Automated Quantification of Pneumonia Infected Volume in Lung CT Images: A Comparison with Subjective Assessment of Radiologists
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Image Preprocessing
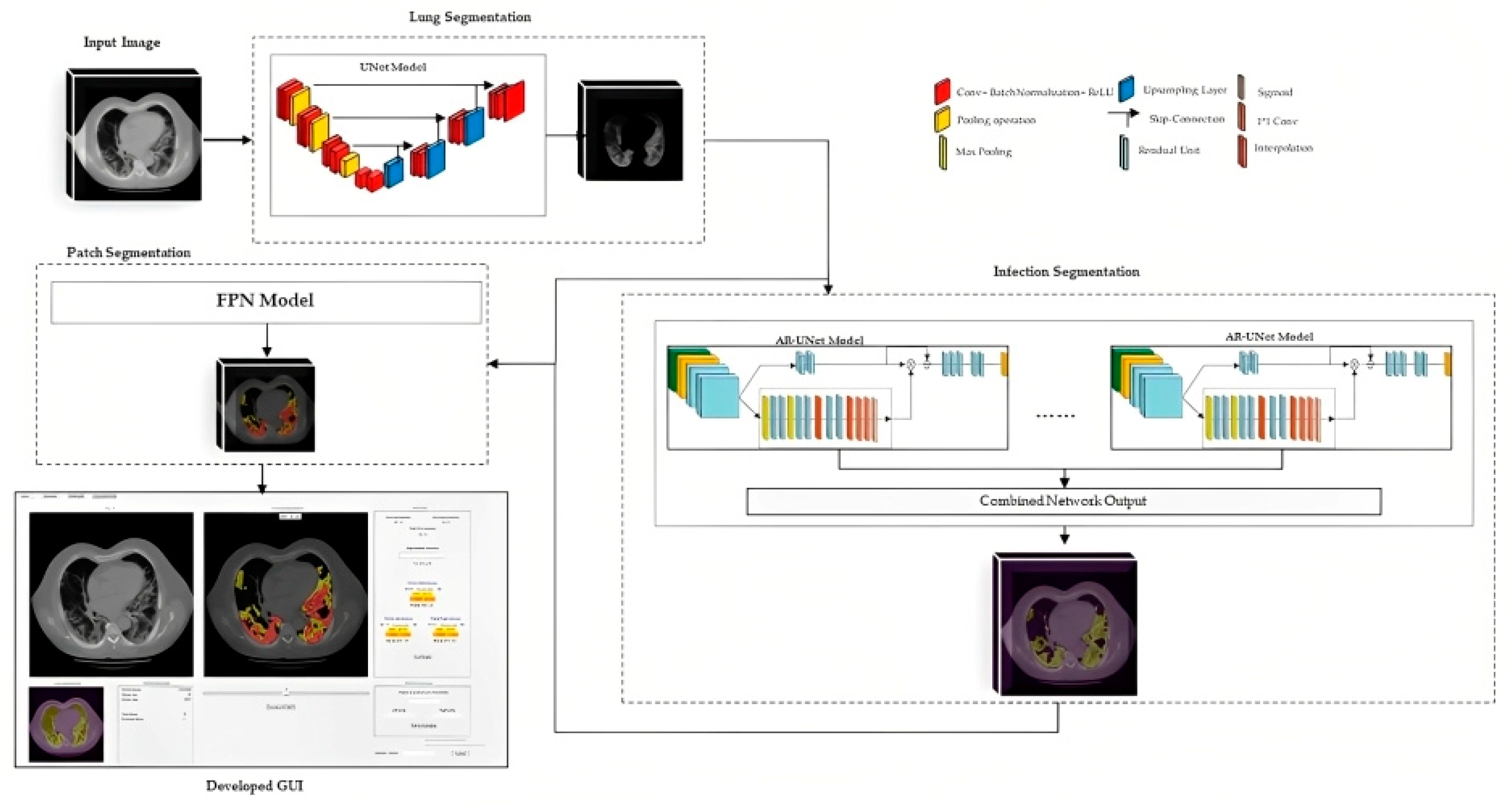
2.3. Image Segmentation Models and Output
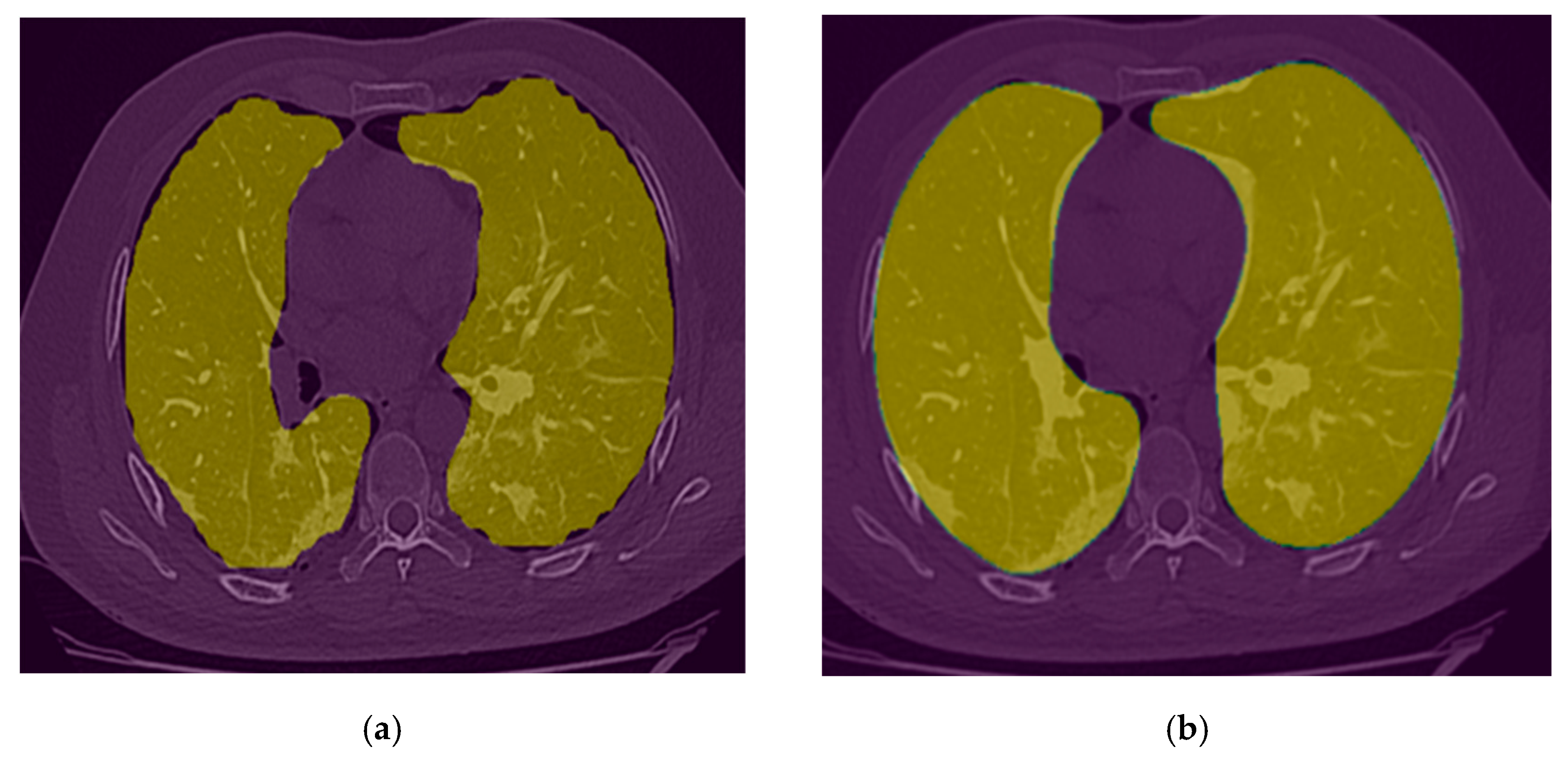
2.3.1. Lung Segmentation
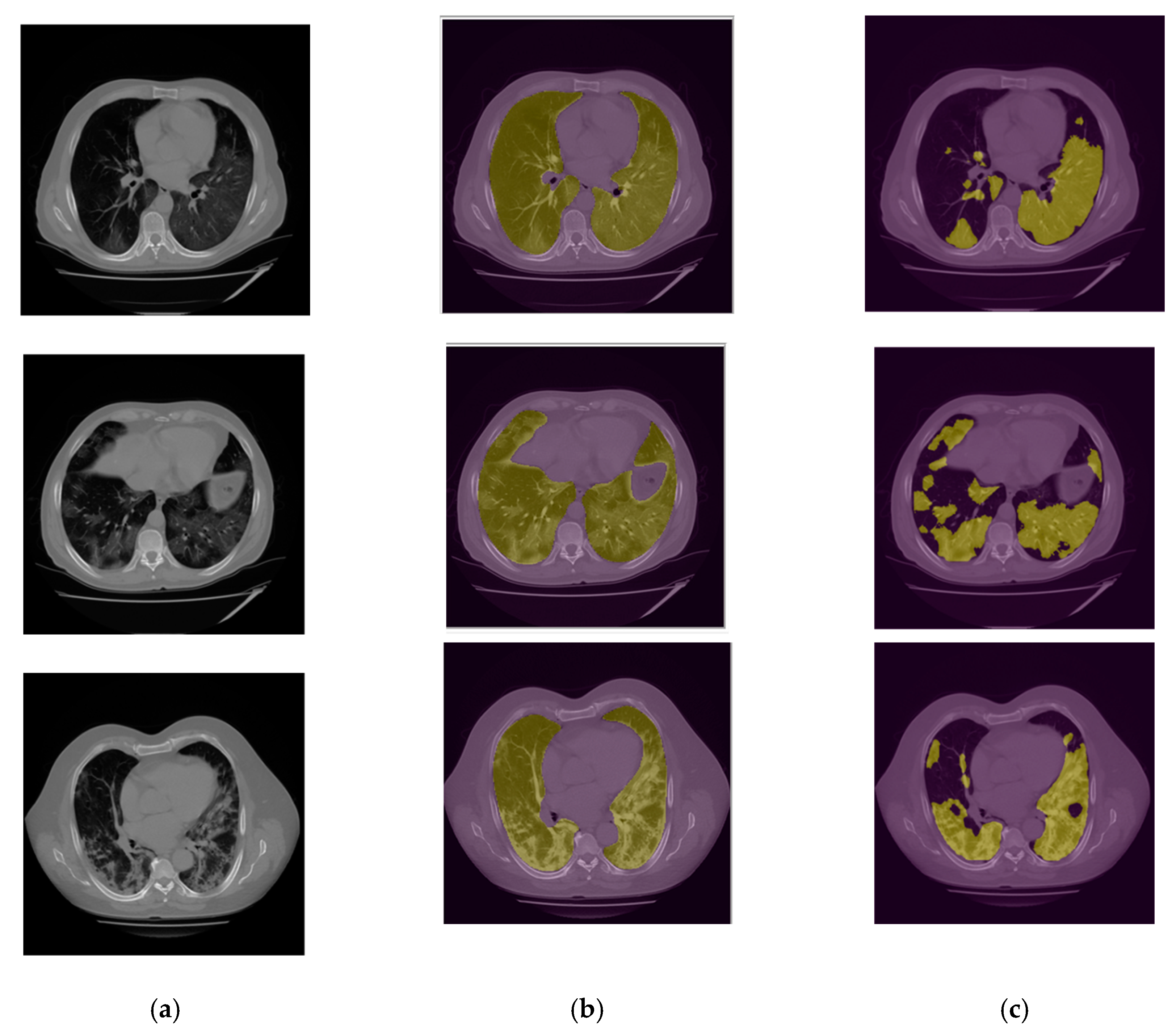
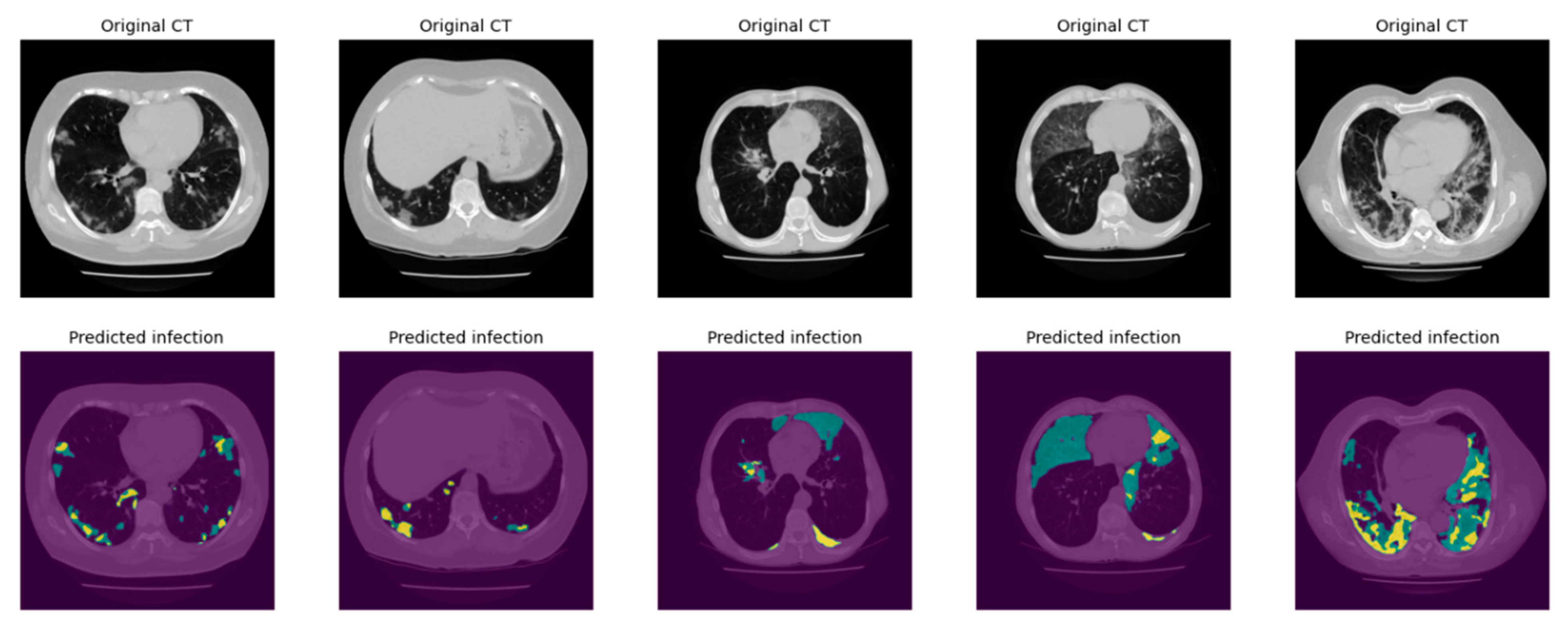
2.3.2. Infection Area Segmentation
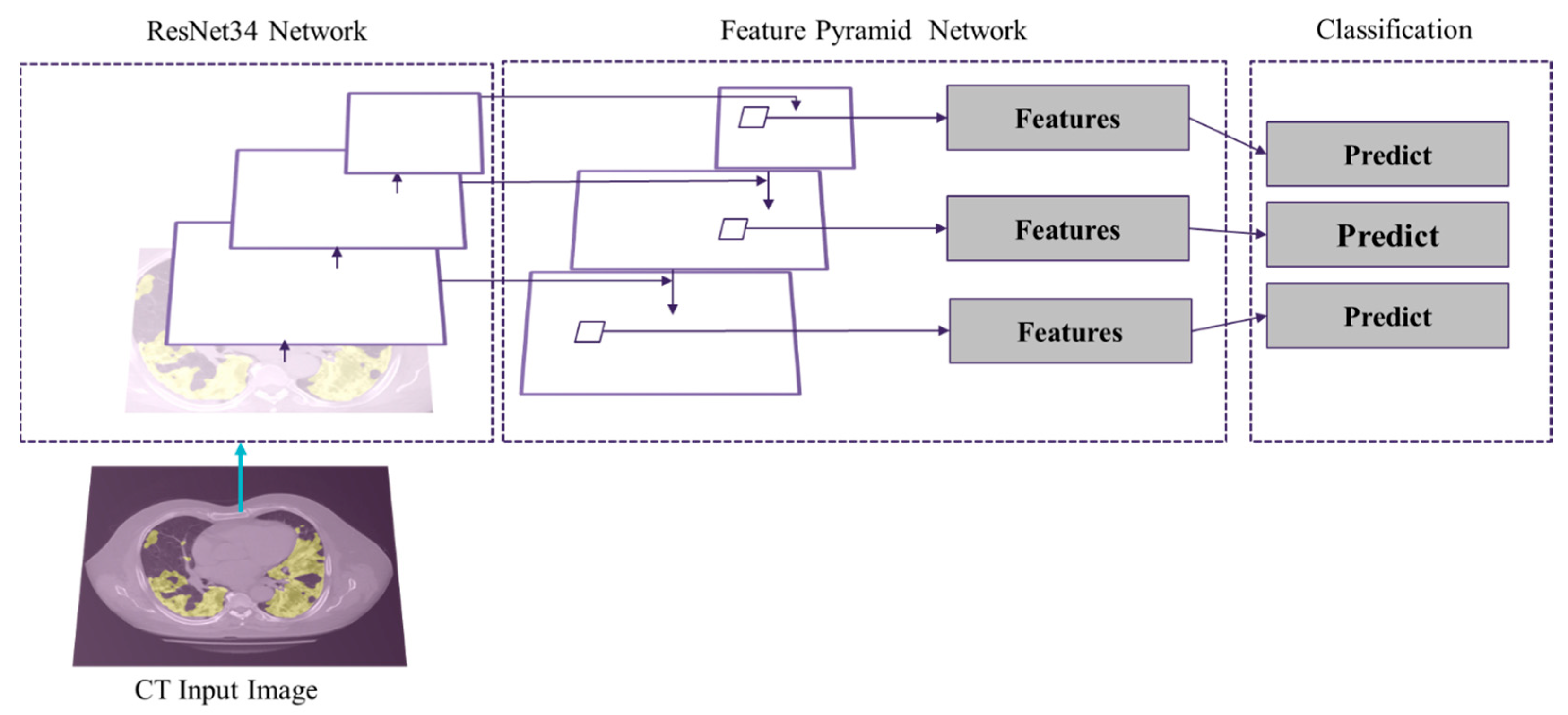
2.3.3. Segmentation of GGO and Consolidation Patches
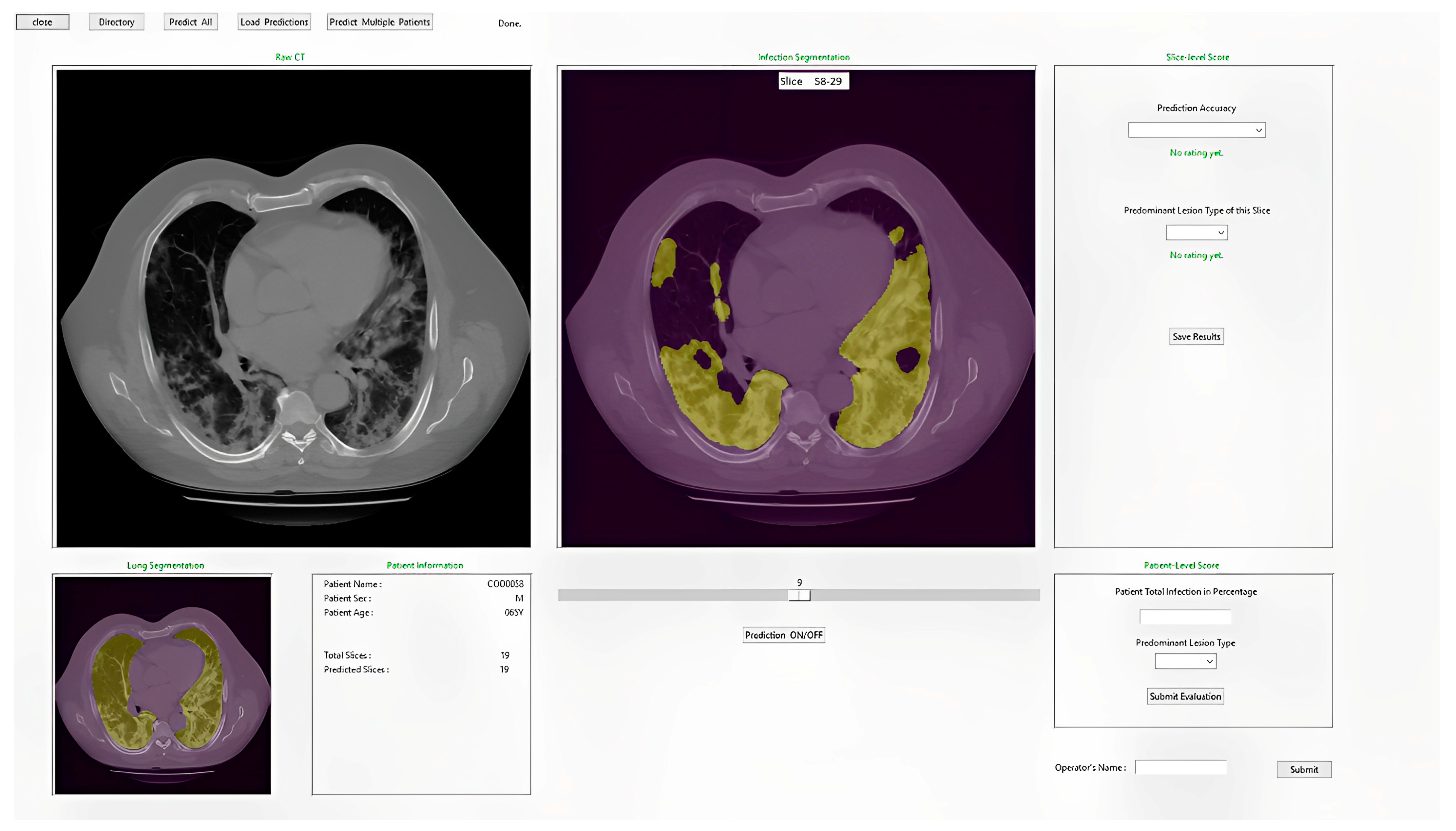
2.3.4. Integrated Model and GUI
2.4. Image Postprocessing and Correction
2.5. Evaluation
3. Results
- (1)
- From Figure 10a, we observe that in 34% (27/80) of testing cases, the difference between the DL model generated diseased region segmentation and radiologist’s estimation is less than 5% (indicating the accuracy > 95%).
- (2)
- In 55% (44/80) of testing cases, the difference between the DL model generated diseased region segmentation and radiologist’s estimation is less than 10% (or accuracy > 90%).
- (3)
- In 90% (72/80) of testing cases, the difference between the DL model generated diseased region segmentation and radiologist’s estimation is less than 30% (or accuracy > 70%).
- (4)
- From Figure 10b, we observe that in 73% (58/80) of testing cases, radiologists rated a score of 3 or higher indicating an acceptable lung and disease-infection region segmentation results generated by the DL model.
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Müller, N.L.; Staples, C.A.; Miller, R.R.; Abboud, R.T. “Density mask”: An objective method to quantitate emphysema using computed tomography. Chest 1988, 94, 782–787. [Google Scholar] [CrossRef]
- Karimi, R.; Tornling, G.; Forsslund, H.; Mikko, M.; Wheelock, M.; Nyrén, S.; Sköld, C.M. Lung density on high resolution computer tomography (HRCT) reflects degree of inflammation in smokers. Respir. Res. 2014, 15, 23. [Google Scholar] [CrossRef] [Green Version]
- Ciotti, M.; Ciccozzi, M.; Terrinoni, A.; Jiang, W.C.; Wang, C.B.; Bernardini, S. The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci. 2020, 57, 365–388. [Google Scholar] [CrossRef]
- Heidari, M.; Mirniaharikandehei, S.; Khuzani, A.Z.; Danala, G.; Qiu, Y.; Zheng, B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inform. 2020, 144, 104284. [Google Scholar] [CrossRef]
- Fan, D.-P.; Zhou, T.; Ji, G.-P.; Zhou, Y.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Inf-net: Automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging 2020, 39, 2626–2637. [Google Scholar] [CrossRef]
- Wang, S.; Kang, B.; Ma, J.; Zeng, X.; Xiao, M.; Guo, J.; Cai, M.; Yang, J.; Li, Y.; Meng, X.; et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). Eur. Radiol. 2021, 31, 6096–6104. [Google Scholar] [CrossRef]
- Porwal, P.; Pachade, S.; Kokare, M.; Deshmukh, G.; Son, J.; Bae, W.; Liu, L.; Wang, J.; Liu, X.; Gao, L.; et al. IDRid: Diabetic retinopathy—Segmentation and grading challenge. Med. Image Anal. 2020, 59, 101561. [Google Scholar] [CrossRef]
- Shi, T.; Jiang, H.; Zheng, B. C2MA-Net: Cross-modal cross-attention network for acute ischemic stroke lesion segmentation based on CT perfusion scans. IEEE Trans. Biomed. Eng. 2022, 69, 108–118. [Google Scholar] [CrossRef]
- Jones, M.A.; Islam, W.; Faiz, R.; Chen, X.; Zheng, B. Applying artificial intelligence technology to assist with breast cancer diagnosis and prognosis prediction. Front. Oncol. 2022, 12, 980793. [Google Scholar] [CrossRef]
- Islam, W.; Jones, M.; Faiz, R.; Sadeghipour, N.; Qiu, Y.; Zheng, B. Improving performance of breast lesion classification using a ResNet50 model optimized with a novel attention mechanism. Tomography 2022, 8, 2411–2425. [Google Scholar] [CrossRef]
- Wu, Y.-H.; Gao, S.-H.; Mei, J.; Xu, J.; Fan, D.-P.; Zhang, R.-G.; Cheng, M.-M. JCS: An explainable COVID-19 diagnosis system by joint classification and segmentation. IEEE Trans. Image Process. 2021, 30, 3113–3126. [Google Scholar] [CrossRef]
- Abbas, A.; Abdelsamea, M.M.; Gaber, M.M. 4S-DT: Self-supervised super sample decomposition for transfer learning with application to COVID-19 detection. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 2798–2808. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Zhuang, Y.; Rahman, M.F.; Wen, Y.; Pokojovy, M.; McCaffrey, P.; Vo, A.; Walser, E.; Moen, S.; Xu, H.; Tseng, T.L. An interpretable multi-task system for clinically applicable COVID-19 diagnosis using CXR. J. X-Ray Sci. Technol. 2022, 30, 847–862. [Google Scholar] [CrossRef]
- Clement, J.C.; Ponnusamy, V.; Sriharipriya, K.C.; Nandakumar, R. A survey on mathematical, machine learning and deep learning models for COVID-19 transmission and diagnosis. IEEE Rev. Biomed. Eng. 2022, 15, 325–340. [Google Scholar]
- Roberts, M.; Driggs, D.; Thorpe, M.; Gilbey, J.; Yeung, M.; Ursprung, S.; Aviles-Rivero, A.I.; Etmann, C.; McCague, C.; Beer, L.; et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021, 3, 199–217. [Google Scholar] [CrossRef]
- Tian, X.; Huang, R.Y. Standardization of imaging methods for machine learning in neuro-oncology. Neuro-Oncol. Adv. 2020, 2 (Suppl. S4), iv49–iv55. [Google Scholar]
- Simard, P.Y.; Steinkraus, D.; Platt, J.C. Best practices for convolutional neural networks applied to visual document analysis. In Proceedings of the Seventh International Conference on Document Analysis and Recognition, IEEE, Edinburgh, UK, 6 August 2003. [Google Scholar] [CrossRef]
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and flexible image augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef] [Green Version]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Yakubovskiy, P. Segmentation Models; GitHub Repository: San Francisco, CA, USA, 2019. [Google Scholar]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2017; pp. 379–387. [Google Scholar]
- Li, C.; Liu, Y.; Yin, H.; Li, Y.; Guo, Q.; Zhang, L.; Du, P. Attention residual U-Net for building segmentation in aerial images. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium, 11–16 July 2021. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, Y.; Zhao, K.; Tian, Y. A comprehensive survey of loss functions in machine learning. Ann. Data Sci. 2022, 9, 187–212. [Google Scholar] [CrossRef]
- Park, S.C.; Tan, J.; Wang, X.; Lederman, D.; Leader, J.K.; Kim, S.H.; Zheng, B. Computer-aided detection of early interstitial lung diseases using low-dose CT images. Phys. Med. Biol. 2011, 56, 1139–1153. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef] [Green Version]
- Pu, J.; Leader, J.K.; Zheng, B.; Knollmann, F.; Fuhrman, C.; Sciurba, F.C.; Gur, D. A computational geometry approach to automated pulmonary fissure segmentation in CT examinations. IEEE Trans. Med. Imaging 2008, 28, 710–719. [Google Scholar]




| Loss Function | Augmentation | Dropout | |
|---|---|---|---|
| Model 1 | Binary Cross Entropy | 5 times | 0 |
| Model 2 | Tversky | 10 times | 0 |
| Model 3 | Tversky | 10 times | 0.10 |
| Model 4 | Binary Cross Entropy | 10 times | 0 |
| Model 5 | Binary Focal Loss | 5 times | 0.10 |
| Radiologists\Model | A | C |
|---|---|---|
| A | 61 | 2 |
| C | 10 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirniaharikandehei, S.; Abdihamzehkolaei, A.; Choquehuanca, A.; Aedo, M.; Pacheco, W.; Estacio, L.; Cahui, V.; Huallpa, L.; Quiñonez, K.; Calderón, V.; et al. Automated Quantification of Pneumonia Infected Volume in Lung CT Images: A Comparison with Subjective Assessment of Radiologists. Bioengineering 2023, 10, 321. https://doi.org/10.3390/bioengineering10030321
Mirniaharikandehei S, Abdihamzehkolaei A, Choquehuanca A, Aedo M, Pacheco W, Estacio L, Cahui V, Huallpa L, Quiñonez K, Calderón V, et al. Automated Quantification of Pneumonia Infected Volume in Lung CT Images: A Comparison with Subjective Assessment of Radiologists. Bioengineering. 2023; 10(3):321. https://doi.org/10.3390/bioengineering10030321
Chicago/Turabian StyleMirniaharikandehei, Seyedehnafiseh, Alireza Abdihamzehkolaei, Angel Choquehuanca, Marco Aedo, Wilmer Pacheco, Laura Estacio, Victor Cahui, Luis Huallpa, Kevin Quiñonez, Valeria Calderón, and et al. 2023. "Automated Quantification of Pneumonia Infected Volume in Lung CT Images: A Comparison with Subjective Assessment of Radiologists" Bioengineering 10, no. 3: 321. https://doi.org/10.3390/bioengineering10030321
APA StyleMirniaharikandehei, S., Abdihamzehkolaei, A., Choquehuanca, A., Aedo, M., Pacheco, W., Estacio, L., Cahui, V., Huallpa, L., Quiñonez, K., Calderón, V., Gutierrez, A. M., Vargas, A., Gamero, D., Castro-Gutierrez, E., Qiu, Y., Zheng, B., & Jo, J. A. (2023). Automated Quantification of Pneumonia Infected Volume in Lung CT Images: A Comparison with Subjective Assessment of Radiologists. Bioengineering, 10(3), 321. https://doi.org/10.3390/bioengineering10030321









