Multi-Class Wound Classification via High and Low-Frequency Guidance Network
Abstract
:1. Introduction
- Wound classification is an important task in the field of medical image processing. By fully exploring the potential of deep learning in medical image processing, our research is crucial for correctly and quickly classifying different types of wounds, helping doctors develop appropriate treatment plans and improve treatment effects.
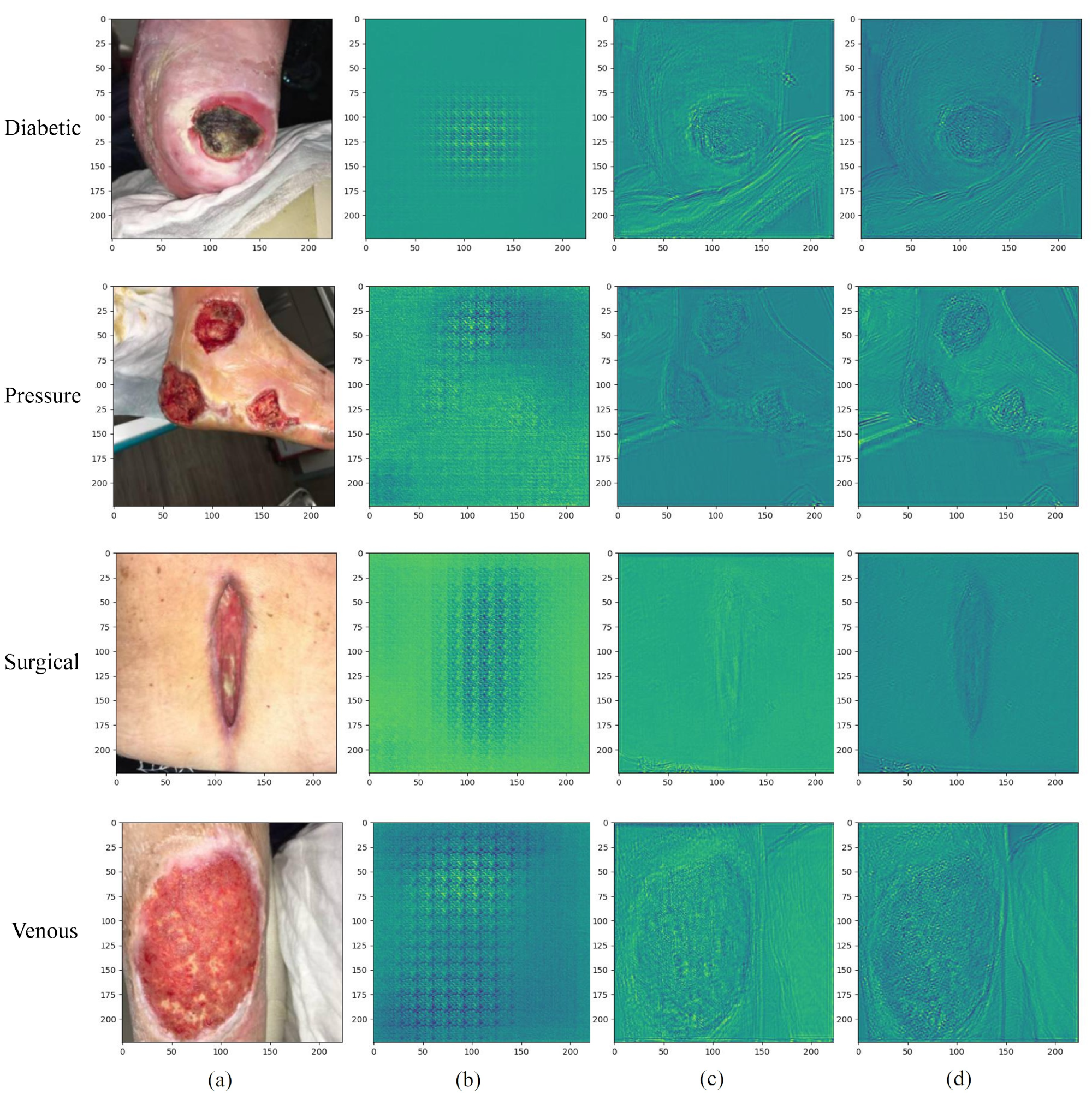
- Wound images contain abundant high- and low-frequency information, which is an important basis for diagnosing wound types. The high-frequency part usually contains the details and texture of the image, while the low-frequency part reflects more of the overall structural characteristics such as wound size, shape, and location. By processing high- and low-frequency information separately, it is expected to better capture the details and overall characteristics of wound images and improve the accuracy of classification.
- The use of a single network in previous studies may be difficult to capture information at different scales at the same time. We adopt a dual-branch structure that can effectively fuse features at different scales, including high- and low-frequency information, to more comprehensively capture the details and overall structure of the wound image [31]. This helps cope with diversity wounds and improves the generalization performance of the network.
- Wound images are often interfered by factors such as illumination, occlusion, and noise, which affect the performance of traditional methods. Our work aims to improve the adaptability to complex images by processing high- and low-frequency information separately, thereby improving the robustness of wound classification.
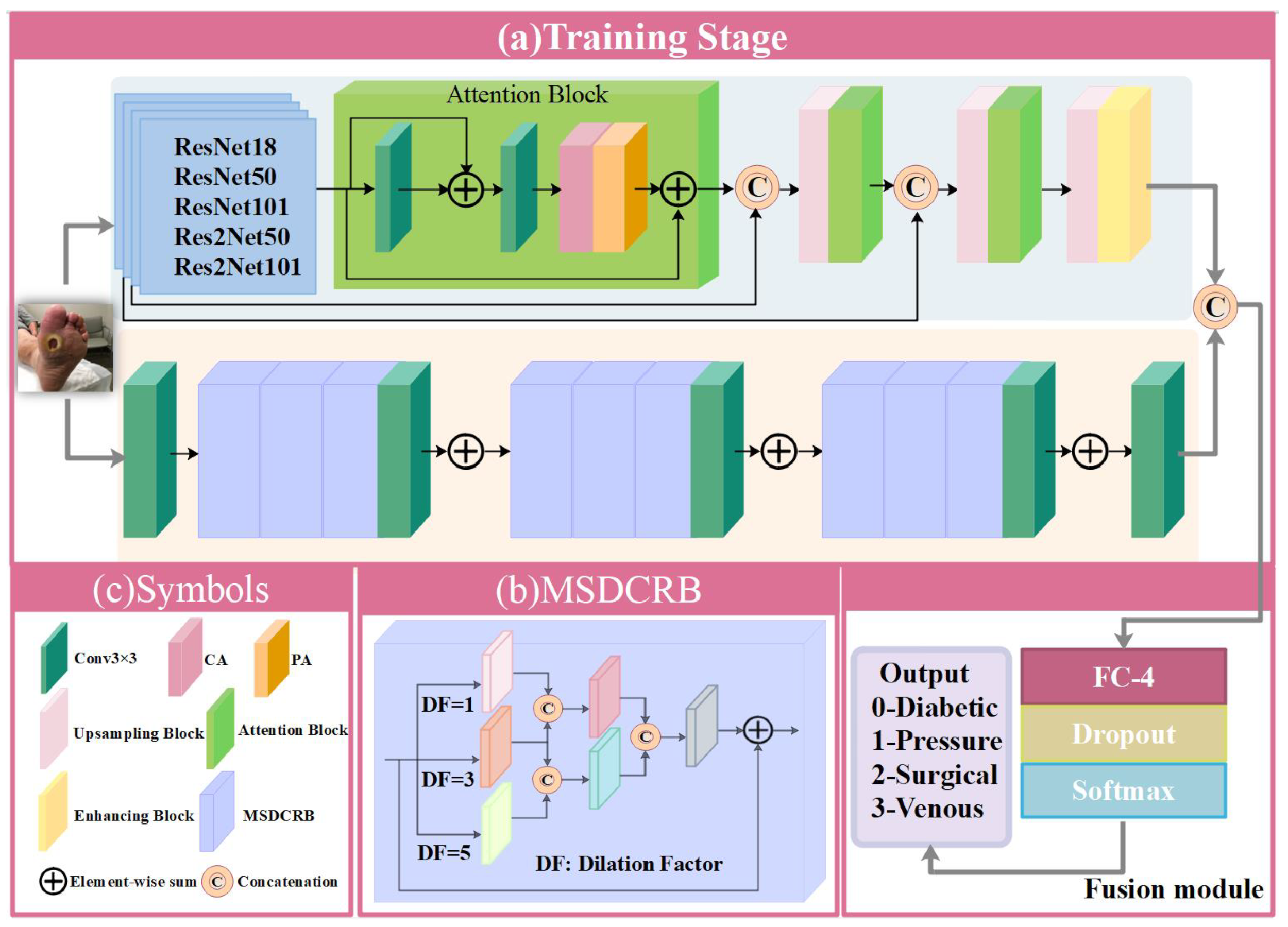
- We propose a novel HLG-Net for multi-class wound classification, and it contains two main branches: High-Frequency Network (HF-Net) and Low-Frequency Network (LF-Net), where the HF-Net focuses on high-frequency information extraction, while the LF-Net can explore low-frequency information of the image. Extensive experiments demonstrate that our HLG-Net can outperform other state-of-the-art methods on multi-class wound classification performance.
- We utilize pre-trained models ResNet and Res2Net as the backbones of the HF-Net, respectively. Both ResNet and Res2Net can allow the network to capture more complex and informative features that contain high-frequency details and texture information. Moreover, feature attention module and enhancing block are employed to improve the representation capability of HF-Net and multi-scale feature extraction, respectively.
- In LF-Net, we adopt a novel Multi-Stream Dilated Convolutional Residual Block (MSDCRB) as the core of feature extraction. A larger receptive field introduced by multi-stream dilation convolution layers can capture global information of wound images, thus pushing HLG-Net to achieve a better classification performance.
- To take full advantage of feature streams from different branches HF-Net and LF-Net, we propose a fusion module to fuse them together and output the final classification result by softmax layer with 4 output channels. The features extracted from each branch are flattened and concatenated to create the final feature vector, which is fed into the multilayer perceptron for wound classification.
2. Materials and Methods
2.1. Transfer Learning
2.2. Proposed Architecture
2.2.1. HF-Net
2.2.2. LF-Net
2.2.3. Fusion Module
2.3. Loss Function
3. Experiment
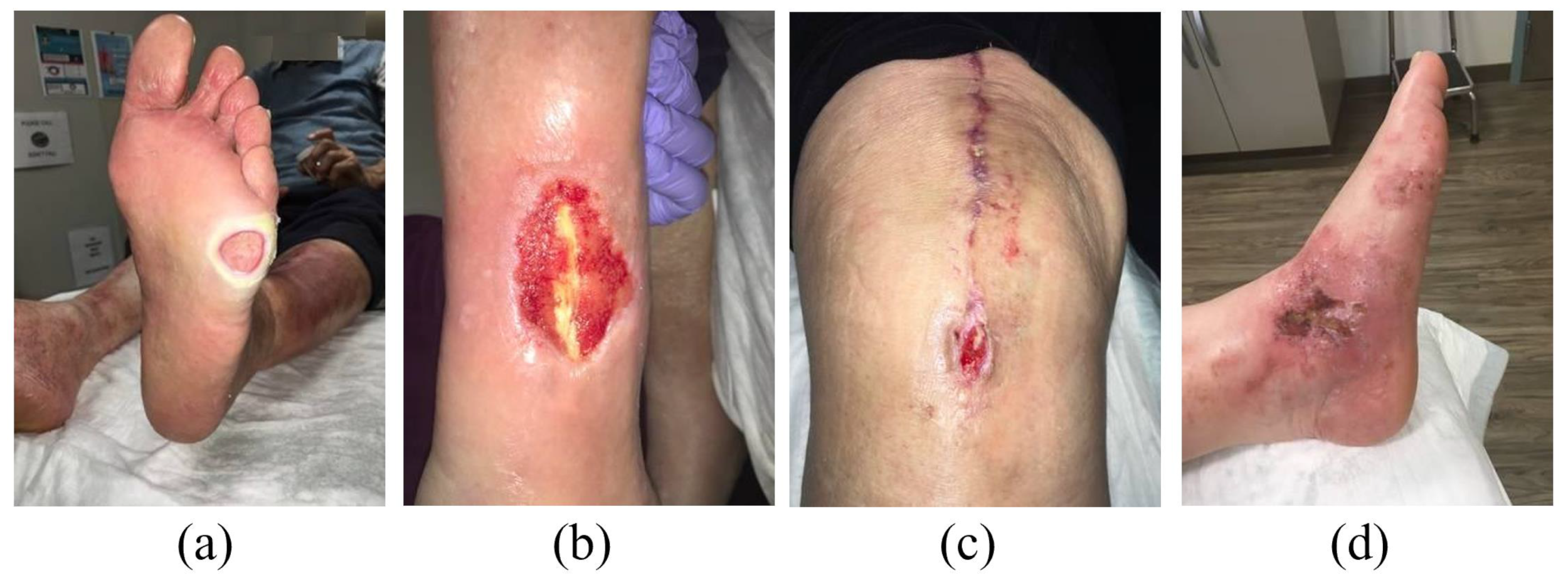
3.1. Dataset: AZH Wound and Vascular Center Database
3.2. Data Preprocessing
3.3. Training Details
3.4. Evaluation Metrics
4. Results
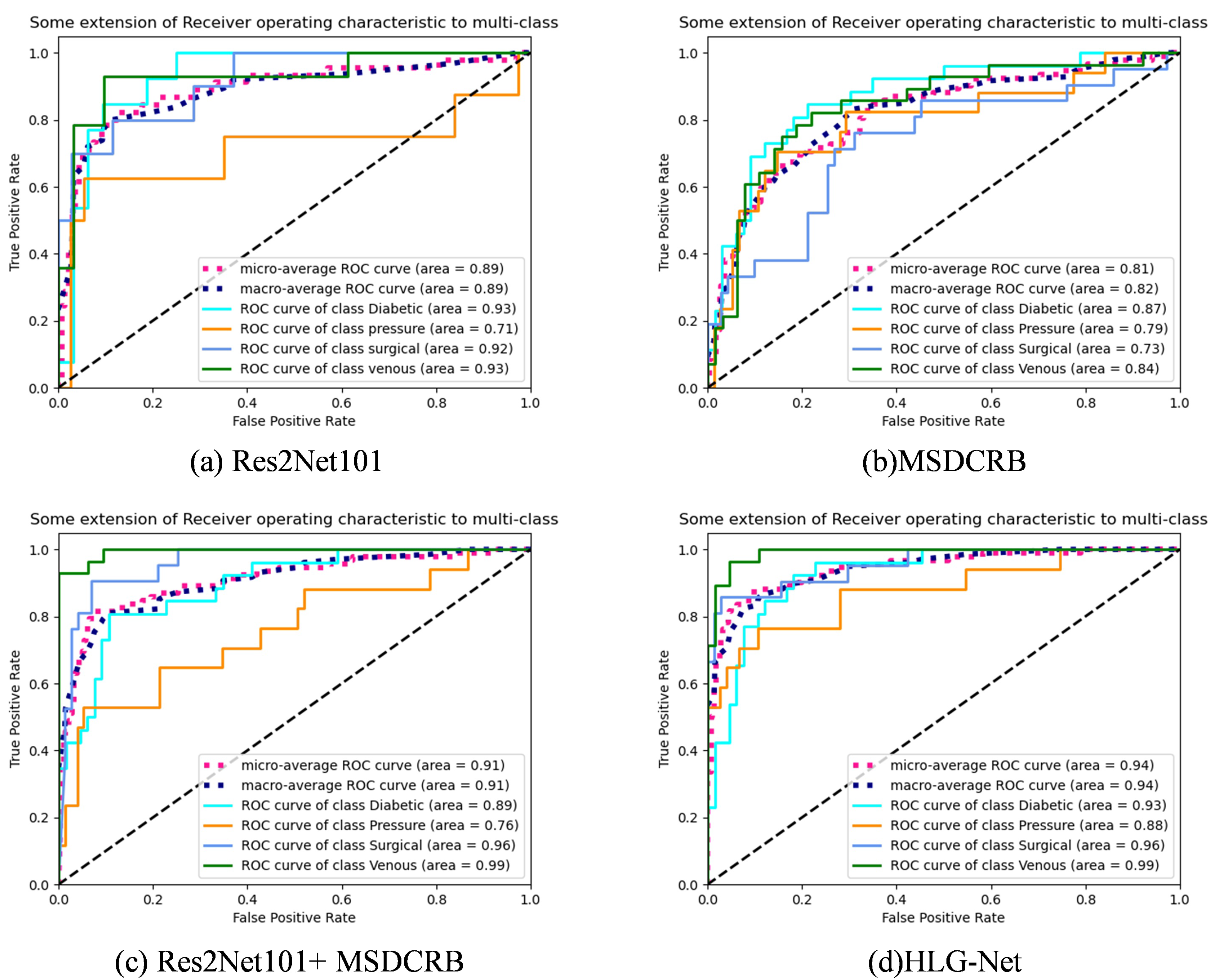
4.1. Ablation Experiment Analysis
- HF-Net: In the first step, we use the models ResNet18, ResNet50, ResNet101, Res2Net50 and Res2Net101 pre-trained on the ImageNet dataset for four-class classification.
- LF-Net: Subsequently, only the MSDCRB is employed for four-class classification of wounds.
- HF-Net + LF-Net: Finally, the HF-Net responsible for extracting high-frequency information and the LF-Net responsible for extracting low-frequency information are combined for the four-class classification. Furthermore, Comparing the classification performance of AE-Res2Net50 + MSDCRB and AE-Res2Net101 + MSDCRB, which are obtained by adding attention and enhancement modules to HF-Net.
4.2. Hyperparameter Experimental Analysis
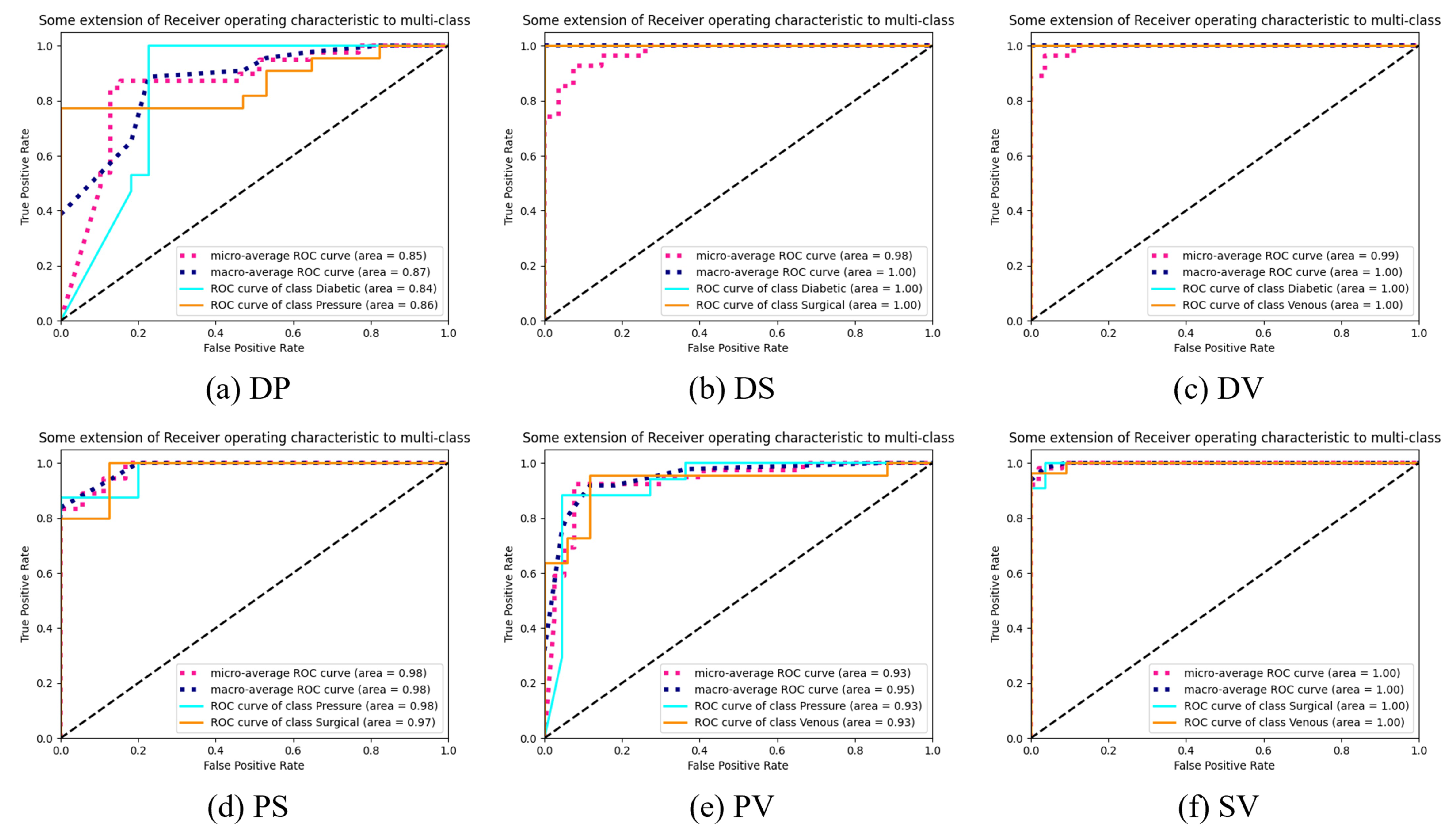
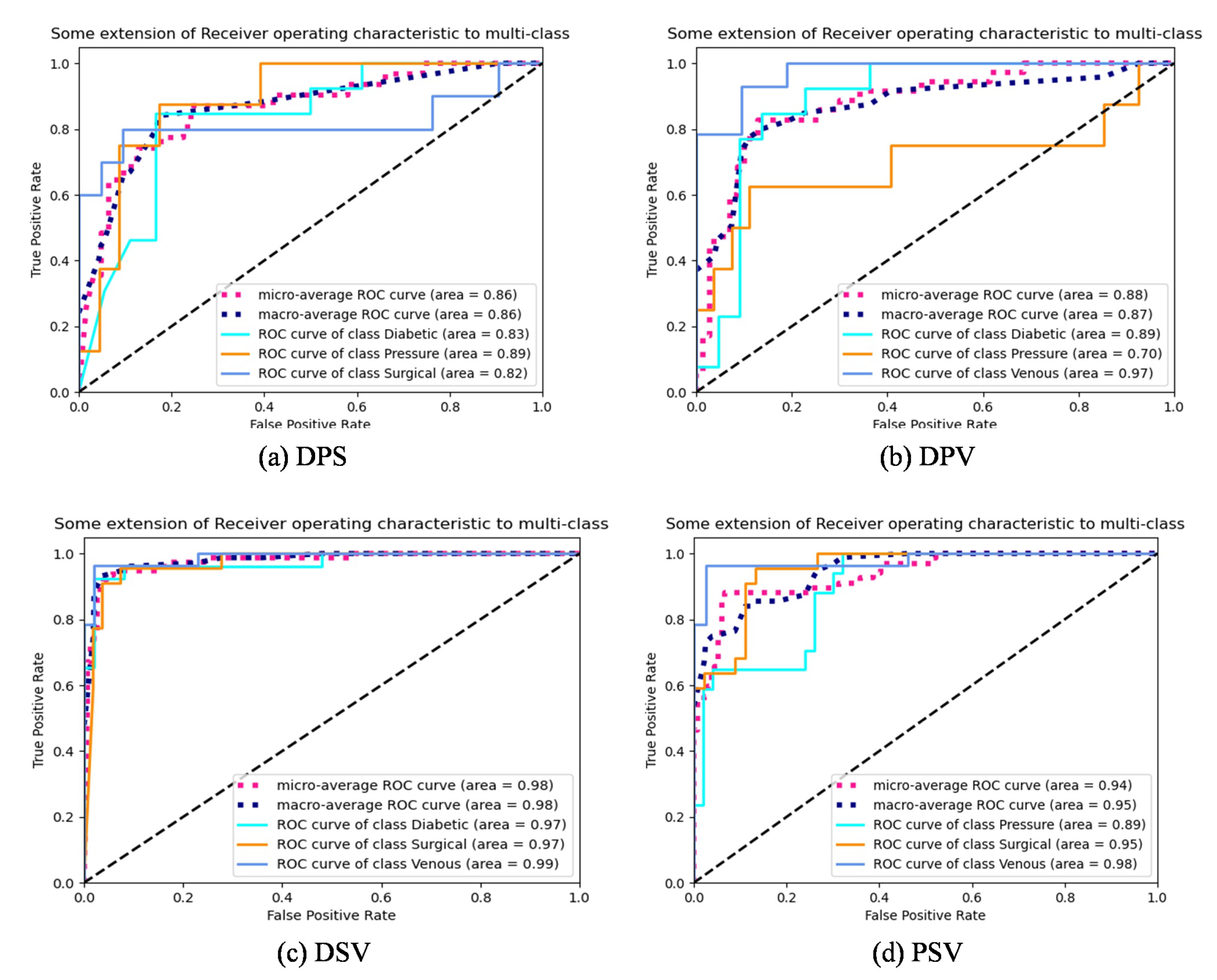
4.3. Classification Result Analysis
5. Discussion
5.1. Comparison of HLG-Net Model for Different Classification Tasks
5.2. Comparison with Previous Works
- Compared with existing literature, we use a high- and low-frequency dual-branch classification network. This structure contributes a new perspective. Through the high and low frequency information of the wound image, we are able to more comprehensively capture the characteristics of the input wound image. Table 9 demonstrates that HLG-Net significantly improves the classification performance.
- In the experiments, HLG-Net deliberately excludes normal skin and background images, which is different from other works that usually focus on binary classification problems containing normal and abnormal samples. This exclusion allows HLG-Net to focus on handling the most complex classification tasks involving only wounds, which is more suitable for practical medical applications.
- The method of Anisuzzaman et al. [30] involves using wound images and the location of the wound, and requires different models in different tasks to achieve the highest classification accuracy. In contrast, HLG-Net can achieve relatively good results for multiple mixed classes using only wound images and a single model, which is more consistent with practical applications.
- Compared with other models that require complex data preprocessing in wound image classification tasks, HLG-Net shows good performance by directly using the whole wound image for training. The simplified data processing method makes our model more versatile and applicable, reducing complex pre-processing steps. Therefore, HLG-Net effectively meets the current demand for a more accurate and versatile wound classification method.
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, R.; Tian, D.; Xu, D.; Qian, W.; Yao, Y. A Survey of Wound Image Analysis Using Deep Learning: Classification, Detection, and Segmentation. IEEE Access 2022, 10, 79502–79515. [Google Scholar] [CrossRef]
- Adam, M.; Ng, E.Y.; Tan, J.H.; Heng, M.L.; Tong, J.W.; Acharya, U.R. Computer Aided Diagnosis of Diabetic Foot Using Infrared Thermography: A Review. Comput. Biol. Med. 2017, 91, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.H.; Hachiuma, R.; Alavi, A.; Brungel, R.; Cassidy, B.; Goyal, M.; Zhu, H.; Ruckert, J.; Olshansky, M.; Huang, X.; et al. Deep Learning in Diabetic Foot Ulcers Detection: A Comprehensive Evaluation. Comput. Biol. Med. 2021, 135, 104596. [Google Scholar] [CrossRef] [PubMed]
- Al-Garaawi, N.; Ebsim, R.; Alharan, A.F.; Yap, M.H. Diabetic Foot Ulcer Classification Using Mapped Binary Patterns and Convolutional Neural Networks. Comput. Biol. Med. 2022, 140, 105055. [Google Scholar] [CrossRef] [PubMed]
- Bergqvist, D.; Lindholm, C.; Nelzén, O. Chronic Leg Ulcers: The Impact of Venous Disease. J. Vasc. Surg. 1999, 29, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Ruckley, C. Socioeconomic Impact of Chronic Venous Insufficiency and Leg Ulcers. Angiology 1997, 48, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Zahia, S.; Sierra-Sosa, D.; Garcia-Zapirain, B.; Elmaghraby, A. Tissue Classification and Segmentation of Pressure Injuries Using Convolutional Neural Networks. Comput. Methods Programs Biomed. 2018, 159, 51–58. [Google Scholar] [CrossRef]
- Serena, T.E.; Hanft, J.R.; Snyder, R. The Lack of Reliability of Clinical Examination in the Diagnosis of Wound Infection: Preliminary Communication. Int. J. Low. Extrem. Wounds 2008, 7, 32–35. [Google Scholar] [CrossRef]
- Monstrey, S.; Hoeksema, H.; Verbelen, J.; Pirayesh, A.; Blondeel, P. Assessment of Burn Depth and Burn Wound Healing Potential. Burns 2008, 34, 761–769. [Google Scholar] [CrossRef]
- Wang, C.; Yan, X.; Smith, M.; Kochhar, K.; Rubin, M.; Warren, S.M.; Wrobel, J.; Lee, H. A Unified Framework for Automatic Wound Segmentation and Analysis with Deep Convolutional Neural Networks. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2415–2418. [Google Scholar] [CrossRef]
- Garcia-Zapirain, B.; Shalaby, A.; El-Baz, A.; Elmaghraby, A. Automated Framework for Accurate Segmentation of Pressure Ulcer Images. Comput. Biol. Med. 2017, 90, 137–145. [Google Scholar] [CrossRef]
- Monroy, B.; Sanchez, K.; Arguello, P.; Estupiñán, J.; Bacca, J.; Correa, C.V.; Valencia, L.; Castillo, J.C.; Mieles, O.; Arguello, H.; et al. Automated Chronic Wounds Medical Assessment and Tracking Framework Based on Deep Learning. Comput. Biol. Med. 2023, 165, 107335. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Gao, S.H.; Cheng, M.M.; Zhao, K.; Zhang, X.Y.; Yang, M.H.; Torr, P. Res2Net: A New Multi-scale Backbone Architecture. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, C.; Zhang, Z.; Zhu, Y.; Lin, H.; Zhang, Z.; Sun, Y.; He, T.; Mueller, J.; Manmatha, R.; et al. ResNeSt: Split-Attention Networks. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), New Orleans, LA, USA, 18–24 June 2022; pp. 2735–2745. [Google Scholar] [CrossRef]
- Duta, I.C.; Liu, L.; Zhu, F.; Shao, L. Improved Residual Networks for Image and Video Recognition. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 9415–9422. [Google Scholar] [CrossRef]
- Abubakar, A.; Ugail, H.; Bukar, A.M. Assessment of Human Skin Burns: A Deep Transfer Learning Approach. J. Med. Biol. Eng. 2020, 40, 321–333. [Google Scholar] [CrossRef]
- Wang, Y.; Ke, Z.; He, Z.; Chen, X.; Zhang, Y.; Xie, P.; Li, T.; Zhou, J.; Li, F.; Yang, C.; et al. Real-Time Burn Depth Assessment Using Artificial Networks: A Large-Scale, Multicentre Study. Burns 2020, 46, 1829–1838. [Google Scholar] [CrossRef]
- Chauhan, J.; Goyal, P. BPBSAM: Body Part-Specific Burn Severity Assessment Model. Burns 2020, 46, 1407–1423. [Google Scholar] [CrossRef]
- Bhansali, R.; Kumar, R. BurnNet: An Efficient Deep Learning Framework for Accurate Dermal Burn Classification. medRxiv 2021. [Google Scholar] [CrossRef]
- Shenoy, V.; Foster, E.; Aalami, L.; Majeed, B.; Aalami, O. Deepwound: Automated Postoperative Wound Assessment and Surgical Site Surveillance through Convolutional Neural Networks, 2018. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018. [Google Scholar]
- Alzubaidi, L.; Fadhel, M.A.; Oleiwi, S.R.; Al-Shamma, O.; Zhang, J. DFU_QUTNet: Diabetic Foot Ulcer Classification Using Novel Deep Convolutional Neural Network. Multimed. Tools Appl. 2020, 79, 15655–15677. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Rostami, B.; Niezgoda, J.; Gopalakrishnan, S.; Yu, Z. Multiclass Burn Wound Image Classification Using Deep Convolutional Neural Networks. arXiv 2021, arXiv:2103.01361. [Google Scholar]
- Goyal, M.; Reeves, N.D.; Davison, A.K.; Rajbhandari, S.; Spragg, J.; Yap, M.H. DFUNet: Convolutional Neural Networks for Diabetic Foot Ulcer Classification. IEEE Trans. Emerg. Top. Comput. Intell. 2020, 4, 728–739. [Google Scholar] [CrossRef]
- Rostami, B.; Anisuzzaman, D.M.; Wang, C.; Gopalakrishnan, S.; Niezgoda, J.; Yu, Z. Multiclass Wound Image Classification Using an Ensemble Deep CNN-based Classifier. Comput. Biol. Med. 2021, 134, 104536. [Google Scholar] [CrossRef] [PubMed]
- Anisuzzaman, D.M.; Patel, Y.; Rostami, B.; Niezgoda, J.; Gopalakrishnan, S.; Yu, Z. Multi-Modal Wound Classification Using Wound Image and Location by Deep Neural Network. Sci. Rep. 2022, 12, 20057. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, J.; Xiao, H.; Jin, X.; Yan, S.; Feng, J. Dual Path Networks. arXiv 2017, arXiv:1707.01629. [Google Scholar]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009. [Google Scholar] [CrossRef]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How Transferable Are Features in Deep Neural Networks? In Proceedings of the Advances in Neural Information Processing Systems 27: Annual Conference on Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 3320–3328. [Google Scholar]
- Lu, S.; Lu, Z.; Zhang, Y.D. Pathological Brain Detection Based on AlexNet and Transfer Learning. J. Comput. Sci. 2019, 30, 41–47. [Google Scholar] [CrossRef]
- Lan, T.; Li, Z.; Chen, J. FusionSegNet: Fusing Global Foot Features and Local Wound Features to Diagnose Diabetic Foot. Comput. Biol. Med. 2023, 152, 106456. [Google Scholar] [CrossRef]
- Shi, W.; Caballero, J.; Huszar, F.; Totz, J.; Aitken, A.P.; Bishop, R.; Rueckert, D.; Wang, Z. Real-Time Single Image and Video Super-Resolution Using an Efficient Sub-Pixel Convolutional Neural Network. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 1874–1883. [Google Scholar] [CrossRef]
- Qu, Y.; Chen, Y.; Huang, J.; Xie, Y. Enhanced Pix2pix Dehazing Network. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 8152–8160. [Google Scholar] [CrossRef]
- Yu, F.; Koltun, V. Multi-Scale Context Aggregation by Dilated Convolutions. arXiv 2016, arXiv:1511.07122. [Google Scholar]
- Wang, C.; Xing, X.; Yao, G.; Su, Z. Single Image Deraining via Deep Shared Pyramid Network. Vis. Comput. 2021, 37, 1851–1865. [Google Scholar] [CrossRef]
- Kornblith, S.; Shlens, J.; Le, Q.V. Do Better ImageNet Models Transfer Better? In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 2656–2666. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, H.; Fu, M.; Chen, J.; Wang, X.; Wang, K. A Two-branch Neural Network for Non-homogeneous Dehazing via Ensemble Learning. arXiv 2021, arXiv:2104.08902. [Google Scholar]
- Qin, X.; Wang, Z.; Bai, Y.; Xie, X.; Jia, H. FFA-Net: Feature Fusion Attention Network for Single Image Dehazing. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 11908–11915. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, K.; Li, K.; Wang, L.; Zhong, B.; Fu, Y. Image Super-Resolution Using Very Deep Residual Channel Attention Networks. In Computer Vision—ECCV 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 11211, pp. 294–310. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6230–6239. [Google Scholar] [CrossRef]
- Luo, J.; Bu, Q.; Zhang, L.; Feng, J. Global Feature Fusion Attention Network For Single Image Dehazing. In Proceedings of the 2021 IEEE International Conference on Multimedia & Expo Workshops (ICMEW), Shenzhen, China, 5–9 July 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Cassidy, B.; Kendrick, C.; Reeves, N.D.; Pappachan, J.M.; O’Shea, C.; Armstrong, D.G.; Yap, M.H. Diabetic Foot Ulcer Grand Challenge 2021: Evaluation and Summary. In Diabetic Foot Ulcers Grand Challenge; Springer International Publishing: Cham, Switzerland, 2022; Volume 13183, pp. 90–105. [Google Scholar] [CrossRef]
- Goyal, M.; Reeves, N.; Rajbhandari, S.; Ahmad, N.; Wang, C.; Yap, M.H. Recognition of Ischaemia and Infection in Diabetic Foot Ulcers: Dataset and Techniques. Comput. Biol. Med. 2020, 117, 103616. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, X.; Wang, T.; Lei, B. Aggregating Deep Convolutional Features for Melanoma Recognition in Dermoscopy Images. In Machine Learning in Medical Imaging; Wang, Q., Shi, Y., Suk, H.I., Suzuki, K., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 10541, pp. 238–246. [Google Scholar] [CrossRef]
- Wu, W.; Liu, H.; Li, L.; Long, Y.; Wang, X.; Wang, Z.; Li, J.; Chang, Y. Application of Local Fully Convolutional Neural Network Combined with YOLO v5 Algorithm in Small Target Detection of Remote Sensing Image. PLoS ONE 2021, 16, e0259283. [Google Scholar] [CrossRef]
- Hossin, M.; Sulaiman, M.N. A Review on Evaluation Metrics for Data Classification Evaluations. Int. J. Data Min. Knowl. Manag. Process. 2015, 5, 1–11. [Google Scholar] [CrossRef]
- Hüsers, J.; Moelleken, M.; Richter, M.L.; Przysucha, M.; Malihi, L.; Busch, D.; Götz, N.A.; Heggemann, J.; Hafer, G.; Wiemeyer, S.; et al. An Image Based Object Recognition System for Wound Detection and Classification of Diabetic Foot and Venous Leg Ulcers. In Studies in Health Technology and Informatics; Séroussi, B., Weber, P., Dhombres, F., Grouin, C., Liebe, J.D., Pelayo, S., Pinna, A., Rance, B., Sacchi, L., Ugon, A., et al., Eds.; IOS Press: Amsterdam, The Netherlands, 2022. [Google Scholar] [CrossRef]



| Abbreviation | Description |
|---|---|
| Diabetic wounds | D |
| Pressure wounds | P |
| Surgical wounds | S |
| Venous wounds | V |
| Class | Training | Validation | Test | Total |
|---|---|---|---|---|
| Diabetic | 108 | 31 | 15 | 154 |
| Pressure | 71 | 19 | 10 | 100 |
| Surgical | 90 | 26 | 12 | 128 |
| Venous | 110 | 31 | 15 | 156 |
| Network | Accuracy | Precision | Recall | F1-Score | |
|---|---|---|---|---|---|
| HF-Net | ResNet18 | 68.48% | 62.20% | 61.60% | 61.80% |
| ResNet50 | 71.74% | 67.30% | 57.70% | 61.40% | |
| ResNet101 | 73.91% | 71.90% | 66.80% | 68.50% | |
| Res2Net50 | 72.83% | 68.70% | 60.50% | 63.10% | |
| Res2Net101 | 75.56% | 78.70% | 69.00% | 71.70% | |
| LF-Net | MSDCRB | 60.87% | 54.10% | 48.20% | 50.10% |
| HF-Net + LF-Net | ResNet18 + MSDCRB | 70.65% | 67.60% | 57.10% | 61.60% |
| ResNet50 + MSDCRB | 72.83% | 75.80% | 61.30% | 67.60% | |
| ResNet101 + MSDCRB | 76.09% | 80.00% | 63.20% | 69.40% | |
| Res2Net50 + MSDCRB | 78.26% | 81.60% | 67.60% | 71.80% | |
| Res2Net101 + MSDCRB | 80.43% | 74.90% | 75.70% | 74.60% | |
| AE-Res2Net50 + MSDCRB | 79.35% | 80.10% | 70.60% | 75.00% | |
| HLG-Net | 82.61% | 79.10% | 75.80% | 77.20% | |
| (Group,Block) | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| (1,2) | 73.33% | 71.50% | 60.70% | 64.20% |
| (2,2) | 77.17% | 75.90% | 68.50% | 71.70% |
| (2,3) | 81.52% | 76.10% | 78.70% | 76.80% |
| (3,2) | 79.35% | 73.40% | 70.50% | 71.00% |
| (3,3) | 82.61% | 79.10% | 75.80% | 77.20% |
| (4,3) | 79.35% | 73.40% | 70.50% | 71.00% |
| (4,4) | 77.17% | 75.90% | 68.50% | 71.70% |
| Dropout | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| 0 | 76.09% | 76.90% | 64.00% | 66.00% |
| 10% | 77.78% | 75.30% | 69.00% | 71.00% |
| 20% | 81.52% | 77.90% | 74.50% | 76.10% |
| 30% | 82.61% | 79.10% | 75.80% | 77.20% |
| 40% | 81.52% | 77.90% | 74.50% | 76.10% |
| Binary | Accuracy | Precision | Recall | F1-Score | |
|---|---|---|---|---|---|
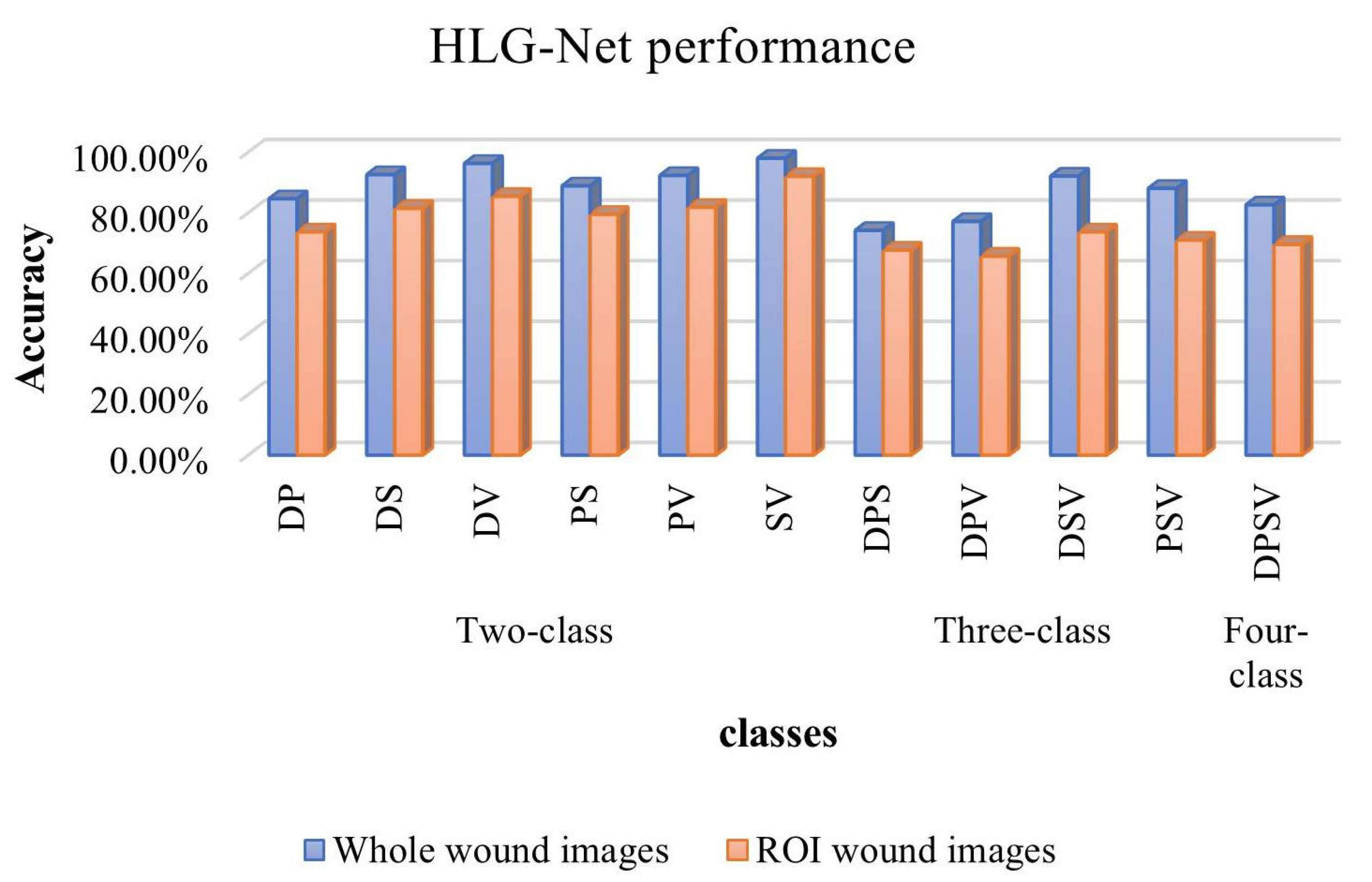
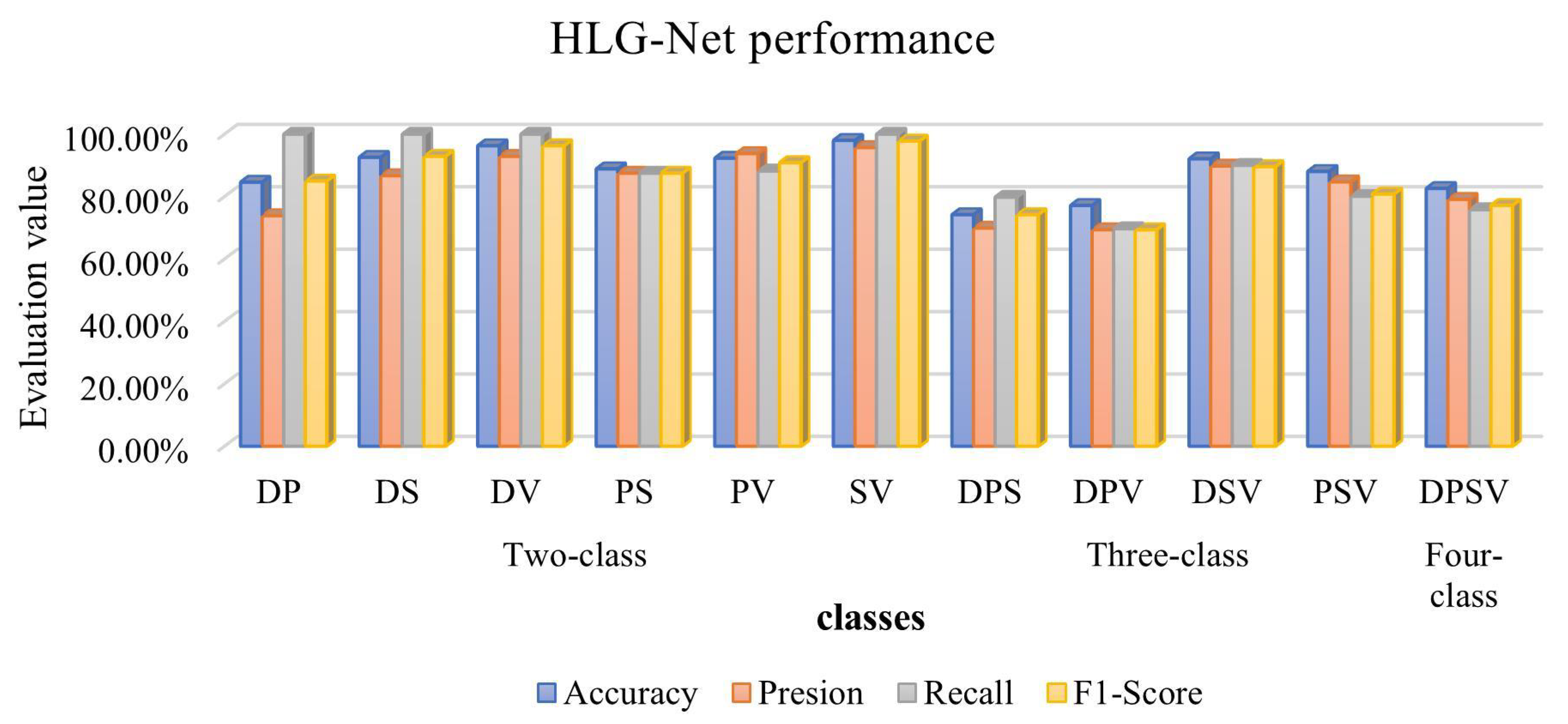
| Whole images | DP | 84.62% | 73.90% | 100% | 85.00% |
| DS | 92.59% | 86.70% | 100% | 92.90% | |
| DV | 96.30% | 92.90% | 100% | 96.30% | |
| PS | 88.89% | 87.50% | 87.50% | 87.50% | |
| PV | 92.31% | 93.80% | 88.20% | 90.90% | |
| SV | 98.00% | 95.70% | 100% | 97.80% | |
| ROI images | DP | 73.68% | 63.60% | 87.50% | 73.70% |
| DS | 81.48% | 100% | 61.50% | 76.20% | |
| DV | 85.45% | 95.00% | 73.10% | 82.60% | |
| PS | 79.49% | 71.40% | 88.20% | 78.90% | |
| PV | 81.82% | 70.00% | 87.50% | 77.80% | |
| SV | 92.00% | 90.90% | 90.90% | 90.90% | |
| Three-Class | Accuracy | Precision | Recall | F1-Score | |
|---|---|---|---|---|---|
| Whole images | DPS | 74.19% | 69.90% | 79.80% | 74.20% |
| DPV | 77.14% | 69.40% | 69.70% | 69.40% | |
| DSV | 92.11% | 89.80% | 90.00% | 89.60% | |
| PSV | 88.06% | 84.70% | 80.10% | 80.80% | |
| ROI images | DPS | 67.69% | 54.40% | 58.10% | 56.20% |
| DPV | 65.71% | 75.00% | 69.20% | 61.10% | |
| DSV | 73.68% | 77.50% | 58.00% | 66.40% | |
| PSV | 70.97% | 69.70% | 70.40% | 70.97% | |
| Four-Class | Accuracy | Precision | Recall | F1-Score | |
|---|---|---|---|---|---|
| Whole images | DPSV | 82.61% | 79.10% | 75.80% | 77.20% |
| ROI images | DPSV | 69.57% | 64.50% | 61.90% | 62.00% |
| Classes | Evaluation Metrics | References | Previous | Present | |||
|---|---|---|---|---|---|---|---|
| Models | Dataset | Result | Model | Result | |||
| DPSV | Accuracy | Anisuzzaman et al. [30] | VGG16 + LSTM | AZH | 79.35% | HLG-Net | 82.61% |
| DSV | Accuracy | VGG19 + MLP | 90.67% | 92.11% | |||
| PSV | Accuracy | VGG16 + MLP | 86.23% | 88.06% | |||
| DPS | Accuracy | VGG16 + LSTM | 72.95% | 74.19% | |||
| DPV | Accuracy | VGG19 + MLP | 80.99% | 77.14% | |||
| DP | Accuracy | VGG16 + MLP | 85.00% | 84.62% | |||
| DS | Accuracy | VGG16 + MLP | 88.64% | 92.59% | |||
| DV | Accuracy | VGG16 + MLP | 92.59% | 96.30% | |||
| PS | Accuracy | VGG16 + MLP | 81.58% | 88.89% | |||
| PV | Accuracy | VGG16 + MLP VGG19 + MLP | 89.58% | 92.31% | |||
| SV | Accuracy | VGG19 + MLP | 98.08% | 98.00% | |||
| DSV | Accuracy | Rostami et al. [29] | Ensemble DCNN- based classifier | AZH | 91.90% | 92.11% | |
| SV | Accuracy | 96.40% | 98.00% | ||||
| DV | Precision Recall | Jens et al. [52] | YoloV5m6 | 435 diabetic foot ulcers and 450 venous leg ulcers | 94.20% 83.70 | 92.90% 100% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Yi, W.; Dong, L.; Kong, L.; Liu, M.; Zhao, Y.; Hui, M.; Chu, X. Multi-Class Wound Classification via High and Low-Frequency Guidance Network. Bioengineering 2023, 10, 1385. https://doi.org/10.3390/bioengineering10121385
Guo X, Yi W, Dong L, Kong L, Liu M, Zhao Y, Hui M, Chu X. Multi-Class Wound Classification via High and Low-Frequency Guidance Network. Bioengineering. 2023; 10(12):1385. https://doi.org/10.3390/bioengineering10121385
Chicago/Turabian StyleGuo, Xiuwen, Weichao Yi, Liquan Dong, Lingqin Kong, Ming Liu, Yuejin Zhao, Mei Hui, and Xuhong Chu. 2023. "Multi-Class Wound Classification via High and Low-Frequency Guidance Network" Bioengineering 10, no. 12: 1385. https://doi.org/10.3390/bioengineering10121385
APA StyleGuo, X., Yi, W., Dong, L., Kong, L., Liu, M., Zhao, Y., Hui, M., & Chu, X. (2023). Multi-Class Wound Classification via High and Low-Frequency Guidance Network. Bioengineering, 10(12), 1385. https://doi.org/10.3390/bioengineering10121385







