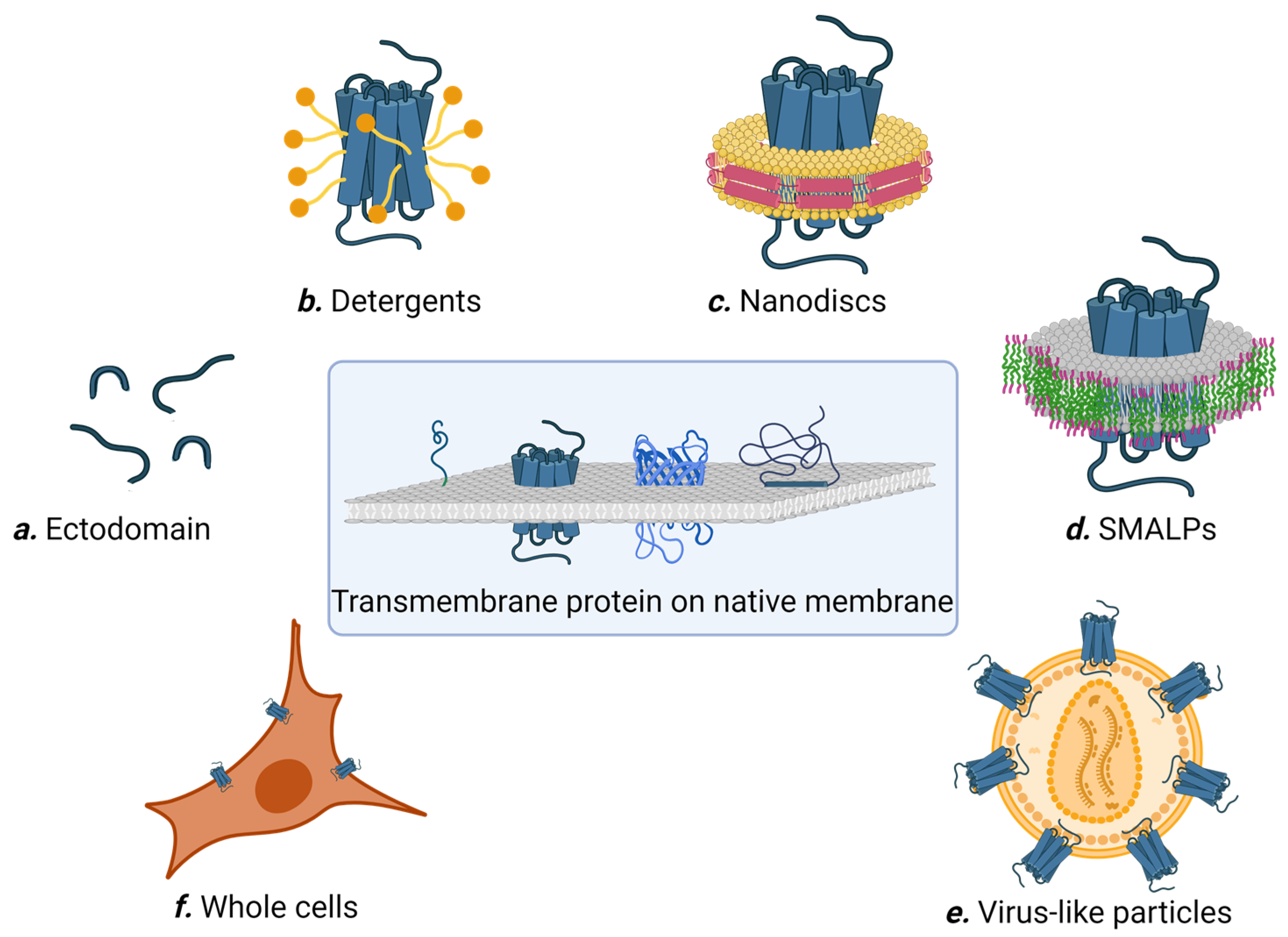
:1. Introduction
2. Formats of Multi-Pass Membrane Proteins for Affinity Selection
2.1. Soluble Extracellular Loop Fragment
2.2. Detergents
2.3. Nanodiscs
2.4. SMALPs
2.5. Virus-Like Particles (VLPs)
2.6. Whole Cells
3. Conclusions
| Name | Drug | Target | Method | Library | Approval | Note | Refs |
|---|---|---|---|---|---|---|---|
| Tositumomab-I131-2013 | Bexxar | CD20 | WC | Hybridoma | 2003 # | Labeled with I131 | [73,74] |
| Rituximab | MabThera, Rituxan | CD20 | WC | Hybridoma | 1997 | CD20-binding mediated by 2B8 | [75] |
| Ibritumomab tiuxetan | Zevalin | CD20 | WC | Hybridoma | 2002 | Rituximab with tiuxetan attached | [76] |
| Ofatumumab (HuMax-CD20) | Arzerra | CD20 | WC | Hybridoma | 2009 | Fully human antibody from transgenic mice | [77] |
| Obinutuzumab (GA101) | Gazyva, Gazyvaro | CD20 | ECL | n/a | 2013 | Glycoengineered murine antibody B-ly1. | [78] |
| Ocrelizumab | OCREVUS | CD20 | n/a | n/a | 2017 | Fc engineered, humanized from 2H7 | [79] |
| Ublituximab (LFB-R603) | BRIUMVI | CD20 | n/a | Hybridoma | 2022 | Glycoengineered | [80,81] |
| Mosunetuzumab (CD20-TDB) | Lunsumio | CD20 | n/a | n/a | 2022 | CD20xCD3 bispecific, CD20-binding mediated by 2H7 | [82] |
| Epcoritamab (GEN3013) | EPKINLY | CD20 | WC | Hybridoma | 2023 | CD20xCD3 bispecific, CD20-binding mediated by 7D8 | [83,84] |
| Glofitamab (RG6026) | Columvi | CD20 | n/a | n/a | 2023 | CD20xCD3 (2:1 format) bispecific | [85] |
| Mogamulizumab (KW-0761) | Poteligeo | CCR4 | ECL | Hybridoma | 2018 | Glycoengineered | [86,87] |
| Talquetamab (JNJ-64407564) | TALVEY | GPRC-5D | WC | Hybridoma | 2023 | GPRC-5DxCD3 bispecific | [88,89] |
| Target | Format | Library | Name | Affinity | Function | Refs |
|---|---|---|---|---|---|---|
| CD20 | Extracellular loop | Naïve human scFv phage library | G7 | KD~64 nM | Marker on B cells | [33] |
| ORF3a | Extracellular loop | Naïve human scFv phage library | N3aB02 3aCA03 | KD~nM | Viroporin of SARS-CoV-2 | [34] |
| NorC | Detergent | Yeast library from immunized camel | ICab3 | Kd~nM | Multidrug efflux transporter of Staphylococcus aureus | [35,90] |
| BamA | Detergent | Phage library from immunized alpaca | 21 clones | Kd~nM | Insertase of Gram-negative bacteria | [36] |
| TM287/288 | Detergent | Phage library from immunized alpacas | 29 families | KD~nM-pM | ATP-binding cassette transporter | [37] |
| MOMP | Detergent | Synthetic nanobody phage library | 5 sybodies | n/a | Major outer membrane protein of Legionella pneumophila serogroup 6 | [37] |
| NTCP | Detergent | Mouse hybridoma | N6HB426-20 | IC50~10 nM | Sodium taurocholate cotransporting polypeptide; HBV/HDV entry receptor | [38] |
| TM287/288 | Detergent | Synthetic nanobody phage library | 40 sybodies | IC50 62 nM | ATP-binding cassette transporter | [39] |
| GlyT1 | Detergent | Synthetic nanobody phage library | 7 sybodies | KD~pM-μM | Glycine transporter 1; roles in diseases of the central and peripheral nervous system | [39] |
| ENT1 | Detergent | Synthetic nanobody phage library | Sb_ENT1#1 | KD 40 nM | Equilibrative nucleoside transporter 1; roles in ischemia; biomarker of pancreatic cancer | [39] |
| APJ | Nanodiscs | Phage library from immunized camel | JN241 | Kd 83 pM | Human Apelin Receptor; mediates fluid homeostasis and cardiovascular function | [42] |
| Influenza Matrix-2 | Nanodiscs | Phage library from immunized shark | AM2H10 | KD 78 nM | Proton channel; required for virus uncoating in endosomes | [43] |
| ASIC1a | Nanodiscs | Naïve human scFv phage library | ASC06-IgG1 | Kd 7.9 nM | Key ASIC protein activated in neuronal injury | [44] |
| Mj0480 | Nanodiscs | Synthetic Fab phage library | 14 clones | KD~nM | YidC homolog from Methanocaldococcus jannaschii | [45] |
| CorA | Nanodiscs | Synthetic Fab phage library | 10 clones | KD~nM | Magnesium ion channel from Thermotoga maritima | [45] |
| VSD4-NavAb | Nanodiscs | Mouse hybridoma | 141B8 | n/a | Voltage-sensor domain 4 of human Nav1.7 fused to voltage-gated sodium channel from Acrobacter butzleri | [46] |
| GLUT4 | VLP | ScFv phage library from immunized chicken | 29 clones | Kd pM–nM | Glucose transporter type 4; roles in diabetes and obesity | [59] |
| CLDN6 | VLP | ScFv phage library from immunized chicken | 6 mAbs | KD pM–nM | Claudin 6; tumor-associated antigen | [61] |
| CLDN6 | VLP | Mouse hybridoma | Polyclonal sera | n/a | Claudin 6; tumor-associated antigen | [60] |
| GCGR | VLP | Fab phage library from immunized llamas | 10 VH families | KD~nM | GPCR glucagon receptor; roles in metabolism and homeostasis | [62] |
| Alexandrium minutum | Whole cell | Pre-immune nanobody phage library | 4 clones | n/a | Toxic species of dinoflagellates that can cause paralytic shellfish poisoning | [63] |
| AGS cells | Whole cell | Semisynthetic human scFv phage library | 14 clones | n/a | Cells isolated from patient with gastric cancer | [64] |
| EGFR | Whole cell | Synthetic peptide phage library | 11 peptides | IC50~µM | Epidermal growth factor receptor; roles in regulation of cell proliferation, differentiation, and migration | [65] |
| CCR6 | Whole cells | Mouse hybridoma | 1C6 | IC50 10 nM | C-C chemokine receptor type 6; roles in maintaining leukocyte homeostasis and inflammation | [66] |
| CD20 | Whole cells | Fab phage library from immunized chicken | 4 mAbs | EC50 12–30 nM | Cluster of differentiate 20; marker on B cells | [67,68] |
| CCR5 | Whole cell | Naïve scFv phage libraries | 5 mAbs | KD~4 nM | C-C chemokine receptor type 5; co-receptor of HIV | [69] |
| hMOR | Whole cell | Naïve scFv yeast libraries | 2 clones | KD~nM | Human GPCR mu opioid receptor | [70] |
| GLP-1 R | Whole cells | Synthetic GPCR-focused scFv phage library | TB01-3 | IC50~5 nM | Glucagon-like peptide-1 receptor; receptor for incretin GLP-1 | [71] |
| Fzd7 | Whole cells | Synthetic Fab phage library | 3 clones | Kd~pM | Human Frizzled-7; roles in the Wnt signaling pathway | [72] |
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhao, H.; Dodds, M.; Tasch, M.; Gewe, M.; Martinez, A.; Hutton, M.; Keeney, K.; Pollock, A.; Jester, B.W.; Khuong, N.; et al. Using synthetic activity to design ultra-potent antibody cocktails. bioRxiv 2022. [Google Scholar] [CrossRef]
- Mankarious, S.; Lee, M.; Fischer, S.; Pyun, K.H.; Ochs, H.D.; Oxelius, V.A.; Wedgwood, R.J. The half-lives of IgG subclasses and specific antibodies in patients with primary immunodeficiency who are receiving intravenously administered immunoglobulin. J. Lab. Clin. Med. 1988, 112, 634–640. [Google Scholar] [PubMed]
- Griffin, M.P.; Yuan, Y.; Takas, T.; Domachowske, J.B.; Madhi, S.A.; Manzoni, P.; Simoes, E.A.F.; Esser, M.T.; Khan, A.A.; Dubovsky, F.; et al. Single-Dose Nirsevimab for Prevention of RSV in Preterm Infants. N. Engl. J. Med. 2020, 383, 415–425. [Google Scholar] [CrossRef] [PubMed]
- van Erp, E.A.; Luytjes, W.; Ferwerda, G.; van Kasteren, P.B. Fc-Mediated Antibody Effector Functions During Respiratory Syncytial Virus Infection and Disease. Front. Immunol. 2019, 10, 548. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.H.; Jung, S.T. Boosting therapeutic potency of antibodies by taming Fc domain functions. Exp. Mol. Med. 2019, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Pang, X.; Zhong, T.; Qu, T.; Chen, N.; Ma, S.; He, X.; Xia, D.; Wang, M.; Xia, M.; et al. Penpulimab, an Fc-Engineered IgG1 Anti-PD-1 Antibody, With Improved Efficacy and Low Incidence of Immune-Related Adverse Events. Front. Immunol. 2022, 13, 924542. [Google Scholar] [CrossRef]
- Coppo, P.; Joly, B.S.; French Reference Center for Thrombotic, M. Caplacizumab: A game changer also in pregnancy-associated immune-mediated thrombotic thrombocytopenic purpura? Br. J. Haematol. 2023, 202, 725–727. [Google Scholar] [CrossRef]
- Picod, A.; Benhamou, Y.; Bouzid, R.; Veyradier, A.; Coppo, P. More on the use of frontline caplacizumab in immune-mediated thrombotic thrombocytopenic purpura. Blood Adv. 2023, 7, 2678–2680. [Google Scholar] [CrossRef]
- Simeon, R.; Chen, Z. In vitro-engineered non-antibody protein therapeutics. Protein Cell 2018, 9, 3–14. [Google Scholar] [CrossRef]
- Nagorsen, D.; Kufer, P.; Baeuerle, P.A.; Bargou, R. Blinatumomab: A historical perspective. Pharmacol. Ther. 2012, 136, 334–342. [Google Scholar] [CrossRef]
- Aldoss, I.; Bargou, R.C.; Nagorsen, D.; Friberg, G.R.; Baeuerle, P.A.; Forman, S.J. Redirecting T cells to eradicate B-cell acute lymphoblastic leukemia: Bispecific T-cell engagers and chimeric antigen receptors. Leukemia 2017, 31, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Scully, M.; Cataland, S.R.; Peyvandi, F.; Coppo, P.; Knobl, P.; Kremer Hovinga, J.A.; Metjian, A.; de la Rubia, J.; Pavenski, K.; Callewaert, F.; et al. Caplacizumab Treatment for Acquired Thrombotic Thrombocytopenic Purpura. N. Engl. J. Med. 2019, 380, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Unverdorben, F.; Hutt, M.; Seifert, O.; Kontermann, R.E. A Fab-Selective Immunoglobulin-Binding Domain from Streptococcal Protein G with Improved Half-Life Extension Properties. PLoS ONE 2015, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Siegemund, M.; Oak, P.; Hansbauer, E.M.; Allersdorfer, A.; Utschick, K.; Winter, A.; Grasmuller, C.; Galler, G.; Mayer, J.P.; Weiche, B.; et al. Pharmacokinetic Engineering of OX40-Blocking Anticalin Proteins Using Monomeric Plasma Half-Life Extension Domains. Front. Pharmacol. 2021, 12, 759337. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Feng, Y.; Cai, H.; Jia, D.; Li, H.; Tao, Z.; Zhong, Y.; Li, Z.; Shi, Q.; Wan, L.; et al. Endogenous IgG-based affinity-controlled release of TRAIL exerts superior antitumor effects. Theranostics 2018, 8, 2459–2476. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Yang, Y.; Zhang, D.; Du, J.; Zhu, X.; Liu, Y.; Yang, F.; Lin, J. A bispecific immunotoxin (IHPP) with a long half-life targeting HER2 and PDGFRbeta exhibited improved efficacy against HER2-positive tumors in a mouse xenograft model. Int. J. Pharm. 2021, 592, 120037. [Google Scholar] [CrossRef] [PubMed]
- Steiner, D.; Werz, F.W.; Sonderegger, I.; Goulotti-Georgieva, M.; Villemagne, D.; Phillips, D.J.; Forrer, P.; Stumpp, M.T.; Zitt, C.; Binz, H.K. Half-life extension using serum albumin-binding DARPin domains. Protein Eng. Des. Sel. 2017, 30, 1. [Google Scholar] [CrossRef]
- Fishburn, C.S. The pharmacology of PEGylation: Balancing PD with PK to generate novel therapeutics. J. Pharm. Sci. 2008, 97, 4167–4183. [Google Scholar] [CrossRef]
- Bech, E.M.; Kaiser, A.; Bellmann-Sickert, K.; Nielsen, S.S.R.; Sorensen, K.K.; Elster, L.; Hatzakis, N.; Pedersen, S.L.; Beck-Sickinger, A.G.; Jensen, K.J. Half-Life Extending Modifications of Peptide YY3-36 Direct Receptor-Mediated Internalization. Mol Pharm. 2019, 16, 3665–3677. [Google Scholar] [CrossRef]
- Kearney, J.F.; Radbruch, A.; Liesegang, B.; Rajewsky, K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J. Immunol. 1979, 123, 1548–1550. [Google Scholar] [CrossRef]
- Stapleton, S.; O’ Kennedy, R.; Tully, E. IMMUNOASSAYS|Production of Antibodies. In Encyclopedia of Analytical Science, 2nd ed.; Worsfold, P., Townshend, A., Poole, C., Eds.; Elsevier: Oxford, UK, 2005; pp. 306–316. [Google Scholar]
- Clackson, T.; Hoogenboom, H.R.; Griffiths, A.D.; Winter, G. Making antibody fragments using phage display libraries. Nature 1991, 352, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Shusta, E.V.; Holler, P.D.; Kieke, M.C.; Kranz, D.M.; Wittrup, K.D. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat. Biotechnol. 2000, 18, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Seelig, B. mRNA display for the selection and evolution of enzymes from in vitro-translated protein libraries. Nat. Protoc. 2011, 6, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Eguchi, M.; Kito, S.; Fujino, T.; Hayashi, G.; Murakami, H. cDNA TRAP display for rapid and stable in vitro selection of antibody-like proteins. Chem. Commun. Camb. 2021, 57, 2416–2419. [Google Scholar] [CrossRef] [PubMed]
- Ishizawa, T.; Kawakami, T.; Reid, P.C.; Murakami, H. TRAP display: A high-speed selection method for the generation of functional polypeptides. J. Am. Chem. Soc. 2013, 135, 5433–5440. [Google Scholar] [CrossRef]
- Zeng, Y.; Woolley, M.; Chockalingam, K.; Thomas, B.; Arora, S.; Hook, M.; Chen, Z. Click display: A rapid and efficient in vitro protein display method for directed evolution. Nucleic Acids Res. 2023, 51, e89. [Google Scholar] [CrossRef] [PubMed]
- Hie, B.L.; Shanker, V.R.; Xu, D.; Bruun, T.U.J.; Weidenbacher, P.A.; Tang, S.; Wu, W.; Pak, J.E.; Kim, P.S. Efficient evolution of human antibodies from general protein language models. Nat. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Verkuil, R.; Kabeli, O.; Du, Y.; Wicky, B.I.M.; Milles, L.F.; Dauparas, J.; Baker, D.; Ovchinnikov, S.; Sercu, T.; Rives, A. Language models generalize beyond natural proteins. bioRxiv 2022. [Google Scholar] [CrossRef]
- Hie, B.; Candido, S.; Lin, Z.; Kabeli, O.; Rao, R.; Smetanin, N.; Sercu, T.; Rives, A. A high-level programming language for generative protein design. bioRxiv 2022. [Google Scholar] [CrossRef]
- Carlander, D.; Stalberg, J.; Larsson, A. Chicken antibodies: A clinical chemistry perspective. Ups. J. Med. Sci. 1999, 104, 179–189. [Google Scholar] [CrossRef]
- Ching, K.H.; Collarini, E.J.; Abdiche, Y.N.; Bedinger, D.; Pedersen, D.; Izquierdo, S.; Harriman, R.; Zhu, L.; Etches, R.J.; van de Lavoir, M.C.; et al. Chickens with humanized immunoglobulin genes generate antibodies with high affinity and broad epitope coverage to conserved targets. MAbs 2018, 10, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Shams, N.; Khoshtinat Nikkhoi, S.; Gu, Z.; Rahbarizadeh, F. Isolation and characterization of human anti-CD20 single-chain variable fragment (scFv) from a Naive human scFv library. Med. Oncol. 2022, 39, 177. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.H.; Patton, E.; Munro, C.A.; Corzo-Leon, D.E.; Porter, A.J.; Palliyil, S. Monoclonal Human Antibodies That Recognise the Exposed N and C Terminal Regions of the Often-Overlooked SARS-CoV-2 ORF3a Transmembrane Protein. Viruses 2021, 13, 2201. [Google Scholar] [CrossRef]
- Kumar, S.; Mahendran, I.; Athreya, A.; Ranjan, R.; Penmatsa, A. Isolation and structural characterization of a Zn(2+)-bound single-domain antibody against NorC, a putative multidrug efflux transporter in bacteria. J. Biol. Chem. 2020, 295, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Hartmann, J.B.; Jakob, R.P.; Zahn, M.; Zimmermann, I.; Maier, T.; Seeger, M.A.; Hiller, S. Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J. Biomol. Nmr. 2019, 73, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Egloff, P.; Zimmermann, I.; Arnold, F.M.; Hutter, C.A.J.; Morger, D.; Opitz, L.; Poveda, L.; Keserue, H.A.; Panse, C.; Roschitzki, B.; et al. Engineered peptide barcodes for in-depth analyses of binding protein libraries. Nat. Methods 2019, 16, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Takemori, T.; Sugimoto-Ishige, A.; Nishitsuji, H.; Futamura, Y.; Harada, M.; Kimura-Someya, T.; Matsumoto, T.; Honma, T.; Tanaka, M.; Yaguchi, M.; et al. Establishment of a Monoclonal Antibody against Human NTCP That Blocks Hepatitis B Virus Infection. J. Virol. 2022, 96, e0168621. [Google Scholar] [CrossRef]
- Zimmermann, I.; Egloff, P.; Hutter, C.A.; Arnold, F.M.; Stohler, P.; Bocquet, N.; Hug, M.N.; Huber, S.; Siegrist, M.; Hetemann, L.; et al. Synthetic single domain antibodies for the conformational trapping of membrane proteins. eLife 2018, 7, e34317. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Sligar, S.G. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010, 584, 1721–1727. [Google Scholar] [CrossRef]
- Denisov, I.G.; Sligar, S.G. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 481–486. [Google Scholar] [CrossRef]
- Ma, Y.; Ding, Y.; Song, X.; Ma, X.; Li, X.; Zhang, N.; Song, Y.; Sun, Y.; Shen, Y.; Zhong, W.; et al. Structure-guided discovery of a single-domain antibody agonist against human apelin receptor. Sci. Adv. 2020, 6, eaax7379. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Ding, W.; Zhu, L.; Zhou, Y.; Dong, Y.; Li, L.; Liu, J.; Wang, Y.; Li, Z.; Zhu, L.; et al. Screening and characterization of inhibitory vNAR targeting nanodisc-assembled influenza M2 proteins. iScience 2023, 26, 105736. [Google Scholar] [CrossRef] [PubMed]
- Qiang, M.; Dong, X.; Zha, Z.; Zuo, X.K.; Song, X.L.; Zhao, L.; Yuan, C.; Huang, C.; Tao, P.; Hu, Q.; et al. Selection of an ASIC1a-blocking combinatorial antibody that protects cells from ischemic death. Proc. Natl. Acad. Sci. USA 2018, 115, E7469–E7477. [Google Scholar] [CrossRef] [PubMed]
- Dominik, P.K.; Borowska, M.T.; Dalmas, O.; Kim, S.S.; Perozo, E.; Keenan, R.J.; Kossiakoff, A.A. Conformational Chaperones for Structural Studies of Membrane Proteins Using Antibody Phage Display with Nanodiscs. Structure 2016, 24, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Gardill, B.; Huang, J.; Tu, L.; Van Petegem, F.; Oxenoid, K.; Thomson, C.A. Nanodisc technology facilitates identification of monoclonal antibodies targeting multi-pass membrane proteins. Sci. Rep. 2020, 10, 1130. [Google Scholar] [CrossRef] [PubMed]
- Jamshad, M.; Grimard, V.; Idini, I.; Knowles, T.J.; Dowle, M.R.; Schofield, N.; Sridhar, P.; Lin, Y.P.; Finka, R.; Wheatley, M.; et al. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res. 2015, 8, 774–789. [Google Scholar] [CrossRef]
- Swainsbury, D.J.K.; Scheidelaar, S.; Foster, N.; van Grondelle, R.; Killian, J.A.; Jones, M.R. The effectiveness of styrene-maleic acid (SMA) copolymers for solubilisation of integral membrane proteins from SMA-accessible and SMA-resistant membranes. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2133–2143. [Google Scholar] [CrossRef]
- Cao, Y.; Fang, J.; Shi, Y.; Wang, H.; Chen, X.; Liu, Y.; Zhu, Z.; Cao, Y.; Hong, Z.; Chai, Y. Screening potential P-glycoprotein inhibitors by combination of a detergent-free membrane protein extraction with surface plasmon resonance biosensor. Acta Pharm. Sin. B 2022, 12, 3113–3123. [Google Scholar] [CrossRef]
- Sarkar, K.; Joedicke, L.; Westwood, M.; Burnley, R.; Wright, M.; McMillan, D.; Byrne, B. Modulation of PTH1R signaling by an ECD binding antibody results in inhibition of β-arrestin 2 coupling. Sci. Rep. 2019, 9, 14432. [Google Scholar] [CrossRef]
- Sarkar, K.; Joedicke, L.; Westwood, M.; Burnley, R.; Wright, M.; McMillan, D.; Byrne, B. Modulation of PTH1R signaling by an extracellular binding antibody. Vitam. Horm. 2022, 120, 109–132. [Google Scholar] [CrossRef]
- Velappan, N.; Micheva-Viteva, S.; Adikari, S.H.; Waldo, G.S.; Lillo, A.M.; Bradbury, A.R.M. Selection and verification of antibodies against the cytoplasmic domain of M2 of influenza, a transmembrane protein. MAbs 2020, 12, 1843754. [Google Scholar] [CrossRef] [PubMed]
- Grime, R.L.; Goulding, J.; Uddin, R.; Stoddart, L.A.; Hill, S.J.; Poyner, D.R.; Briddon, S.J.; Wheatley, M. Single molecule binding of a ligand to a G-protein-coupled receptor in real time using fluorescence correlation spectroscopy, rendered possible by nano-encapsulation in styrene maleic acid lipid particles. Nanoscale 2020, 12, 11518–11525. [Google Scholar] [CrossRef] [PubMed]
- Willis, S.; Davidoff, C.; Schilling, J.; Wanless, A.; Doranz, B.J.; Rucker, J. Virus-like particles as quantitative probes of membrane protein interactions. Biochemistry 2008, 47, 6988–6990. [Google Scholar] [CrossRef] [PubMed]
- Krementsov, D.N.; Rassam, P.; Margeat, E.; Roy, N.H.; Schneider-Schaulies, J.; Milhiet, P.E.; Thali, M. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic 2010, 11, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Bach, P.; Kamphuis, E.; Odermatt, B.; Sutter, G.; Buchholz, C.J.; Kalinke, U. Vesicular stomatitis virus glycoprotein displaying retrovirus-like particles induce a type I IFN receptor-dependent switch to neutralizing IgG antibodies. J. Immunol. 2007, 178, 5839–5847. [Google Scholar] [CrossRef] [PubMed]
- Bach, P.; Tschape, J.A.; Kopietz, F.; Braun, G.; Baade, J.K.; Wiederhold, K.H.; Staufenbiel, M.; Prinz, M.; Deller, T.; Kalinke, U.; et al. Vaccination with Abeta-displaying virus-like particles reduces soluble and insoluble cerebral Abeta and lowers plaque burden in APP transgenic mice. J. Immunol. 2009, 182, 7613–7624. [Google Scholar] [CrossRef] [PubMed]
- Stitz, J.; Buchholz, C.J.; Engelstadter, M.; Uckert, W.; Bloemer, U.; Schmitt, I.; Cichutek, K. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology 2000, 273, 16–20. [Google Scholar] [CrossRef]
- Tucker, D.F.; Sullivan, J.T.; Mattia, K.-A.; Fisher, C.R.; Barnes, T.; Mabila, M.N.; Wilf, R.; Sulli, C.; Pitts, M.; Payne, R.J.; et al. Isolation of state-dependent monoclonal antibodies against the 12-transmembrane domain glucose transporter 4 using virus-like particles. Proc. Natl. Acad. Sci. USA 2018, 115, E4990–E4999. [Google Scholar] [CrossRef]
- Schneider, I.C.; Hartmann, J.; Braun, G.; Stitz, J.; Klamp, T.; Bihi, M.; Sahin, U.; Buchholz, C.J. Displaying Tetra-Membrane Spanning Claudins on Enveloped Virus-Like Particles for Cancer Immunotherapy. Biotechnol. J. 2018, 13, e1700345. [Google Scholar] [CrossRef]
- Screnci, B.; Stafford, L.J.; Barnes, T.; Shema, K.; Gilman, S.; Wright, R.; Al Absi, S.; Phillips, T.; Azuelos, C.; Slovik, K.; et al. Antibody specificity against highly conserved membrane protein Claudin 6 driven by single atomic contact point. iScience 2022, 25, 105665. [Google Scholar] [CrossRef]
- van der Woning, B.; De Boeck, G.; Blanchetot, C.; Bobkov, V.; Klarenbeek, A.; Saunders, M.; Waelbroeck, M.; Laeremans, T.; Steyaert, J.; Hultberg, A.; et al. DNA immunization combined with scFv phage display identifies antagonistic GCGR specific antibodies and reveals new epitopes on the small extracellular loops. MAbs 2016, 8, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Mazzega, E.; Beran, A.; Cabrini, M.; de Marco, A. In vitro isolation of nanobodies for selective Alexandrium minutum recognition: A model for convenient development of dedicated immuno-reagents to study and diagnostic toxic unicellular algae. Harmful Algae 2019, 82, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Nikfarjam, S.; Tohidkia, M.R.; Mehdipour, T.; Soleimani, R.; Rahimi, A.A.R.; Nouri, M. Successful Application of Whole Cell Panning for Isolation of Phage Antibody Fragments Specific to Differentiated Gastric Cancer Cells. Adv. Pharm. Bull. 2019, 9, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Furman, O.; Zaporozhets, A.; Tobi, D.; Bazylevich, A.; Firer, M.A.; Patsenker, L.; Gellerman, G.; Lubin, B.C.R. Novel Cyclic Peptides for Targeting EGFR and EGRvIII Mutation for Drug Delivery. Pharmaceutics 2022, 14, 1505. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Melero, S.; Garcia-Maceira, F.I.; Garcia-Maceira, T.; Luna-Guerrero, V.; Montero-Penalvo, G.; Tunez-Finana, I.; Paz-Rojas, E. Amino terminal recognition by a CCR6 chemokine receptor antibody blocks CCL20 signaling and IL-17 expression via beta-arrestin. BMC Biotechnol. 2021, 21, 41. [Google Scholar] [CrossRef] [PubMed]
- Chockalingam, K.; Peng, Z.; Vuong, C.N.; Berghman, L.R.; Chen, Z. Golden Gate assembly with a bi-directional promoter (GBid): A simple, scalable method for phage display Fab library creation. Sci. Rep. 2020, 10, 2888. [Google Scholar] [CrossRef] [PubMed]
- Chockalingam, K.; Kumar, A.; Song, J.; Chen, Z. Chicken-derived CD20 antibodies with potent B-cell depletion activity. Br. J. Haematol. 2022, 199, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Shimoni, M.; Herschhorn, A.; Britan-Rosich, Y.; Kotler, M.; Benhar, I.; Hizi, A. The isolation of novel phage display-derived human recombinant antibodies against CCR5, the major co-receptor of HIV. Viral Immunol. 2013, 26, 277–290. [Google Scholar] [CrossRef]
- Yang, Z.; Wan, Y.; Tao, P.; Qiang, M.; Dong, X.; Lin, C.-W.; Yang, G.; Zheng, T.; Lerner, R.A. A cell–cell interaction format for selection of high-affinity antibodies to membrane proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 14971–14978. [Google Scholar] [CrossRef]
- Liu, Q.; Garg, P.; Hasdemir, B.; Wang, L.; Tuscano, E.; Sever, E.; Keane, E.; Hernandez, A.G.L.; Yuan, T.Z.; Kwan, E.; et al. Functional GLP-1R antibodies identified from a synthetic GPCR-focused library demonstrate potent blood glucose control. MAbs 2021, 13, 1893425. [Google Scholar] [CrossRef]
- Philpott, D.N.; Gomis, S.; Wang, H.; Atwal, R.; Kelil, A.; Sack, T.; Morningstar, B.; Burnie, C.; Sargent, E.H.; Angers, S.; et al. Rapid On-Cell Selection of High-Performance Human Antibodies. ACS Cent. Sci. 2022, 8, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.S.; Zasadny, K.R.; Francis, I.R.; Fenner, M.C.; Ross, C.W.; Milik, A.W.; Estes, J.; Tuck, M.; Regan, D.; Fisher, S.; et al. Iodine-131-anti-B1 radioimmunotherapy for B-cell lymphoma. J. Clin. Oncol. 1996, 14, 1974–1981. [Google Scholar] [CrossRef] [PubMed]
- Stashenko, P.; Nadler, L.M.; Hardy, R.; Schlossman, S.F. Characterization of a human B lymphocyte-specific antigen. J. Immunol. 1980, 125, 1678–1685. [Google Scholar] [CrossRef] [PubMed]
- Reff, M.; Carner, K.; Chambers, K.; Chinn, P.; Leonard, J.; Raab, R.; Newman, R.; Hanna, N.; Anderson, D. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood 1994, 83, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Alcindor, T.; Witzig, T.E. Radioimmunotherapy with yttrium-90 ibritumomab tiuxetan for patients with relapsed CD20+ B-cell non-Hodgkin’s lymphoma. Curr. Treat. Options Oncol. 2002, 3, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B. Ofatumumab. MAbs 2009, 1, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Mossner, E.; Brunker, P.; Moser, S.; Puntener, U.; Schmidt, C.; Herter, S.; Grau, R.; Gerdes, C.; Nopora, A.; van Puijenbroek, E.; et al. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell-mediated B-cell cytotoxicity. Blood 2010, 115, 4393–4402. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.; Chan, A.C.; Crowley, C.W.; Lowman, H.B.; Nakamura, G.R.; Presta, L.G. Immunoglobulin Variants and Uses Thereof. International Application No. PCT/US2003/040426. WO/2004/056312. 8 July 2004. [Google Scholar]
- de Romeuf, C.; Dutertre, C.A.; Le Garff-Tavernier, M.; Fournier, N.; Gaucher, C.; Glacet, A.; Jorieux, S.; Bihoreau, N.; Behrens, C.K.; Beliard, R.; et al. Chronic lymphocytic leukaemia cells are efficiently killed by an anti-CD20 monoclonal antibody selected for improved engagement of FcgammaRIIIA/CD16. Br. J. Haematol. 2008, 140, 635–643. [Google Scholar] [CrossRef]
- Baritaki, S.; Militello, L.; Malaponte, G.; Spandidos, D.A.; Salcedo, M.; Bonavida, B. The anti-CD20 mAb LFB-R603 interrupts the dysregulated NF-kappaB/Snail/RKIP/PTEN resistance loop in B-NHL cells: Role in sensitization to TRAIL apoptosis. Int. J. Oncol. 2011, 38, 1683–1694. [Google Scholar] [CrossRef]
- Sun, L.L.; Ellerman, D.; Mathieu, M.; Hristopoulos, M.; Chen, X.; Li, Y.; Yan, X.; Clark, R.; Reyes, A.; Stefanich, E.; et al. Anti-CD20/CD3 T cell-dependent bispecific antibody for the treatment of B cell malignancies. Sci. Transl. Med. 2015, 7, 287ra70. [Google Scholar] [CrossRef]
- Teeling, J.L.; French, R.R.; Cragg, M.S.; van den Brakel, J.; Pluyter, M.; Huang, H.; Chan, C.; Parren, P.W.; Hack, C.E.; Dechant, M.; et al. Characterization of new human CD20 monoclonal antibodies with potent cytolytic activity against non-Hodgkin lymphomas. Blood 2004, 104, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Engelberts, P.J.; Hiemstra, I.H.; de Jong, B.; Schuurhuis, D.H.; Meesters, J.; Beltran Hernandez, I.; Oostindie, S.C.; Neijssen, J.; van den Brink, E.N.; Horbach, G.J.; et al. DuoBody-CD3xCD20 induces potent T-cell-mediated killing of malignant B cells in preclinical models and provides opportunities for subcutaneous dosing. EBioMedicine 2020, 52, 102625. [Google Scholar] [CrossRef] [PubMed]
- Cremasco, F.; Menietti, E.; Speziale, D.; Sam, J.; Sammicheli, S.; Richard, M.; Varol, A.; Klein, C.; Umana, P.; Bacac, M.; et al. Cross-linking of T cell to B cell lymphoma by the T cell bispecific antibody CD20-TCB induces IFNγ/CXCL10-dependent peripheral T cell recruitment in humanized murine model. PLoS ONE 2021, 16, e0241091. [Google Scholar] [CrossRef] [PubMed]
- Niwa, R.; Shoji-Hosaka, E.; Sakurada, M.; Shinkawa, T.; Uchida, K.; Nakamura, K.; Matsushima, K.; Ueda, R.; Hanai, N.; Shitara, K. Defucosylated chimeric anti-CC chemokine receptor 4 IgG1 with enhanced antibody-dependent cellular cytotoxicity shows potent therapeutic activity to T-cell leukemia and lymphoma. Cancer Res. 2004, 64, 2127–2133. [Google Scholar] [CrossRef] [PubMed]
- Ollila, T.A.; Sahin, I.; Olszewski, A.J. Mogamulizumab: A new tool for management of cutaneous T-cell lymphoma. Onco Targets Ther. 2019, 12, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Pillarisetti, K.; Edavettal, S.; Mendonca, M.; Li, Y.; Tornetta, M.; Babich, A.; Majewski, N.; Husovsky, M.; Reeves, D.; Walsh, E.; et al. A T-cell-redirecting bispecific G-protein-coupled receptor class 5 member D x CD3 antibody to treat multiple myeloma. Blood 2020, 135, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Chari, A.; Minnema, M.C.; Berdeja, J.G.; Oriol, A.; van de Donk, N.; Rodriguez-Otero, P.; Askari, E.; Mateos, M.V.; Costa, L.J.; Caers, J.; et al. Talquetamab, a T-Cell-Redirecting GPRC5D Bispecific Antibody for Multiple Myeloma. N. Engl. J. Med. 2022, 387, 2232–2244. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Athreya, A.; Gulati, A.; Nair, R.M.; Mahendran, I.; Ranjan, R.; Penmatsa, A. Structural basis of inhibition of a transporter from Staphylococcus aureus, NorC, through a single-domain camelid antibody. Commun. Biol. 2021, 4, 836. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, B.; Chockalingam, K.; Chen, Z. Methods for Engineering Binders to Multi-Pass Membrane Proteins. Bioengineering 2023, 10, 1351. https://doi.org/10.3390/bioengineering10121351
Thomas B, Chockalingam K, Chen Z. Methods for Engineering Binders to Multi-Pass Membrane Proteins. Bioengineering. 2023; 10(12):1351. https://doi.org/10.3390/bioengineering10121351
Chicago/Turabian StyleThomas, Benjamin, Karuppiah Chockalingam, and Zhilei Chen. 2023. "Methods for Engineering Binders to Multi-Pass Membrane Proteins" Bioengineering 10, no. 12: 1351. https://doi.org/10.3390/bioengineering10121351





