Screening for Osteoporosis from Blood Test Data in Elderly Women Using a Machine Learning Approach
Abstract
:1. Introduction
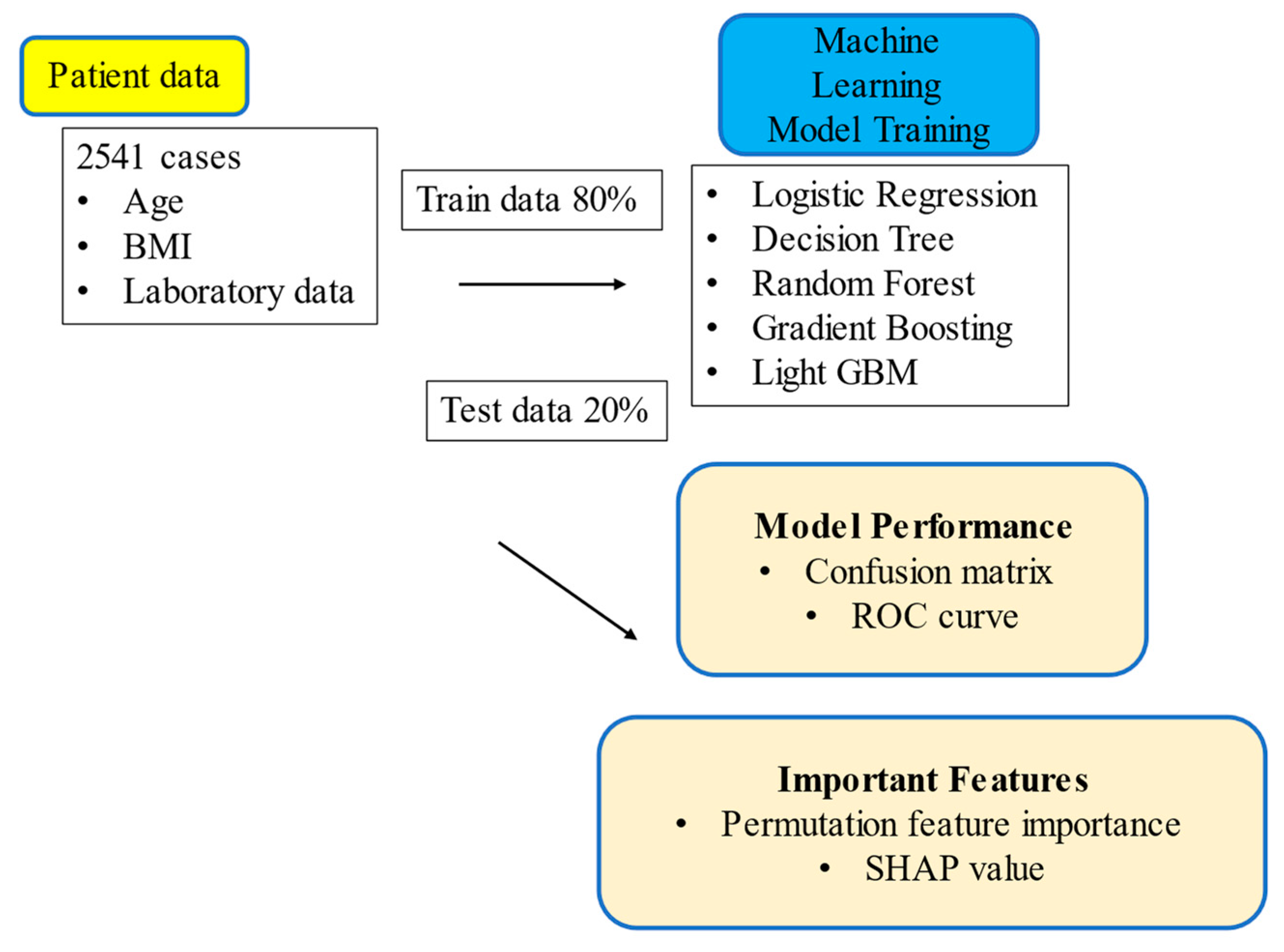
2. Materials and Methods
2.1. Ethical Approval
2.2. Data Collection
2.3. Machine Learning
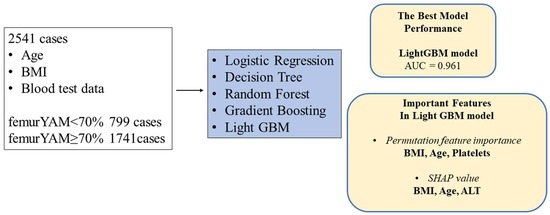
3. Results
3.1. Study Participants
3.2. Prediction of Osteoporosis in each Machine Learning Model
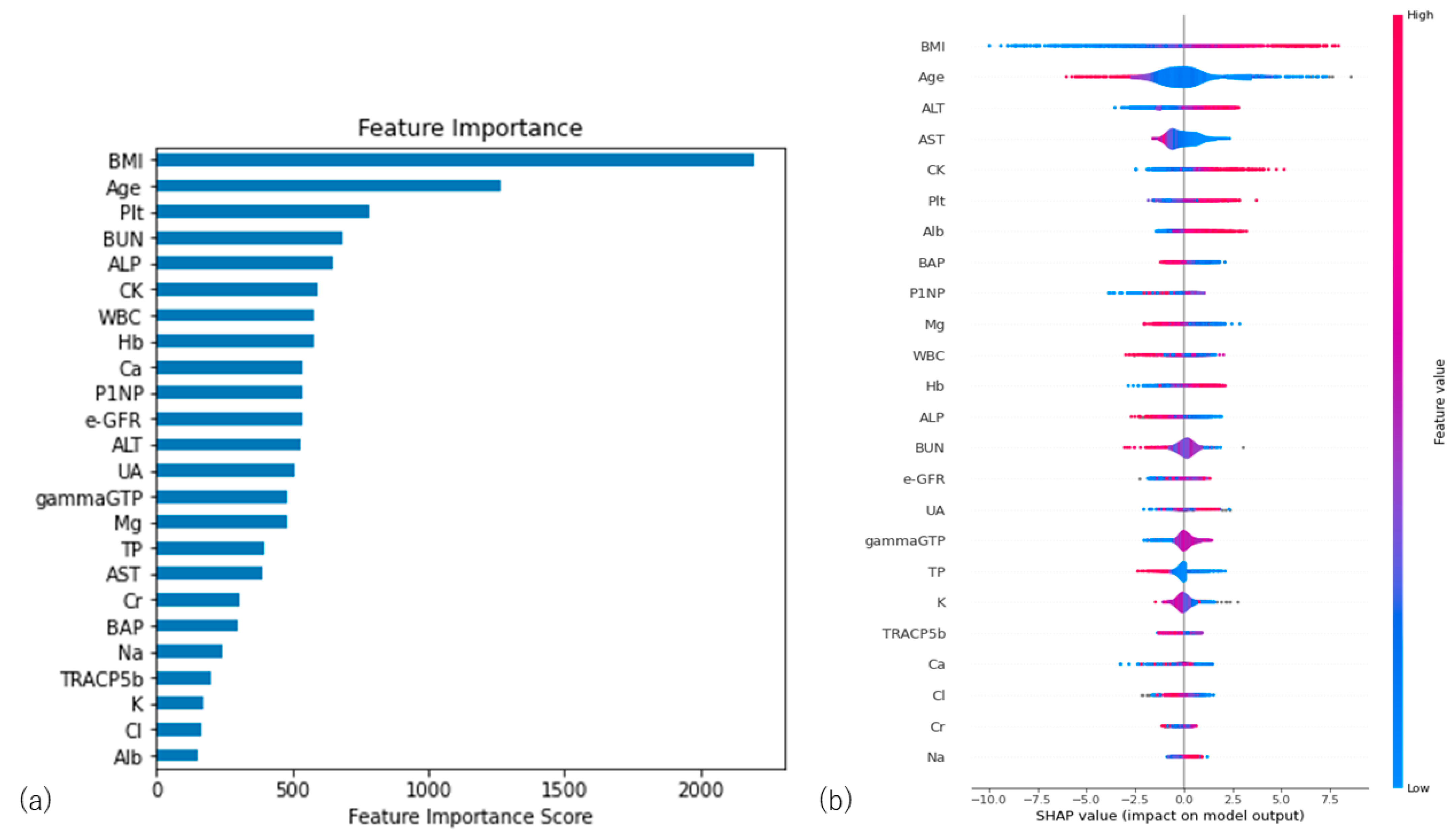
3.3. Importance Values of the Predictors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferizi, U.; Honig, S.; Chang, G. Artificial intelligence, osteoporosis, and fragility fractures. Curr. Opin. Rheumatol. 2019, 31, 368–375. [Google Scholar] [CrossRef]
- Peck, W.A. Consensus development conference: Diagnosis, prophylaxis, and treatment of osteoporosis. Am. J. Med. 1993, 94, 646–650. [Google Scholar]
- Kanis, J.A. Diagnosis of osteoporosis and assessment of fracture risk. Lancet 2002, 359, 1929–1936. [Google Scholar] [CrossRef]
- Kanis, J.A.; McCloskey, E.V.; Johansson, H.; Strom, O.; Borgstrom, F.; Oden, A. National Osteoporosis Guideline Group Case finding for the management of osteoporosis with FRAX®—Assessment and intervention thresholds for the UK. Osteoporos. Int. 2008, 19, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Asch, S.M. Machine Learning and Prediction in Medicine—Beyond the Peak of Inflated Expectations. New Engl. J. Med. 2017, 376, 2507–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, M. Artificial Intelligence Is Becoming Natural. Cell 2018, 173, 531–533. [Google Scholar] [CrossRef] [Green Version]
- Madelin, G.; Poidevin, F.; Makrymallis, A.; Regatte, R.R. Classification of sodium MRI data of cartilage using machine learning. Magn. Reson. Med. 2015, 74, 1435–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruse, C.; Eiken, P.; Vestergaard, P. Machine Learning Principles Can Improve Hip Fracture Prediction. Calcif. Tissue Int. 2017, 100, 348–360. [Google Scholar] [CrossRef]
- Kruse, C.; Eiken, P.; Vestergaard, P. Clinical fracture risk evaluated by hierarchical agglomerative clustering. Osteoporos. Int. 2017, 28, 819–832. [Google Scholar] [CrossRef]
- Villamor, E.; Monserrat, C.; Del Río, L.; Romero-Martín, J.; Rupérez, M. Prediction of osteoporotic hip fracture in postmenopausal women through patient-specific FE analyses and machine learning. Comput. Methods Programs Biomed. 2020, 193, 105484. [Google Scholar] [CrossRef] [PubMed]
- Shioji, M.; Yamamoto, T.; Ibata, T.; Tsuda, T.; Adachi, K.; Yoshimura, N. Artificial neural networks to predict future bone mineral density and bone loss rate in Japanese postmenopausal women. BMC Res. Notes 2017, 10, 590. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-B.; Hu, J.-S.; Tan, K.; Zhang, W.; Xu, T.-H.; Yang, L. Application of machine learning model to predict osteoporosis based on abdominal computed tomography images of the psoas muscle: A retrospective study. BMC Geriatr. 2022, 22, 796. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Matsuo, K.; Aihara, H.; Nakai, T.; Morishita, A.; Tohma, Y.; Kohmura, E. Machine Learning to Predict In-Hospital Morbidity and Mortality after Traumatic Brain Injury. J. Neurotrauma 2020, 37, 202–210. [Google Scholar] [CrossRef]
- Brunelli, A.; Rocco, G. Internal validation of risk models in lung resection surgery: Bootstrap versus training-and-test sampling. J. Thorac. Cardiovasc. Surg. 2006, 131, 1243–1247. [Google Scholar] [CrossRef] [Green Version]
- Crandall, C.J.; Ensrud, K.E. Osteoporosis Screening in Younger Postmenopausal Women. JAMA 2020, 323, 367–368. [Google Scholar] [CrossRef] [PubMed]
- Koh, L.K.H.; Ben Sedrine, W.; Torralba, T.P.; Kung, A.; Fujiwara, S.; Chan, S.P.; Huang, Q.R.; Rajatanavin, R.; Tsai, K.-S.; Park, H.M.; et al. A Simple Tool to Identify Asian Women at Increased Risk of Osteoporosis. Osteoporos. Int. 2001, 12, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Bui, H.M.; Ha, M.H.; Pham, H.G.; Dao, T.P.; Nguyen, T.-T.T.; Nguyen, M.L.; Vuong, N.T.; Hoang, X.H.T.; Do, L.T.; Dao, T.X.; et al. Predicting the risk of osteoporosis in older Vietnamese women using machine learning approaches. Sci. Rep. 2022, 12, 20160. [Google Scholar] [CrossRef]
- Erjiang, E.; Wang, T.; Yang, L.; Dempsey, M.; Brennan, A.; Yu, M.; Chan, W.P.; Whelan, B.; Silke, C.; O’Sullivan, M.; et al. Machine Learning Can Improve Clinical Detection of Low BMD: The DXA-HIP Study. J. Clin. Densitom. 2020, 24, 527–537. [Google Scholar] [CrossRef]
- Ou Yang, W.Y.; Lai, C.C.; Tsou, M.T.; Hwang, L.C. Development of Machine Learning Models for Prediction of Osteoporosis from Clinical Health Examination Data. Int. J. Environ. Res. Public Health 2021, 18, 7635. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Xu, Y.; Cheng, Q.; Jiang, S.; Wang, Q.; Xiao, Y.; Ma, C.; Yan, J.; Wang, X. LightGBM: Accelerated genomically designed crop breeding through ensemble learning. Genome Biol. 2021, 22, 271. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Suda, K.; Maki, S.; Koda, M.; Harmon, S.M.; Komatsu, M.; Ota, M.; Ushirozako, H.; Minami, A.; Takahata, M.; et al. Efficacy of a machine learning-based approach in predicting neurological prognosis of cervical spinal cord injury patients following urgent surgery within 24 h after injury. J. Clin. Neurosci. 2022, 107, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Ahamad, M.; Aktar, S.; Uddin, J.; Rahman, T.; Alyami, S.A.; Al-Ashhab, S.; Akhdar, H.F.; Azad, A.; Moni, M.A. Early-Stage Detection of Ovarian Cancer Based on Clinical Data Using Machine Learning Approaches. J. Pers. Med. 2022, 12, 1211. [Google Scholar] [CrossRef] [PubMed]
- Tsai, I.-J.; Shen, W.-C.; Lee, C.-L.; Wang, H.-D.; Lin, C.-Y. Machine Learning in Prediction of Bladder Cancer on Clinical Laboratory Data. Diagnostics 2022, 12, 203. [Google Scholar] [CrossRef]
- Breitling, L.P. Liver enzymes and bone mineral density in the general population. J. Clin. Endocrinol. Metab. 2015, 100, 3832–3840. [Google Scholar] [CrossRef]
- Kim, J.; Kim, H.S.; Lee, H.S.; Kwon, Y.-J. The relationship between platelet count and bone mineral density: Results from two independent population-based studies. Arch. Osteoporos. 2020, 15, 43. [Google Scholar] [CrossRef]

| Data for Machine Learning (Abbreviation) | Mean | SD |
|---|---|---|
| Age | 73.5 | 9.1 |
| White blood cells (WBCs: counts/µL) | 5783.5 | 1485.5 |
| Hemoglobin (Hb: g/dL) | 12.7 | 1.2 |
| Platelets (Plt: counts × 104/µL) | 22.7 | 6.1 |
| Total protein (TP: g/dL) | 7.1 | 0.4 |
| Albumin (Alb: g/dL) | 4.1 | 0.3 |
| Aspartate transferase (AST: IU/L) | 22.9 | 7.6 |
| Alanine transaminase (ALT: IU/L) | 17.0 | 9.9 |
| Gamma-glutamyl transpeptidase (gammaGTP: IU/L) | 24.3 | 27.8 |
| Alkaline phosphatase (ALP: IU/L) | 222.2 | 87.1 |
| Calcium (Ca: mg/dL) | 9.4 | 0.4 |
| Creatine kinase (CK: IU/L) | 115.4 | 83.2 |
| Chloride (Cl: mEq/L) | 105.1 | 2.6 |
| Sodium (Na: mEq/L) | 141.1 | 2.2 |
| Potassium (K: mEq/L) | 4.1 | 0.3 |
| Magnesium (Mg: mg/dL) | 2.1 | 0.2 |
| Creatinine (Cr: mg/dL) | 0.7 | 0.4 |
| Blood urea nitrogen (BUN: mg/dL) | 17.8 | 5.8 |
| Uric acid (UA: mg/dL) | 4.8 | 1.3 |
| Tartrate-resistant acid phosphatase 5b (TRACP5b: mU/dL) | 379.6 | 172.0 |
| Bone-Specific Alkaline Phosphatase (BAP: µg/L) | 12.9 | 5.3 |
| Procollagen I Intact N-Terminal (PINP: ng/mL) | 49.6 | 37.9 |
| Estimated Glomerular Filtration Rate (eGFR: mL/min) | 66.5 | 17.6 |
| Body mass index (BMI: kg/m2) | 21.7 | 3.4 |
| Osteoporosis Group (n = 799) (femur YAM < 70%) | Normal Group (n = 1742) (femur YAM ≥ 70%) | p Value | |
|---|---|---|---|
| Data for Machine Learning (Abbreviation) | Mean ± SD | Mean ± SD | |
| Age | 76.5 ± 8.5 | 72.1 ± 9.2 | 9.2 × 10−30 |
| White blood cells (WBCs: counts/µL) | 5703 ± 1571 | 5851 ± 1442 | 0.06 |
| Hemoglobin (Hb: g/dL) | 12.3 ± 1.2 | 12.8 ± 1.2 | 3.9 × 10−28 |
| Platelets (Plt: counts × 104/µL) | 21.0 ± 5.7 | 23.1 ± 6.3 | 0.0001 |
| Total protein (TP: g/dL) | 7.1 ± 0.5 | 7.0 ± 1.4 | 0.02 |
| Albumin (Alb: g/dL) | 4.0 ± 0.3 | 4.1 ± 0.3 | 1.5 × 10−11 |
| Aspartate transferase (AST: IU/L) | 23.1 ± 7.6 | 22.8 ± 7.5 | 0.26 |
| Alanine transaminase (ALT: IU/L) | 15.4 ± 10.3 | 17.8 ± 9.7 | 1.8 × 10−8 |
| Gamma-glutamyl transpeptidase (gammaGTP: IU/L) | 23.4 ± 27.0 | 24.7 ± 28.1 | 0.26 |
| Alkaline phosphatase (ALP: IU/L) | 233.2 ± 108.7 | 217.1 ± 74.6 | 1.37 × 10−5 |
| Calcium (Ca: mg/dL) | 9.4 ± 0.5 | 9.4 ± 0.4 | 0.11 |
| Creatine kinase (CK: IU/L) | 105.9 ± 79.8 | 119.9 ± 84.5 | 8.6 × 10−5 |
| Chloride (Cl: mEq/L) | 104.8 ± 2.9 | 105.2 ± 2.5 | 1.1 × 10−3 |
| Sodium (Na: mEq/L) | 140.8 ± 2.3 | 141.2 ± 2.2 | 8.7 × 10−5 |
| Potassium (K: mEq/L) | 4.1 ± 0.4 | 4.1 ± 0.3 | 0.54 |
| Magnesium (Mg: mg/dL) | 2.1 ± 0.2 | 2.1 ± 0.2 | 0.02 |
| Creatinine (Cr: mg/dL) | 0.8 ± 0.6 | 0.7 ± 0.3 | 5.3 × 10−7 |
| Blood urea nitrogen (BUN: mg/dL) | 18.9 ± 6.4 | 17.3 ± 5.4 | 1.5 × 10−5 |
| Uric acid (UA: mg/dL) | 4.8 ± 1.4 | 4.8 ± 1.3 | 0.71 |
| Tartrate-resistant acid phosphatase 5b (TRACP5b: mU/dL) | 399.1 ± 204.1 | 370.6 ± 154.4 | 1.0 × 10−5 |
| Bone-Specific Alkaline Phosphatase (BAP: µg/L) | 13.3 ± 6.8 | 12.7 ± 4.4 | 0.004 |
| Procollagen I Intact N-Terminal (PINP: ng/mL) | 53.3 ± 44.2 | 47.9 ± 34.4 | 6.7 × 10−5 |
| Estimated Glomerular Filtration Rate (eGFR: mL/min) | 64.0 ± 19.2 | 67.6 ± 16.7 | 2.1 × 10−6 |
| Body mass index (BMI: kg/m2) | 20.1 ± 2.9 | 22.5 ± 3.3 | 3.9 × 10−66 |
| Data for Machine Learning (Abbreviation) | Correlation Coefficient between Femur YAM |
|---|---|
| Age | −0.22 |
| Blood urea nitrogen (BUN: mg/dL) | −0.13 |
| Creatinine (Cr: mg/dL) | −0.1 |
| Alkaline phosphatase (ALP: IU/L) | −0.09 |
| Tartrate-resistant acid phosphatase 5b (TRACP5b: mU/dL) | −0.09 |
| Procollagen I Intact N-Terminal (PINP: ng/mL) | −0.08 |
| Bone-Specific Alkaline Phosphatase (BAP: µg/L) | −0.07 |
| Magnesium (Mg: mg/dL) | −0.05 |
| Total Protein (TP: g/dL) | −0.04 |
| Aspartate Transferase (AST: IU/L) | −0.02 |
| Potassium (K: mEq/L) | −0.01 |
| Uric acid (UA: mg/dL) | −0.01 |
| gamma-glutamyl transpeptidase (gammaGTP: IU/L) | 0.02 |
| Calcium (Ca: mg/dL) | 0.03 |
| White blood cells (WBCs: counts/µL) | 0.04 |
| Platelets (Plt: counts × 104/µL) | 0.08 |
| Creatine kinase (CK: IU/L) | 0.08 |
| Chloride (Cl: mEq/L) | 0.08 |
| Sodium (Na: mEq/L) | 0.08 |
| Estimated Glomerular Filtration Rate (eGFR: mL/min) | 0.09 |
| Alanine transaminase (ALT: IU/L) | 0.11 |
| Albumin (Alb: g/dL) | 0.13 |
| Hemoglobin (Hb: g/dL) | 0.22 |
| Body mass index (BMI: kg/m2) | 0.34 |
| ML Model | Logistic Regression (95% CI) | Decision Tree (95% CI) | Random Forest (95% CI) | Gradient Boosting (95% CI) | Light GBM (95% CI) |
|---|---|---|---|---|---|
| Accuracy | 0.772 (0.768–0.776) | 0.739 (0.735–0.743) | 0.775 (0.769–0.781) | 0.800 (0.794–0.806) | 0.834 (0.827–0.841) |
| Precision | 0.771 (0.768–0.774) | 0.737 (0.734–0.742) | 0.764 (0.760–0.769) | 0.800 (0.795–0.801) | 0.835 (0.827–0.843) |
| Recall | 0.956 (0.952–0.960) | 0.968 (0.966–0.970) | 0.978 (0.976–0.980) | 0.957 (0.954–0.959) | 0.961 (0.959–0.963) |
| F-measure | 0.853 (0.851–0.856) | 0.837 (0.834–0.840) | 0.858 (0.855–0.862) | 0.870 (0.867–0.874) | 0.891 (0.887–0.896) |
| ML Model | Logistic Regression | Decision Tree | Random Forest | Gradient Boosting | Light GBM |
|---|---|---|---|---|---|
| Representative parameters | Penalty: l2 C: 100 Solver: lbfgs | Criterion: gini Max depth: 3 | Max depth: 6 Number of estimators: 300 Scoring: auc | Learning rate: 0.19 Number of estimators: 100 scoring: auc | Number of itertions: 1000 Max depth: 6 Scoring: auc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inui, A.; Nishimoto, H.; Mifune, Y.; Yoshikawa, T.; Shinohara, I.; Furukawa, T.; Kato, T.; Tanaka, S.; Kusunose, M.; Kuroda, R. Screening for Osteoporosis from Blood Test Data in Elderly Women Using a Machine Learning Approach. Bioengineering 2023, 10, 277. https://doi.org/10.3390/bioengineering10030277
Inui A, Nishimoto H, Mifune Y, Yoshikawa T, Shinohara I, Furukawa T, Kato T, Tanaka S, Kusunose M, Kuroda R. Screening for Osteoporosis from Blood Test Data in Elderly Women Using a Machine Learning Approach. Bioengineering. 2023; 10(3):277. https://doi.org/10.3390/bioengineering10030277
Chicago/Turabian StyleInui, Atsuyuki, Hanako Nishimoto, Yutaka Mifune, Tomoya Yoshikawa, Issei Shinohara, Takahiro Furukawa, Tatsuo Kato, Shuya Tanaka, Masaya Kusunose, and Ryosuke Kuroda. 2023. "Screening for Osteoporosis from Blood Test Data in Elderly Women Using a Machine Learning Approach" Bioengineering 10, no. 3: 277. https://doi.org/10.3390/bioengineering10030277
APA StyleInui, A., Nishimoto, H., Mifune, Y., Yoshikawa, T., Shinohara, I., Furukawa, T., Kato, T., Tanaka, S., Kusunose, M., & Kuroda, R. (2023). Screening for Osteoporosis from Blood Test Data in Elderly Women Using a Machine Learning Approach. Bioengineering, 10(3), 277. https://doi.org/10.3390/bioengineering10030277