Bioactive Aspergteroids G–J from Soft-Coral-Associated Symbiotic and Epiphytic Fungus Aspergillus terreus EGF7-0-1
Abstract
1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Fungal Materials, Extraction, and Fermentation
2.3. Isolation
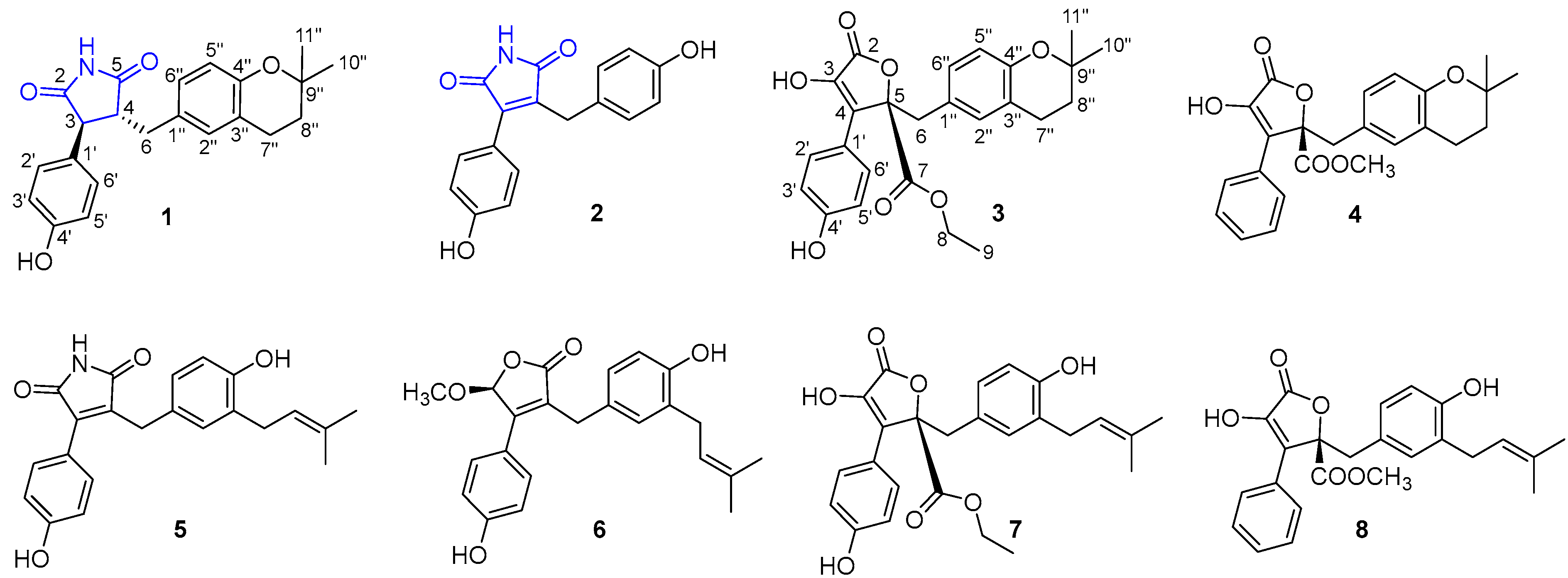
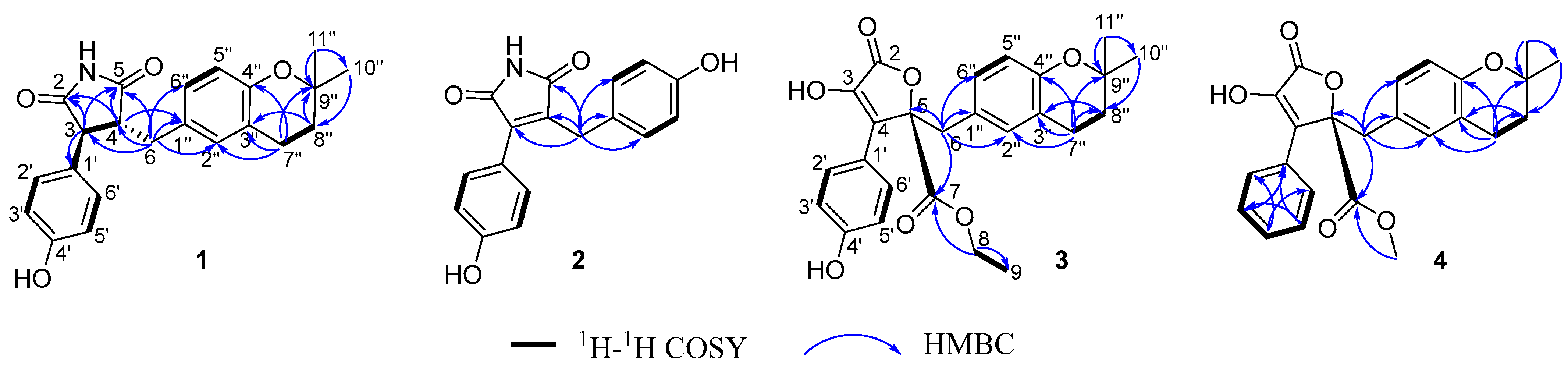
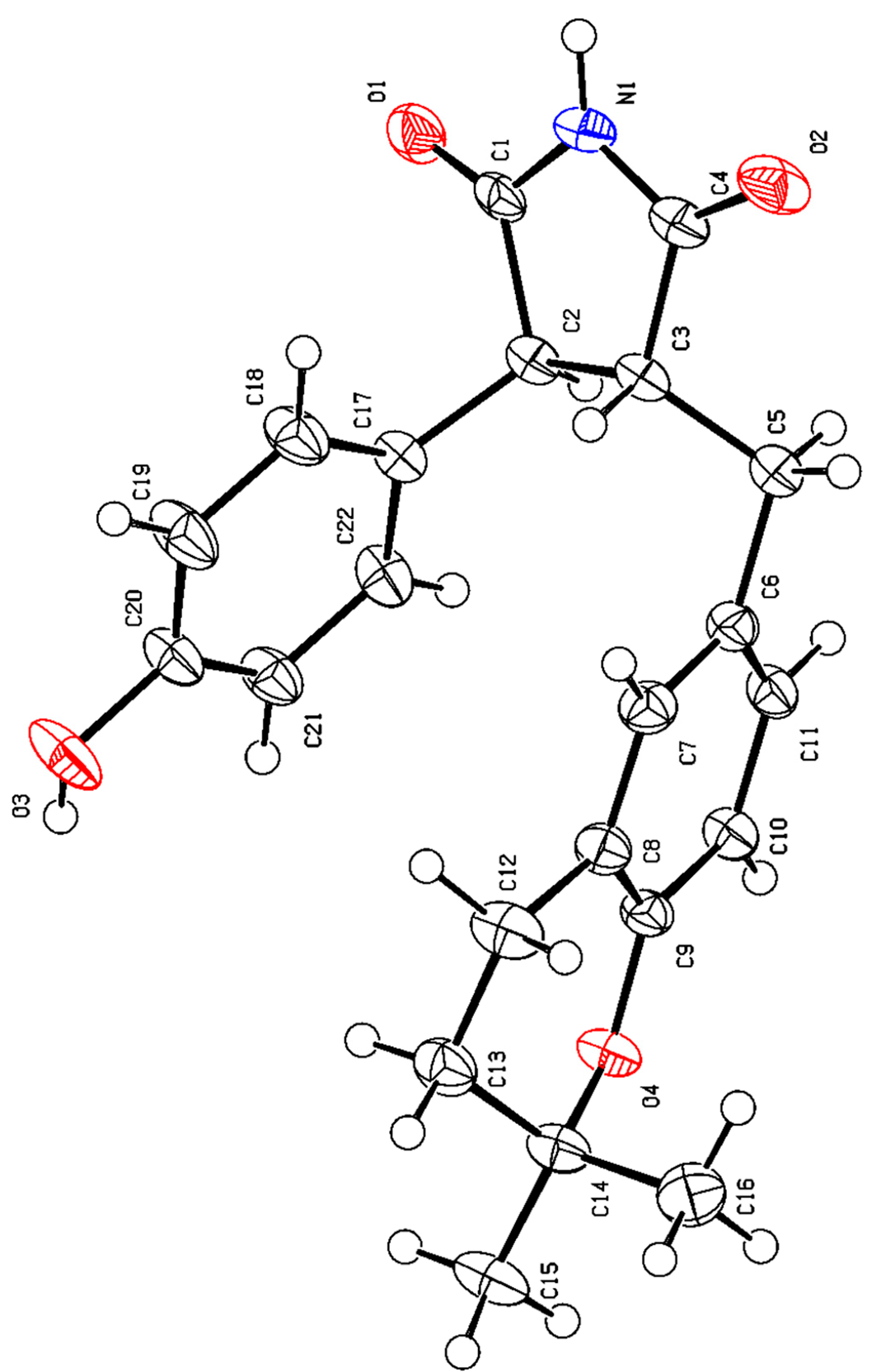
2.4. Structural Characterizations of Compounds 1–8
2.5. Quantum Chemical Calculations
2.6. Cell Viability Assays
2.7. Targets Prediction
2.8. Western Blot Assays
3. Results and Discussion
3.1. Structural Identifications of New Compounds
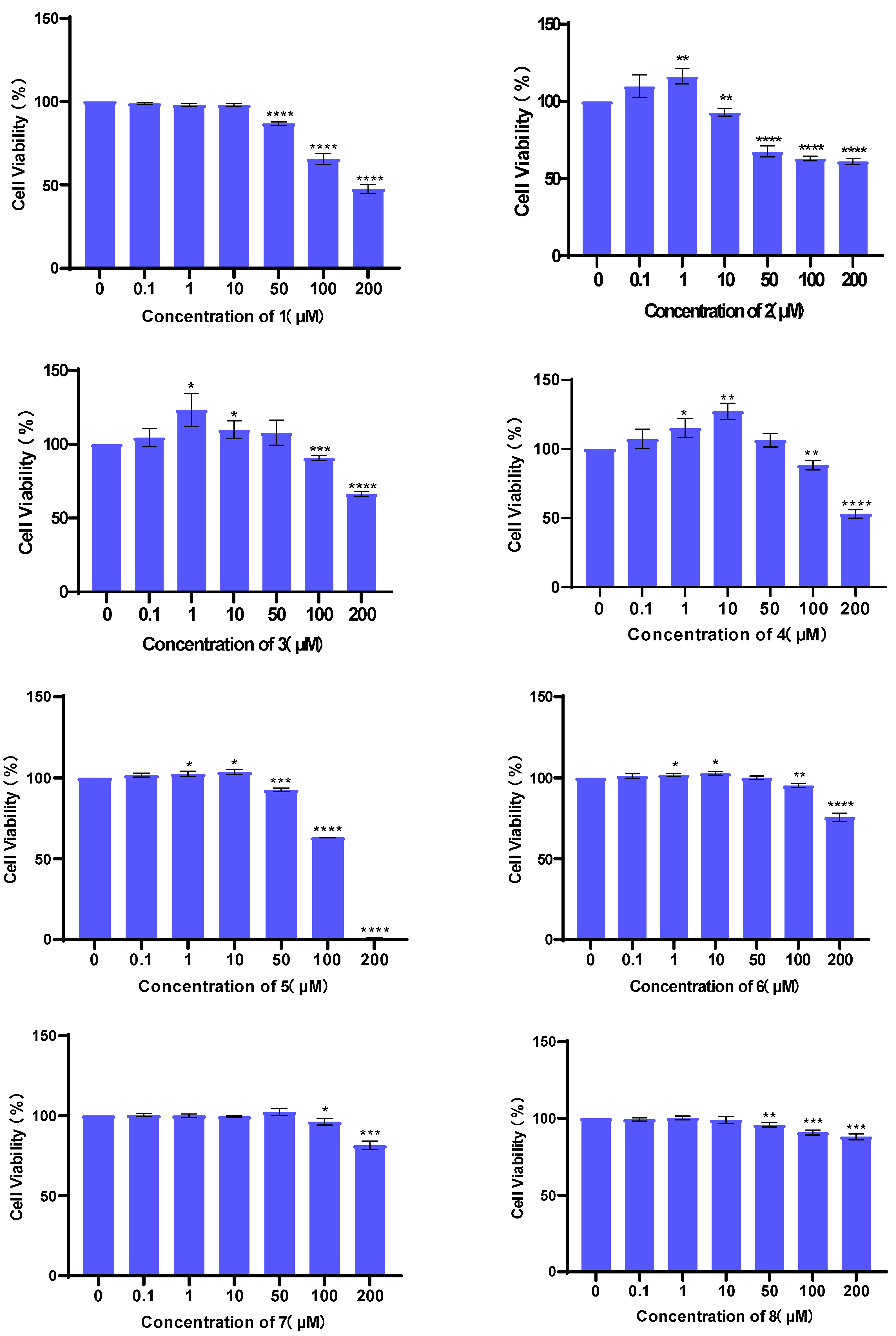
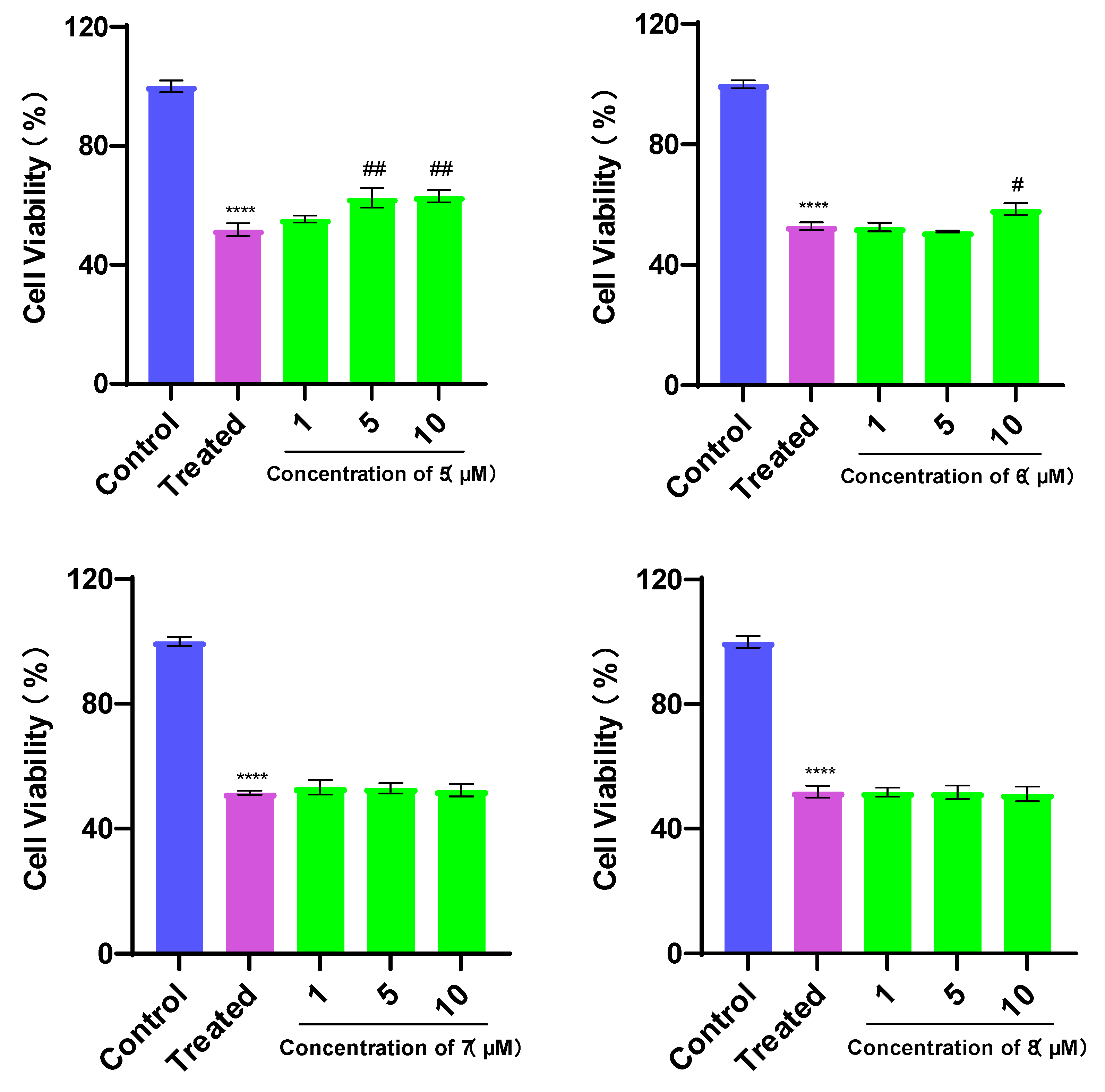
3.2. Evaluation of Cell Viability
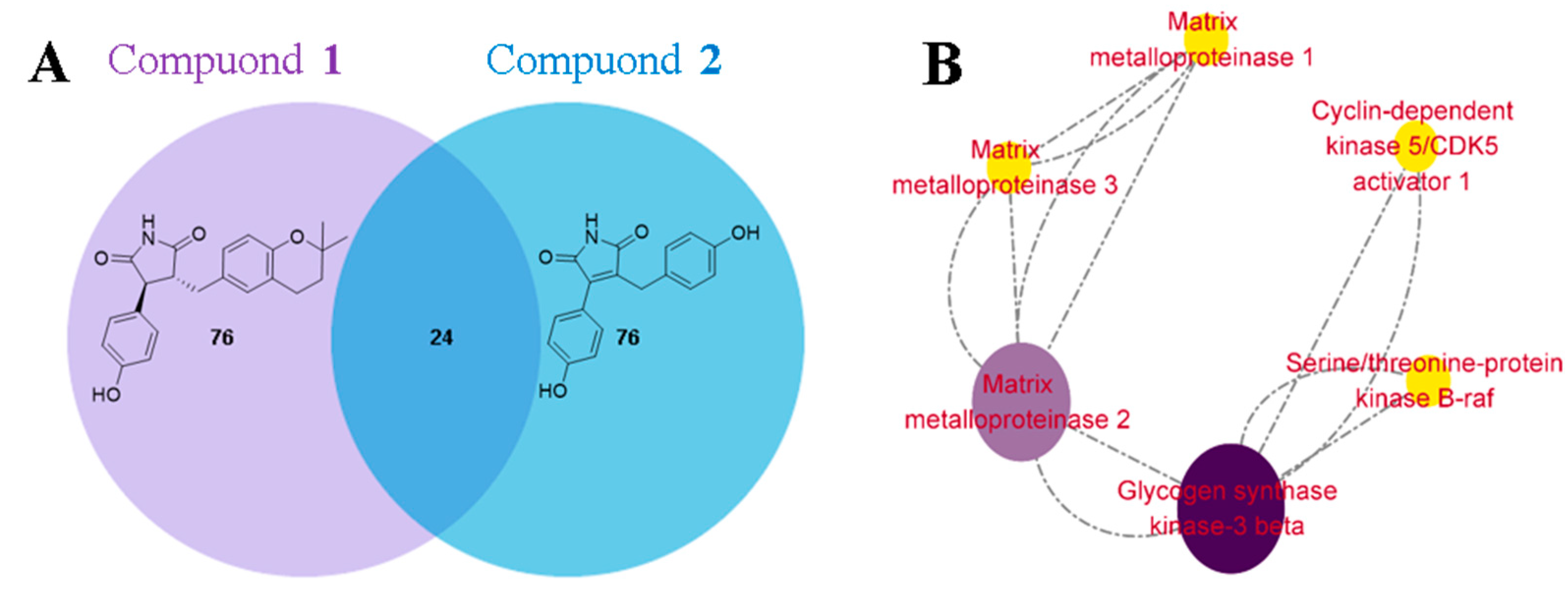
3.3. Evaluation of Cardioprotective Effects
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Read, S.H.; Wild, S.H. Prevention of premature cardiovascular death worldwide. Lancet 2020, 395, 758–760. [Google Scholar] [CrossRef] [PubMed]
- Umbarkar, P.; Tousif, S.; Singh, A.P.; Anderson, J.C.; Zhang, Q.; Tallquist, M.D.; Woodgett, J.; Lal, H. Fibroblast GSK-3α promotes fibrosis via RAF-MEK-ERK pathway in the injured heart. Circ. Res. 2022, 131, 620–636. [Google Scholar] [CrossRef] [PubMed]
- Hosoe, T.; Iizuka, T.; Komai, S.; Wakana, D.; Itabashi, T.; Nozawa, K.; Fukushima, K.; Kawai, K. 4-Benzyl-3-phenyl-5H-furan-2-one, a vasodilator isolated from Malbranchea filamentosa IFM 41300. Phytochemistry 2005, 66, 2776–2779. [Google Scholar] [CrossRef]
- San-Martín, A.; Rovirosa, J.; Vaca, I.; Vergara, K.; Acevedo, L.; Vina, D.; Orallo, F.; Chamy, M.C. New butyrolactine from a marine-derived fungus Aspergillus sp. J. Chil. Chem. Soc. 2011, 56, 625–627. [Google Scholar] [CrossRef]
- Peng, Q.Y.; Chen, W.H.; Lin, X.P.; Xiao, J.; Liu, Y.H.; Zhou, X.F. Butenolides from the Coral-Derived fungus Aspergillius terreus SCSIO41404. Mar. Drugs 2022, 20, 212. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.W.; Lin, Y.; Lu, Y.J.; Zhou, X.F.; Liu, Y.H. Peptides and polyketides isolated from the marine sponge-derived fungus Aspergillus terreus SCSIO 41008. Chin. J. Nat. Med. 2019, 17, 149–154. [Google Scholar] [CrossRef]
- He, F.; Bao, J.; Zhang, X.Y.; Tu, Z.C.; Shi, Y.M.; Qi, S.H. Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J. Nat. Prod. 2013, 76, 1182–1186. [Google Scholar] [CrossRef]
- Hamed, A.; Abdel-Razek, A.S.; Omran, D.A.; EiMetwally, M.M.; Ei-Hosari, D.G.; Frese, M.; Soliman, S.M.; Sewald, N.; Shaaban, M. Terretonin O: A new meroterpenoid from Aspergillus terreus. Nat. Prod. Res. 2019, 34, 965–974. [Google Scholar] [CrossRef]
- Liu, C.; Liu, H.; Wen, Y.Z.; Huang, H.Q.; Hao, J.; Lv, Y.B.; Qin, R.; Yang, X.Z. Aspernolide a inhibits the proliferation of human laryngeal carcinoma cells through the mitochondrial apoptotic and STAT3 signaling pathways. Molecules 2019, 24, 1074. [Google Scholar] [CrossRef]
- Dewi, R.T.; Tachibana, S.; Darmawan, A. Effect on α-glucosidase inhibition and antioxidant activities of butyrolactone derivatives from Aspergillus terreus MC751. Med. Chem. Res. 2014, 23, 454–460. [Google Scholar] [CrossRef]
- Fan, H.; Wei, X.; Si-Tu, M.X.; Lei, Y.H.; Zhou, F.G.; Zhang, C.X. γ-aromatic butenolides of microbial source—A review of their structures, biological activities and biosynthesis. Chem. Biodiv. 2022, 19, e202200208. [Google Scholar] [CrossRef]
- Liao, G.F.; Wu, P.; Xue, J.H.; Liu, L.; Li, H.X.; Wei, X.Y. Asperimides A–D, anti-inflammatory aromatic butenolides from a tropical endophytic fungus Aspergillus terreus. Fitoterapia 2018, 131, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.R.; Elkhayat, E.S.; Mohamed, G.A.; Khedr, A.I.; Fouad, M.A.; Kotb, M.H.; Ross, S.A. Aspernolides F and G, new butyrolactones from the endophytic fungus Aspergillus terreus. Phytochem. Lett. 2015, 14, 84–90. [Google Scholar] [CrossRef]
- Gu, W.; Qiao, C. Furandiones from an endophytic Aspergillus terreus residing in Malus halliana. Chem. Pharm. Bull. 2012, 60, 1474–1477. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.; Sadhukhan, A.; Veerender, M.; Ravikumar, V.; Mohan, E.; Dhanvantri, S.; Sitaramkumar, M.; Babu, J.M.; Vyas, K.; Reddy, G.O. Butyrolactones from Aspergillus terreus. Chem. Pharm. Bull. 2000, 48, 559–562. [Google Scholar] [CrossRef]
- Guo, F.; Li, Z.; Xu, X.; Wang, K.; Shao, M.; Zhao, F.; Wang, H.; Hua, H.; Pei, Y.; Bai, J. Butenolide derivatives from the plant endophytic fungus Aspergillus terreus. Fitoterapia 2016, 113, 44–50. [Google Scholar] [CrossRef]
- Li, L.J.; Li, T.X.; Kong, L.Y.; Yang, M.H. Antioxidant aromatic butenolides from an insect-associated Aspergillus iizukae. Phytochem. Lett. 2016, 16, 134–140. [Google Scholar] [CrossRef]
- Wei, X.; Su, J.C.; Hu, H.S.; He, X.X.; Lin, S.J.; Zhang, D.M.; Ye, W.C.; Chen, M.F.; Lin, H.W.; Zhang, C.X. Probing indole diketopiperazine-based hybrids as environmental-2 induced products from Aspergillus sp. EGF 15-0-3. Org. Lett. 2022, 24, 158–163. [Google Scholar] [CrossRef]
- Cheng, Y.J.; Chen, N.N.; Li, J.; Su, J.C.; Yang, J.Y.; Zhang, C.X.; Lin, H.W.; Zhou, Y.J. Antimicrobial chlorinated carbazole alkaloids from the sponge-associated actinomycete streptomyces diacarni LHW51701. Chin. J. Chem. 2021, 39, 1188–1192. [Google Scholar] [CrossRef]
- Fan, H.; Shi, Z.M.; Lei, Y.H.; Si-Tu, M.X.; Zhou, F.G.; Feng, C.; Wei, X.; Shao, X.H.; Zhang, C.X. Rare carbon-bridged citrinin dimers from the starfish-derived symbiotic fungus Penicillium sp. GGF16-1-2. Mar. Drugs 2022, 20, 443. [Google Scholar] [CrossRef]
- Sun, Y.T.; Liu, J.T.; Li, L.; Gong, C.; Wang, S.P.; Yang, F.; Hua, H.M.; Lin, H.W. New butenolide derivatives from the marine sponge-derived fungus Aspergillus terreus. Bioorg. Med. Chem. Lett. 2018, 28, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Rachada, H.; Pranee, R.; Rungtiwa, C.; Nattawut, B.; Masahiko, I. Butyrolactones from the Fungus Aspergillus terreus BCC 4651. Chem. Pharm. Bull. 2010, 58, 1545–1548. [Google Scholar]
- Nakamura, N.; Hirakawa, A.; Gao, J.J.; Kakuda, H.; Shiro, M.; Komatsu, Y.; Sheu, C.C.; Hattori, M. Five new maleic and succinic acid derivatives from the mycelium of Antrodia camphorata and their cytotoxic effects on LLC tumor cell line. J. Nat. Prod. 2004, 67, 46–48. [Google Scholar] [CrossRef]
- Wu, M.D.; Cheng, M.J.; Wang, B.C.; Yech, Y.J.; Lai, J.T.; Kuo, Y.H.; Yuan, G.F.; Chen, I.S. Maleimide and maleic anhydride derivatives from the mycelia of Antrodia cinnamomea and their nitric oxide inhibitory activities in macrophages. J. Nat. Prod. 2008, 71, 1258–1261. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Li, X.; He, F.; Zhang, X.Y.; Zhu, K.K.; Tao, H.R.; Yu, J.H.; Liu, H.S.; Zhang, H. Asperbutenolide A, an unusual aromatic butenolide dimer with diverse bioactivities from a marine-derived fungus Aspergillus terreus SCAU011. Tetrahedron Lett. 2020, 61, 152193. [Google Scholar] [CrossRef]
- Senoner, T.; Dichtl, W. Oxidative stress in cardiovascular diseases: Still a therapeutic target? Nutrients 2019, 11, 2090. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. Swiss Target Prediction: A web server for target prediction of bioactive small molecules. Nucleic Acids. Res. 2019, 47, 32–38. [Google Scholar]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids. Res. 2019, 47, 607–613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wang, Z.R.; Yao, M.R.; Wang, L.Y.; Yang, Y.Q.; Wang, Q.; Qian, X.; Zhao, Y.; Qiao, J.Q. Dexmedetomidine attenuates myocardial ischemia/reperfusion-induced ferroptosis via AMPK/GSK-3β/Nrf2 axis. Biomed. Pharmacother. 2022, 154, 113572. [Google Scholar] [CrossRef]



| No. | 1 | 2 | 3 | 4 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| δH, mult, (J in Hz) | δC, Type | No. | δH, mult, (J in Hz) | δC, Type | No. | δH, mult, (J in Hz) | δC, Type | No. | δH, mult, (J in Hz) | δC, Type | |
| 2 | 181.3, C | 2 | 174,7 C | 2 | 170.8, C | 2 | 169.8, C | ||||
| 3 | 3.64, d (5.7) | 53.2, CH | 3 | 139.7, C | 3 | 140.3, C | 3 | 141.8, C | |||
| 4 | 3.16, m | 53.6, CH | 4 | 138.2, C | 4 | 125.7, C | 4 | 125.4, C | |||
| 5 | 181.7, C | 5 | 174.0, C | 5 | 86.9, C | 5 | 86.9, C | ||||
| 6 | a 2.88, dd (13.8, 8.5) b 3.13, m | 35.6, CH2 | 6 | 2.80, s | 29.4, CH2 | 6 | 3.43, s | 39.6, CH2 | 6 | 3.44, d (5.20) | 39.4, CH2 |
| 1′ | 121.5, C | 7 | 171.1, C | 7 | 171.3, C | ||||||
| 1′ | 129.7, C | 2′ (6′) | 7.43, d (8.6) | 132.4, CH | 8 | 4.25, q (7.0) | 63.6, CH2 | 1′ | 130.2, C | ||
| 2′ (6′) | 6.75, d (8.6) | 130.2, CH | 3′ (5′) | 6.69, d (8.6) | 116.5, CH | 9 | 1.21, t (7.0) | 14.3, CH3 | 2′ (6′) | 7.66, d (8.5) | 128.6, CH |
| 3′ (5′) | 6.62, d (8.6) | 116.4, CH | 4′ | 160.4, C | 1′ | 123.4, C | 3′ (5′) | 6.44, m | 129.8, CH | ||
| 4′ | 157.8, C | 1″ | 129.8, C | 2′ (6′) | 7.59, d (8.8) | 130.3, CH | 4′ | 7.37, m | 130.2, CH | ||
| 1″ | 129.7, C | 2″ (6″) | 6.99, d (8.6) | 130.3, CH | 3′ (5′) | 6.87, d (8.8) | 116.5, CH | 1″ | 131.8, C | ||
| 2″ | 6.83, s | 131.4, CH | 3″ (5″) | 6.83, d (8.6) | 116.4, CH | 4′ | 159.2, C | 2″ | 6.45, s | 132.6, CH | |
| 3″ | 122.3, C | 4″ | 157.1, C | 1″ | 129.0, C | 3″ | 121.4, C | ||||
| 4″ | 154.1, C | 2″ | 6.49, s | 132.6, CH | 4″ | 154.3, C | |||||
| 5″ | 6.58, d (8.2) | 118.2, CH | 3″ | 121.4, C | 5″ | 6.42, d (8.0) | 117.4, CH | ||||
| 6″ | 6.85, d (8.2) | 129.3, CH | 4″ | 154.3, C | 6″ | 6.48, d (8.0) | 130.2, CH | ||||
| 7″ | 2.66, m | 23.3, CH2 | 5″ | 6.43, d (8.0) | 117.4, CH | 7″ | 2.53, m | 23.1, CH2 | |||
| 8″ | 1.76, t (6.8) | 33.8, CH2 | 6″ | 6.52, d (8.0) | 130.2, CH | 8″ | 1.68, t (6.5) | 33.6, CH2 | |||
| 9″ | 75.2, C | 7″ | 2.57, m | 23.2, CH2 | 9″ | 75.1, C | |||||
| 10″ | 1.28, s | 26.9, CH3 | 8″ | 1.71, t (6.5) | 33.7, CH2 | 10″ | 1.22, s | 26.9, CH3 | |||
| 11″ | 1.28, s | 27.1, CH3 | 9″ | 75.1, C | 11″ | 1.22, s | 27.0, CH3 | ||||
| 10″ | 1.24, s | 26.9, CH3 | -OMe | 3,49, s | 53.9 | ||||||
| 11″ | 1.24, s | 27.0, CH3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, H.; Wang, L.; Zhang, Z.-K.; Wu, P.-P.; He, Y.-P.; Chen, L.-Y.; Wang, Q.; Zhang, C.-X. Bioactive Aspergteroids G–J from Soft-Coral-Associated Symbiotic and Epiphytic Fungus Aspergillus terreus EGF7-0-1. Bioengineering 2023, 10, 805. https://doi.org/10.3390/bioengineering10070805
Fan H, Wang L, Zhang Z-K, Wu P-P, He Y-P, Chen L-Y, Wang Q, Zhang C-X. Bioactive Aspergteroids G–J from Soft-Coral-Associated Symbiotic and Epiphytic Fungus Aspergillus terreus EGF7-0-1. Bioengineering. 2023; 10(7):805. https://doi.org/10.3390/bioengineering10070805
Chicago/Turabian StyleFan, Hao, Li Wang, Ze-Kun Zhang, Ping-Ping Wu, Yu-Pei He, Le-Yi Chen, Qian Wang, and Cui-Xian Zhang. 2023. "Bioactive Aspergteroids G–J from Soft-Coral-Associated Symbiotic and Epiphytic Fungus Aspergillus terreus EGF7-0-1" Bioengineering 10, no. 7: 805. https://doi.org/10.3390/bioengineering10070805
APA StyleFan, H., Wang, L., Zhang, Z.-K., Wu, P.-P., He, Y.-P., Chen, L.-Y., Wang, Q., & Zhang, C.-X. (2023). Bioactive Aspergteroids G–J from Soft-Coral-Associated Symbiotic and Epiphytic Fungus Aspergillus terreus EGF7-0-1. Bioengineering, 10(7), 805. https://doi.org/10.3390/bioengineering10070805







