Assessing Tumorigenicity in Stem Cell-Derived Therapeutic Products: A Critical Step in Safeguarding Regenerative Medicine
Abstract
1. Introduction
2. Consideration When Assessing Tumorigenicity
3. Existing Approaches
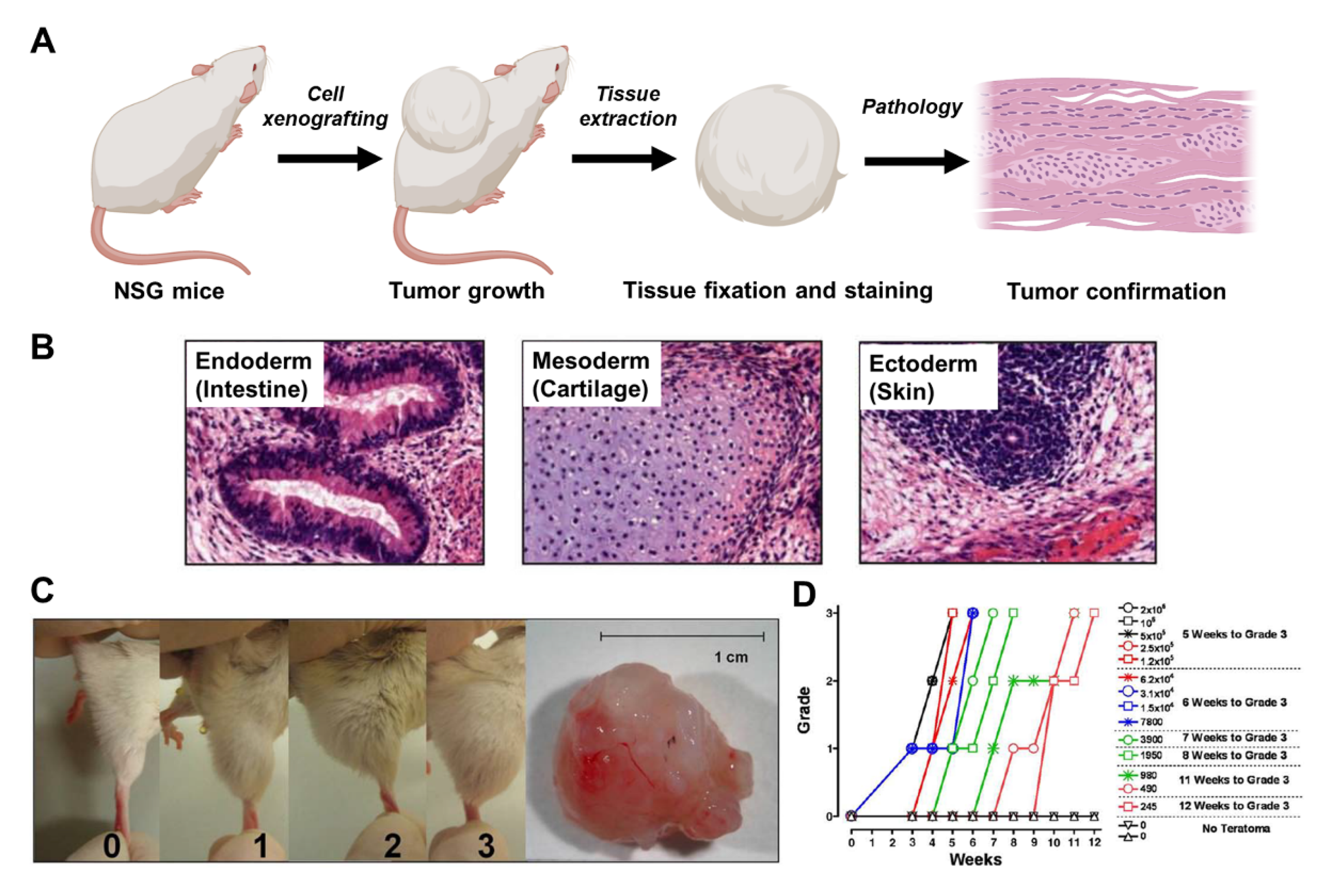
3.1. Animal Model
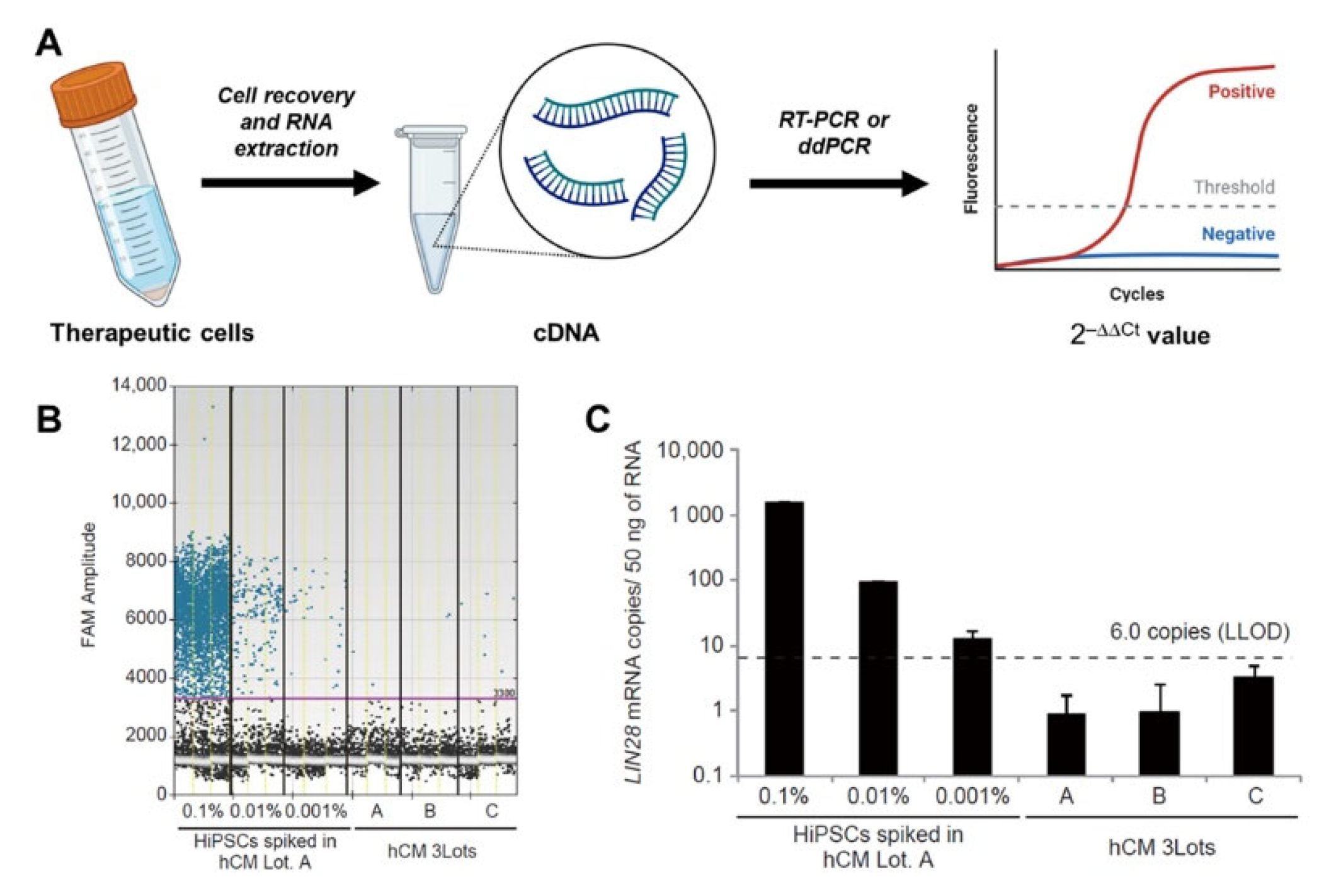
3.2. PCR
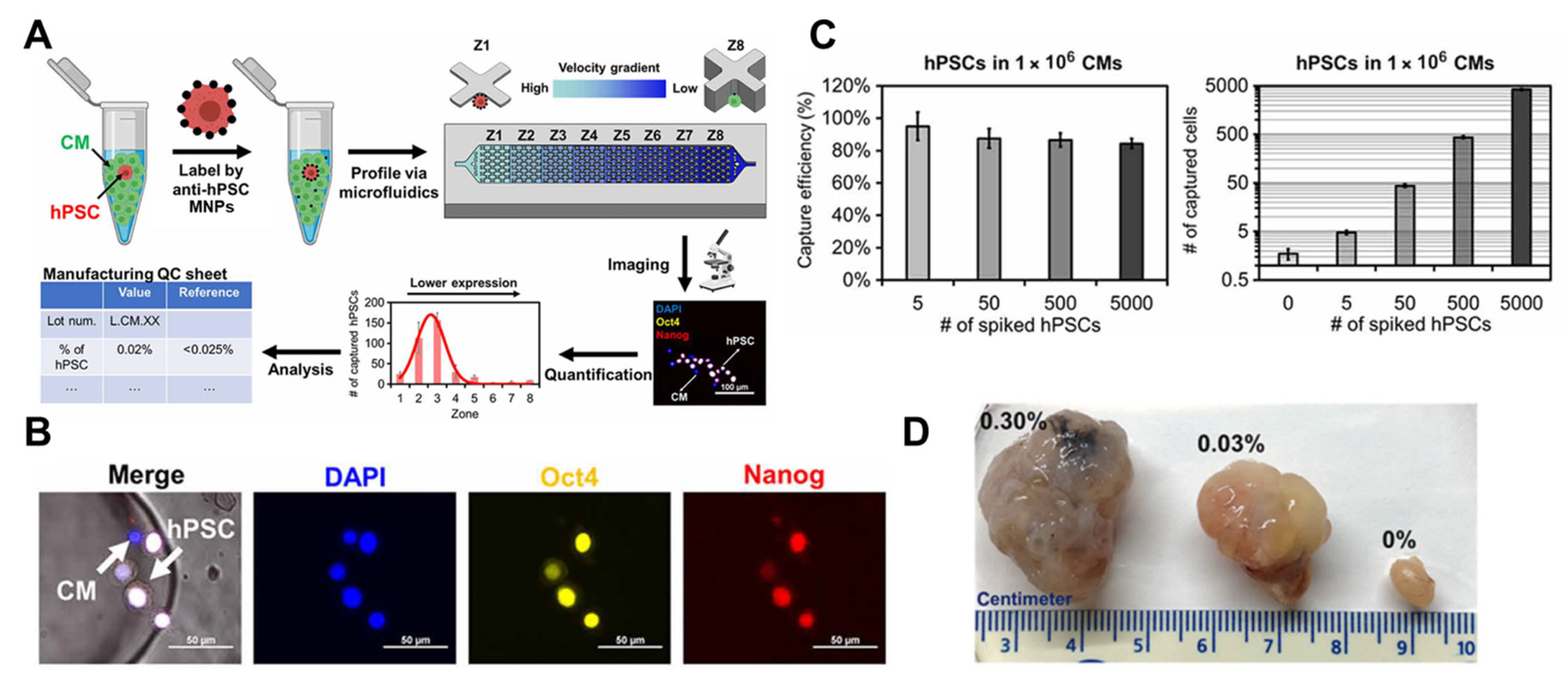
3.3. Cytometry
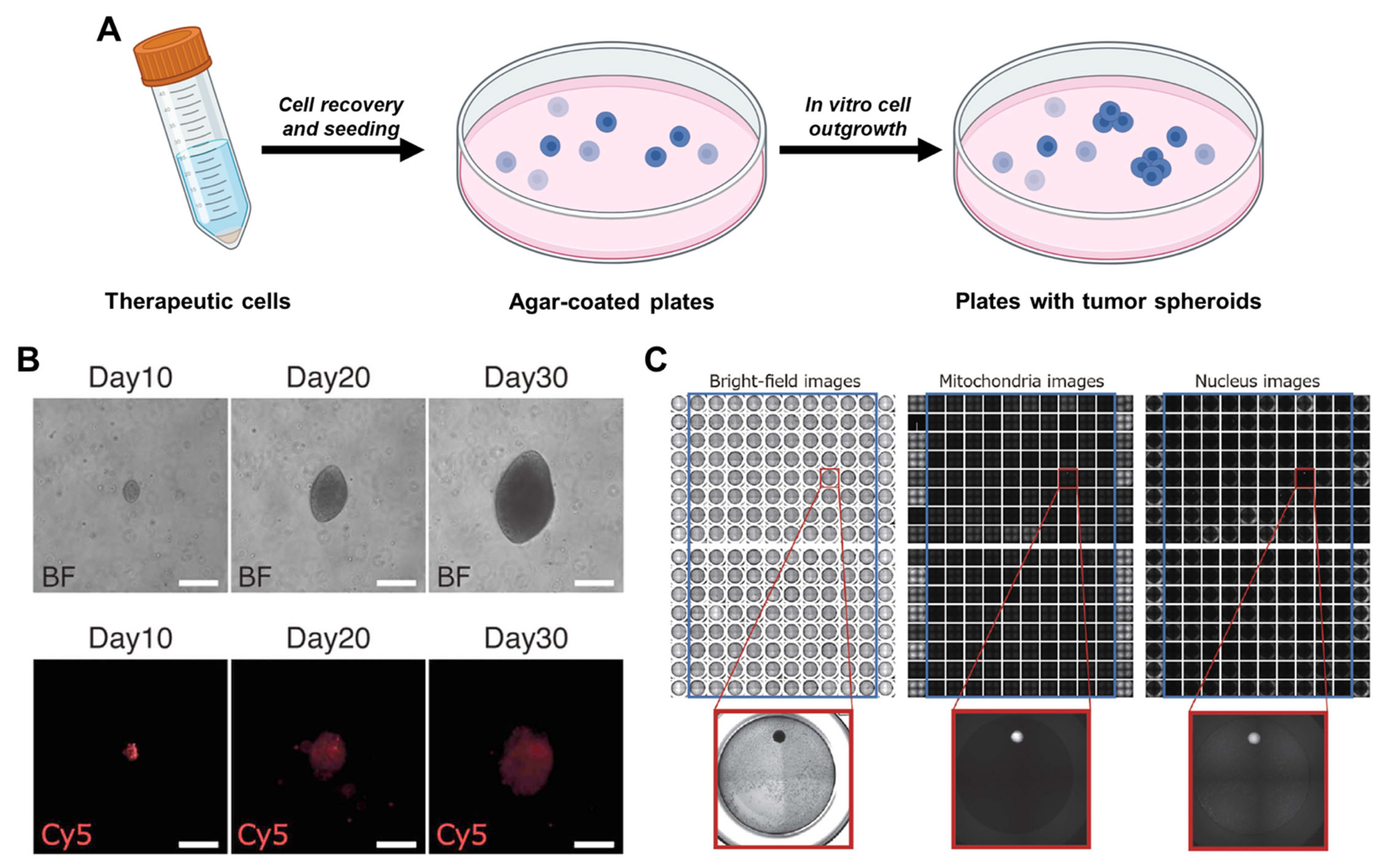
3.4. Soft Agar
4. Outlook and Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alison, M.R.; Poulsom, R.; Forbes, S.; Wright, N.A. An Introduction to Stem Cells. J. Pathol. 2002, 197, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Trounson, A.; DeWitt, N.D. Pluripotent Stem Cells Progressing to the Clinic. Nat. Rev. Mol. Cell Biol. 2016, 17, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Rubin, L.L. Stem Cells and Drug Discovery: The Beginning of a New Era? Cell 2008, 132, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Mandrycky, C.; Wang, Z.; Kim, K.; Kim, D.-H. 3D Bioprinting for Engineering Complex Tissues. Biotechnol. Adv. 2016, 34, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.H.Y.; Yu, F.; Zhu, S.; Wang, Z. 3D Bioprinting for Next-Generation Personalized Medicine. Int. J. Mol. Sci. 2023, 24, 6357. [Google Scholar] [CrossRef] [PubMed]
- Ranga, A.; Gjorevski, N.; Lutolf, M.P. Drug Discovery through Stem Cell-Based Organoid Models. Adv. Drug Deliv. Rev. 2014, 69–70, 19–28. [Google Scholar] [CrossRef]
- Wang, Z.; Samanipour, R.; Kim, K. Organ-on-a-Chip Platforms for Drug Screening and Tissue Engineering. In Biomedical Engineering: Frontier Research and Converging Technologies; Jo, H., Jun, H.-W., Shin, J., Lee, S., Eds.; Biosystems & Biorobotics; Springer International Publishing: Cham, Switzerland, 2016; Volume 9, pp. 209–233. ISBN 978-3-319-21812-0. [Google Scholar]
- Wang, Z.; Calpe, B.; Zerdani, J.; Lee, Y.; Oh, J.; Bae, H.; Khademhosseini, A.; Kim, K. High-Throughput Investigation of Endothelial-to-Mesenchymal Transformation (EndMT) with Combinatorial Cellular Microarrays: High-Throughput Investigation of Endothelial-to-Mesenchymal Transformation (EndMT). Biotechnol. Bioeng. 2016, 113, 1403–1412. [Google Scholar] [CrossRef]
- Keller, G.M. In Vitro Differentiation of Embryonic Stem Cells. Curr. Opin. Cell Biol. 1995, 7, 862–869. [Google Scholar] [CrossRef]
- Odorico, J.S.; Kaufman, D.S.; Thomson, J.A. Multilineage Differentiation from Human Embryonic Stem Cell Lines. Stem Cells 2001, 19, 193–204. [Google Scholar] [CrossRef]
- Murry, C.E.; Keller, G. Differentiation of Embryonic Stem Cells to Clinically Relevant Populations: Lessons from Embryonic Development. Cell 2008, 132, 661–680. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Strategies and New Developments in the Generation of Patient-Specific Pluripotent Stem Cells. Cell Stem Cell 2007, 1, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A More Efficient Method to Generate Integration-Free Human IPS Cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced Pluripotent Stem Cell Technology: A Decade of Progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Mallapaty, S. ‘Reprogrammed’ Stem Cells for Heart Disease Tested in China. Nature 2020, 581, 249–250. [Google Scholar] [CrossRef]
- Sugai, K.; Sumida, M.; Shofuda, T.; Yamaguchi, R.; Tamura, T.; Kohzuki, T.; Abe, T.; Shibata, R.; Kamata, Y.; Ito, S.; et al. First-in-Human Clinical Trial of Transplantation of IPSC-Derived NS/PCs in Subacute Complete Spinal Cord Injury: Study Protocol. Regen. Ther. 2021, 18, 321–333. [Google Scholar] [CrossRef]
- Deinsberger, J.; Reisinger, D.; Weber, B. Global Trends in Clinical Trials Involving Pluripotent Stem Cells: A Systematic Multi-Database Analysis. NPJ Regen. Med. 2020, 5, 15. [Google Scholar] [CrossRef]
- Carvalho, T. Stem Cell–Derived Heart Cells Injected into First Patient. Nat. Med. 2023, 29, 1030–1031. [Google Scholar] [CrossRef]
- Yefroyev, D.A.; Jin, S. Induced Pluripotent Stem Cells for Treatment of Alzheimer’s and Parkinson’s Diseases. Biomedicines 2022, 10, 208. [Google Scholar] [CrossRef]
- Dai, R.; Wang, Z.; Samanipour, R.; Koo, K.; Kim, K. Adipose-Derived Stem Cells for Tissue Engineering and Regenerative Medicine Applications. Stem Cells Int. 2016, 2016, 6737345. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.P.; Alexander, W.S. Haematopoietic Stem Cells: Past, Present and Future. Cell Death Discov. 2017, 3, 17002. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Moore, D.L. Neural Stem Cells: Developmental Mechanisms and Disease Modeling. Cell Tissue Res. 2018, 371, 1–6. [Google Scholar] [CrossRef]
- Blanpain, C.; Horsley, V.; Fuchs, E. Epithelial Stem Cells: Turning over New Leaves. Cell 2007, 128, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Coles, B.L.K.; Labib, M.; Poudineh, M.; Innes, B.T.; Belair-Hickey, J.; Gomis, S.; Wang, Z.; Bader, G.D.; Sargent, E.H.; Kelley, S.O.; et al. A Microfluidic Platform Enables Comprehensive Gene Expression Profiling of Mouse Retinal Stem Cells. Lab. Chip 2021, 21, 4464–4476. [Google Scholar] [CrossRef]
- Díaz-García, D.; Filipová, A.; Garza-Veloz, I.; Martinez-Fierro, M.L. A Beginner’s Introduction to Skin Stem Cells and Wound Healing. Int. J. Mol. Sci. 2021, 22, 11030. [Google Scholar] [CrossRef]
- Robert, A.W.; Marcon, B.H.; Dallagiovanna, B.; Shigunov, P. Adipogenesis, Osteogenesis, and Chondrogenesis of Human Mesenchymal Stem/Stromal Cells: A Comparative Transcriptome Approach. Front. Cell Dev. Biol. 2020, 8, 561. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Das, J.; Labib, M.; Ahmed, S.; Sargent, E.H.; Kelley, S.O. Potential-Responsive Surfaces for Manipulation of Cell Adhesion, Release, and Differentiation. Angew. Chem. Int. Ed. 2019, 58, 14519–14523. [Google Scholar] [CrossRef]
- Somoza, R.A.; Welter, J.F.; Correa, D.; Caplan, A.I. Chondrogenic Differentiation of Mesenchymal Stem Cells: Challenges and Unfulfilled Expectations. Tissue Eng. Part B Rev. 2014, 20, 596–608. [Google Scholar] [CrossRef]
- Poudineh, M.; Wang, Z.; Labib, M.; Ahmadi, M.; Zhang, L.; Das, J.; Ahmed, S.; Angers, S.; Kelley, S.O. Three-Dimensional Nanostructured Architectures Enable Efficient Neural Differentiation of Mesenchymal Stem Cells via Mechanotransduction. Nano Lett. 2018, 18, 7188–7193. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, L.; Labib, M.; Chen, H.; Wei, M.; Poudineh, M.; Green, B.J.; Duong, B.; Das, J.; Ahmed, S.; et al. Peptide-Functionalized Nanostructured Microarchitectures Enable Rapid Mechanotransductive Differentiation. ACS Appl. Mater. Interfaces 2019, 11, 41030–41037. [Google Scholar] [CrossRef] [PubMed]
- Afshari, A.; Shamdani, S.; Uzan, G.; Naserian, S.; Azarpira, N. Different Approaches for Transformation of Mesenchymal Stem Cells into Hepatocyte-like Cells. Stem Cell Res. Ther. 2020, 11, 54. [Google Scholar] [CrossRef]
- Van Heijst, J.W.J.; Ceberio, I.; Lipuma, L.B.; Samilo, D.W.; Wasilewski, G.D.; Gonzales, A.M.R.; Nieves, J.L.; van den Brink, M.R.M.; Perales, M.A.; Pamer, E.G. Quantitative Assessment of T Cell Repertoire Recovery after Hematopoietic Stem Cell Transplantation. Nat. Med. 2013, 19, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Yuan, Z.; Weng, J.; Pei, D.; Du, X.; He, C.; Lai, P. Challenges and Advances in Clinical Applications of Mesenchymal Stromal Cells. J. Hematol. Oncol. 2021, 14, 24. [Google Scholar] [CrossRef]
- Park, B.; Yoo, K.H.; Kim, C. Hematopoietic Stem Cell Expansion and Generation: The Ways to Make a Breakthrough. Blood Res. 2015, 50, 194. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Tang, C.; Rao, M.S.; Weissman, I.L.; Wu, J.C. Tumorigenicity as a Clinical Hurdle for Pluripotent Stem Cell Therapies. Nat. Med. 2013, 19, 998–1004. [Google Scholar] [CrossRef]
- Montilla-Rojo, J.; Bialecka, M.; Wever, K.E.; Mummery, C.L.; Looijenga, L.H.J.; Roelen, B.A.J.; Salvatori, D.C.F. Teratoma Assay for Testing Pluripotency and Malignancy of Stem Cells: Insufficient Reporting and Uptake of Animal-Free Methods—A Systematic Review. Int. J. Mol. Sci. 2023, 24, 3879. [Google Scholar] [CrossRef]
- Han, L.; He, H.; Yang, Y.; Meng, Q.; Ye, F.; Chen, G.; Zhang, J. Distinctive Clinical and Pathologic Features of Immature Teratomas Arising from Induced Pluripotent Stem Cell-Derived Beta Cell Injection in a Diabetes Patient. Stem Cells Dev. 2022, 31, 97–101. [Google Scholar] [CrossRef]
- DeFrancesco, L. Fits and Starts for Geron. Nat. Biotechnol. 2009, 27, 877–878. [Google Scholar] [CrossRef]
- Berkowitz, A.L.; Miller, M.B.; Mir, S.A.; Cagney, D.; Chavakula, V.; Guleria, I.; Aizer, A.; Ligon, K.L.; Chi, J.H. Glioproliferative Lesion of the Spinal Cord as a Complication of “Stem-Cell Tourism”. N. Engl. J. Med. 2016, 375, 196–198. [Google Scholar] [CrossRef]
- Marks, P.W.; Witten, C.M.; Califf, R.M. Clarifying Stem-Cell Therapy’s Benefits and Risks. N. Engl. J. Med. 2017, 376, 1007–1009. [Google Scholar] [CrossRef] [PubMed]
- Marks, P.; Gottlieb, S. Balancing Safety and Innovation for Cell-Based Regenerative Medicine. N. Engl. J. Med. 2018, 378, 954–959. [Google Scholar] [CrossRef] [PubMed]
- FDA. FDA Statement from FDA Commissioner Scott Gottlieb, M.D. on the FDA’s New Policy Steps and Enforcement Efforts to Ensure Proper Oversight of Stem Cell Therapies and Regenerative Medicine 2018; FDA: Silver Spring, MD, USA, 2018.
- Ben-David, U.; Benvenisty, N. The Tumorigenicity of Human Embryonic and Induced Pluripotent Stem Cells. Nat. Rev. Cancer 2011, 11, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.-J.; Goldmann, J.; Löser, P.; Loring, J.F. A Call to Standardize Teratoma Assays Used to Define Human Pluripotent Cell Lines. Cell Stem Cell 2010, 6, 412–414. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Liu, M.; Pan, X.; Zhu, H. Tumorigenicity Risk of IPSCs in Vivo: Nip It in the Bud. Precis. Clin. Med. 2022, 5, pbac004. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Gan, Q.-F.; Golan-Lev, T.; Arora, P.; Yanuka, O.; Oren, Y.S.; Leikin-Frenkel, A.; Graf, M.; Garippa, R.; Boehringer, M.; et al. Selective Elimination of Human Pluripotent Stem Cells by an Oleate Synthesis Inhibitor Discovered in a High-Throughput Screen. Cell Stem Cell 2013, 12, 167–179. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, Y.; Qin, G.; Chen, L.; Chatterjee, T.; Weintraub, N.; Tang, Y. Inhibition of Stearoyl-CoA Desaturase Selectively Eliminates Tumorigenic Nanog-Positive Cells: Improving the Safety of IPS Cell Transplantation to Myocardium. Cell Cycle 2014, 13, 762–771. [Google Scholar] [CrossRef]
- Hattori, F.; Chen, H.; Yamashita, H.; Tohyama, S.; Satoh, Y.; Yuasa, S.; Li, W.; Yamakawa, H.; Tanaka, T.; Onitsuka, T.; et al. Nongenetic Method for Purifying Stem Cell–Derived Cardiomyocytes. Nat. Methods 2010, 7, 61–66. [Google Scholar] [CrossRef]
- Chour, T.; Tian, L.; Lau, E.; Thomas, D.; Itzhaki, I.; Malak, O.; Zhang, J.Z.; Qin, X.; Wardak, M.; Liu, Y.; et al. Method for Selective Ablation of Undifferentiated Human Pluripotent Stem Cell Populations for Cell-Based Therapies. JCI Insight 2021, 6, e142000. [Google Scholar] [CrossRef]
- Lipsitz, Y.Y.; Timmins, N.E.; Zandstra, P.W. Quality Cell Therapy Manufacturing by Design. Nat. Biotechnol. 2016, 34, 393–400. [Google Scholar] [CrossRef]
- Ruella, M.; Xu, J.; Barrett, D.M.; Fraietta, J.A.; Reich, T.J.; Ambrose, D.E.; Klichinsky, M.; Shestova, O.; Patel, P.R.; Kulikovskaya, I.; et al. Induction of Resistance to Chimeric Antigen Receptor T Cell Therapy by Transduction of a Single Leukemic B Cell. Nat. Med. 2018, 24, 1499–1503. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.N.; Fry, T.J. Mechanisms of Resistance to CAR T Cell Therapy. Nat. Rev. Clin. Oncol. 2019, 16, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Gropp, M.; Shilo, V.; Vainer, G.; Gov, M.; Gil, Y.; Khaner, H.; Matzrafi, L.; Idelson, M.; Kopolovic, J.; Zak, N.B.; et al. Standardization of the Teratoma Assay for Analysis of Pluripotency of Human ES Cells and Biosafety of Their Differentiated Progeny. PLoS ONE 2012, 7, e45532. [Google Scholar] [CrossRef] [PubMed]
- Kanemura, H.; Go, M.J.; Shikamura, M.; Nishishita, N.; Sakai, N.; Kamao, H.; Mandai, M.; Morinaga, C.; Takahashi, M.; Kawamata, S. Tumorigenicity Studies of Induced Pluripotent Stem Cell (IPSC)-Derived Retinal Pigment Epithelium (RPE) for the Treatment of Age-Related Macular Degeneration. PLoS ONE 2014, 9, e85336. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gagliardi, M.; Mohamadi, R.M.; Ahmed, S.U.; Labib, M.; Zhang, L.; Popescu, S.; Zhou, Y.; Sargent, E.H.; Keller, G.M.; et al. Ultrasensitive and Rapid Quantification of Rare Tumorigenic Stem Cells in HPSC-Derived Cardiomyocyte Populations. Sci. Adv. 2020, 6, eaay7629. [Google Scholar] [CrossRef]
- FDA. Guidance for Industry: Characterization and Qualification of Cell Substrates and Other Biological Materials Used in the Production of Viral Vaccines for Infectious Disease Indications 2010; FDA: Silver Spring, MD, USA, 2010.
- Ogawa, M.; Ogawa, S.; Bear, C.E.; Ahmadi, S.; Chin, S.; Li, B.; Grompe, M.; Keller, G.; Kamath, B.M.; Ghanekar, A. Directed Differentiation of Cholangiocytes from Human Pluripotent Stem Cells. Nat. Biotechnol. 2015, 33, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Protze, S.I.; Laksman, Z.; Backx, P.H.; Keller, G.M. Human Pluripotent Stem Cell-Derived Atrial and Ventricular Cardiomyocytes Develop from Distinct Mesoderm Populations. Cell Stem Cell 2017, 21, 179–194.e4. [Google Scholar] [CrossRef]
- Protze, S.I.; Liu, J.; Nussinovitch, U.; Ohana, L.; Backx, P.H.; Gepstein, L.; Keller, G.M. Sinoatrial Node Cardiomyocytes Derived from Human Pluripotent Cells Function as a Biological Pacemaker. Nat. Biotechnol. 2017, 35, 56–68. [Google Scholar] [CrossRef]
- Oh, S.; Gu, E.-Y.; Han, J.-S.; Lee, B.-S.; Moon, K.-S.; Kim, Y.-B.; Han, K.-H. Tumorigenicity Assessment of Human Cancer Cell Lines Xenografted on Immunodeficient Mice as Positive Controls of Tumorigenicity Testing. Int. J. Toxicol. 2022, 41, 476–487. [Google Scholar] [CrossRef]
- Szyska, M.; Na, I.-K. Bone Marrow GvHD after Allogeneic Hematopoietic Stem Cell Transplantation. Front. Immunol. 2016, 7, 118. [Google Scholar] [CrossRef]
- Ehx, G.; Somja, J.; Warnatz, H.-J.; Ritacco, C.; Hannon, M.; Delens, L.; Fransolet, G.; Delvenne, P.; Muller, J.; Beguin, Y.; et al. Xenogeneic Graft-Versus-Host Disease in Humanized NSG and NSG-HLA-A2/HHD Mice. Front. Immunol. 2018, 9, 1943. [Google Scholar] [CrossRef] [PubMed]
- Hentze, H.; Soong, P.L.; Wang, S.T.; Phillips, B.W.; Putti, T.C.; Dunn, N.R. Teratoma Formation by Human Embryonic Stem Cells: Evaluation of Essential Parameters for Future Safety Studies. Stem Cell Res. 2009, 2, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Gimotty, P.A.; Van Belle, P.; Elder, D.E.; Murry, T.; Montone, K.T.; Xu, X.; Hotz, S.; Raines, S.; Ming, M.E.; Wahl, P.; et al. Biologic and Prognostic Significance of Dermal Ki67 Expression, Mitoses, and Tumorigenicity in Thin Invasive Cutaneous Melanoma. J. Clin. Oncol. 2005, 23, 8048–8056. [Google Scholar] [CrossRef]
- Parkin, B.; Londoño-Joshi, A.; Kang, Q.; Tewari, M.; Rhim, A.D.; Malek, S.N. Ultrasensitive Mutation Detection Identifies Rare Residual Cells Causing Acute Myelogenous Leukemia Relapse. J. Clin. Investig. 2017, 127, 3484–3495. [Google Scholar] [CrossRef]
- Chakritbudsabong, W.; Chaiwattanarungruengpaisan, S.; Sariya, L.; Pamonsupornvichit, S.; Ferreira, J.N.; Sukho, P.; Gronsang, D.; Tharasanit, T.; Dinnyes, A.; Rungarunlert, S. Exogenous LIN28 Is Required for the Maintenance of Self-Renewal and Pluripotency in Presumptive Porcine-Induced Pluripotent Stem Cells. Front. Cell Dev. Biol. 2021, 9, 709286. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, H.; Wu, T. LIN28: A Cancer Stem Cell Promoter for Immunotherapy in Head and Neck Squamous Cell Carcinoma. Oral Oncol. 2019, 98, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, T.; Yasuda, S.; Matsuyama, S.; Tano, K.; Kusakawa, S.; Sawa, Y.; Kawamata, S.; Sato, Y. Highly Sensitive Droplet Digital PCR Method for Detection of Residual Undifferentiated Cells in Cardiomyocytes Derived from Human Pluripotent Stem Cells. Regen. Ther. 2015, 2, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Middelkamp, M.; Ruck, L.; Krisp, C.; Sumisławski, P.; Mohammadi, B.; Dottermusch, M.; Meister, V.; Küster, L.; Schlüter, H.; Windhorst, S.; et al. Overexpression of Lin28A in Neural Progenitor Cells in Vivo Does Not Lead to Brain Tumor Formation but Results in Reduced Spine Density. Acta Neuropathol. Commun. 2021, 9, 185. [Google Scholar] [CrossRef]
- Lemmens, M.; Perner, J.; Potgeter, L.; Zogg, M.; Thiruchelvam, S.; Müller, M.; Doll, T.; Werner, A.; Gilbart, Y.; Couttet, P.; et al. Identification of Marker Genes to Monitor Residual IPSCs in IPSC-Derived Products. Cytotherapy 2023, 25, 59–67. [Google Scholar] [CrossRef]
- Tsujimoto, H.; Katagiri, N.; Ijiri, Y.; Sasaki, B.; Kobayashi, Y.; Mima, A.; Ryosaka, M.; Furuyama, K.; Kawaguchi, Y.; Osafune, K. In Vitro Methods to Ensure Absence of Residual Undifferentiated Human Induced Pluripotent Stem Cells Intermingled in Induced Nephron Progenitor Cells. PLoS ONE 2022, 17, e0275600. [Google Scholar] [CrossRef]
- Chung, L.; Cogburn, L.A.; Sui, L.; Dashnau, J.L. Development of an Induced Pluripotent Stem Cell–Specific MicroRNA Assay for Detection of Residual Undifferentiated Cells in Natural Killer Cell Therapy Products. Cytotherapy 2022, 24, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Wills, Q.F.; Tipping, A.J.; Datta, K.; Mittal, R.; Goldson, A.J.; Sexton, D.W.; Holmes, C.C. Methods for QPCR Gene Expression Profiling Applied to 1440 Lymphoblastoid Single Cells. Methods 2013, 59, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Picot, J.; Guerin, C.L.; Le Van Kim, C.; Boulanger, C.M. Flow Cytometry: Retrospective, Fundamentals and Recent Instrumentation. Cytotechnology 2012, 64, 109–130. [Google Scholar] [CrossRef] [PubMed]
- Schraivogel, D.; Kuhn, T.M.; Rauscher, B.; Rodríguez-Martínez, M.; Paulsen, M.; Owsley, K.; Middlebrook, A.; Tischer, C.; Ramasz, B.; Ordoñez-Rueda, D.; et al. High-Speed Fluorescence Image–Enabled Cell Sorting. Science 2022, 375, 315–320. [Google Scholar] [CrossRef]
- Holzner, G.; Mateescu, B.; Van Leeuwen, D.; Cereghetti, G.; Dechant, R.; Stavrakis, S.; deMello, A. High-Throughput Multiparametric Imaging Flow Cytometry: Toward Diffraction-Limited Sub-Cellular Detection and Monitoring of Sub-Cellular Processes. Cell Rep. 2021, 34, 108824. [Google Scholar] [CrossRef]
- Bhagwat, N.; Dulmage, K.; Pletcher, C.H.; Wang, L.; DeMuth, W.; Sen, M.; Balli, D.; Yee, S.S.; Sa, S.; Tong, F.; et al. An Integrated Flow Cytometry-Based Platform for Isolation and Molecular Characterization of Circulating Tumor Single Cells and Clusters. Sci. Rep. 2018, 8, 5035. [Google Scholar] [CrossRef]
- Zhu, X.; Suo, Y.; Fu, Y.; Zhang, F.; Ding, N.; Pang, K.; Xie, C.; Weng, X.; Tian, M.; He, H.; et al. In Vivo Flow Cytometry Reveals a Circadian Rhythm of Circulating Tumor Cells. Light Sci. Appl. 2021, 10, 110. [Google Scholar] [CrossRef]
- Mușină, A.-M.; Zlei, M.; Mentel, M.; Scripcariu, D.-V.; Ștefan, M.; Aniţei, M.-G.; Filip, B.; Radu, I.; Gavrilescu, M.-M.; Panuţa, A.; et al. Evaluation of Circulating Tumor Cells in Colorectal Cancer Using Flow Cytometry. J. Int. Med. Res. 2021, 49, 030006052098021. [Google Scholar] [CrossRef]
- Allan, A.L.; Keeney, M. Circulating Tumor Cell Analysis: Technical and Statistical Considerations for Application to the Clinic. J. Oncol. 2010, 2010, 426218. [Google Scholar] [CrossRef]
- Wang, Z.; Sargent, E.H.; Kelley, S.O. Ultrasensitive Detection and Depletion of Rare Leukemic B Cells in T Cell Populations via Immunomagnetic Cell Ranking. Anal. Chem. 2021, 93, 2327–2335. [Google Scholar] [CrossRef]
- Lin, G. Magnetic Particles for Multidimensional in Vitro Bioanalysis. View 2021, 2, 20200076. [Google Scholar] [CrossRef]
- Mohamadi, R.M.; Besant, J.D.; Mepham, A.; Green, B.; Mahmoudian, L.; Gibbs, T.; Ivanov, I.; Malvea, A.; Stojcic, J.; Allan, A.L.; et al. Nanoparticle-Mediated Binning and Profiling of Heterogeneous Circulating Tumor Cell Subpopulations. Angew. Chem. Int. Ed. 2015, 54, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Wang, Z.; Flynn, C.D.; Mahmud, A.; Labib, M.; Wang, H.; Geraili, A.; Li, X.; Zhang, J.; Sargent, E.H.; et al. A High-Dimensional Microfluidic Approach for Selection of Aptamers with Programmable Binding Affinities. Nat. Chem. 2023, 15, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.; Pao, E.; Tseng, P.; Aftab, S.; Kulkarni, R.; Rettig, M.; Di Carlo, D. Quantitative Magnetic Separation of Particles and Cells Using Gradient Magnetic Ratcheting. Small 2016, 12, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.; Wang, Z.; Ahmed, S.U.; Mohamadi, R.M.; Duong, B.; Green, B.; Sargent, E.H.; Kelley, S.O. Tracking the Expression of Therapeutic Protein Targets in Rare Cells by Antibody-Mediated Nanoparticle Labelling and Magnetic Sorting. Nat. Biomed. Eng. 2021, 5, 41–52. [Google Scholar] [CrossRef]
- Wang, Z.; Ahmed, S.; Labib, M.; Wang, H.; Hu, X.; Wei, J.; Yao, Y.; Moffat, J.; Sargent, E.H.; Kelley, S.O. Efficient Recovery of Potent Tumour-Infiltrating Lymphocytes through Quantitative Immunomagnetic Cell Sorting. Nat. Biomed. Eng. 2022, 6, 108–117. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, H.; Lin, S.; Ahmed, S.; Angers, S.; Sargent, E.H.; Kelley, S.O. Nanoparticle Amplification Labeling for High-Performance Magnetic Cell Sorting. Nano Lett. 2022, 22, 4774–4783. [Google Scholar] [CrossRef]
- Ding, J.; Xu, H.; Faiola, F.; Ma’ayan, A.; Wang, J. Oct4 Links Multiple Epigenetic Pathways to the Pluripotency Network. Cell Res. 2012, 22, 155–167. [Google Scholar] [CrossRef]
- Pan, G.; Thomson, J.A. Nanog and Transcriptional Networks in Embryonic Stem Cell Pluripotency. Cell Res. 2007, 17, 42–49. [Google Scholar] [CrossRef]
- Labib, M.; Philpott, D.N.; Wang, Z.; Nemr, C.; Chen, J.B.; Sargent, E.H.; Kelley, S.O. Magnetic Ranking Cytometry: Profiling Rare Cells at the Single-Cell Level. Acc. Chem. Res. 2020, 53, 1445–1457. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Z. Micro-Magnetofluidic System for Rare Cell Analysis: From Principle to Translation. Chemosensors 2023, 11, 335. [Google Scholar] [CrossRef]
- Wang, Z.; Ahmed, S.; Labib, M.; Wang, H.; Wu, L.; Bavaghar-Zaeimi, F.; Shokri, N.; Blanco, S.; Karim, S.; Czarnecka-Kujawa, K.; et al. Isolation of Tumour-Reactive Lymphocytes from Peripheral Blood via Microfluidic Immunomagnetic Cell Sorting. Nat. Biomed. Eng. 2023. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Yasuda, S.; Kusakawa, S.; Kuroda, T.; Futamura, M.; Ogawa, M.; Mochizuki, H.; Kikkawa, E.; Furukawa, H.; Nagaoka, M.; et al. Multisite Studies for Validation and Improvement of a Highly Efficient Culture Assay for Detection of Undifferentiated Human Pluripotent Stem Cells Intermingled in Cell Therapy Products. Cytotherapy 2021, 23, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Horibata, S.; Vo, T.V.; Subramanian, V.; Thompson, P.R.; Coonrod, S.A. Utilization of the Soft Agar Colony Formation Assay to Identify Inhibitors of Tumorigenicity in Breast Cancer Cells. J. Vis. Exp. 2015, 99, e52727. [Google Scholar] [CrossRef]
- Rotem, A.; Janzer, A.; Izar, B.; Ji, Z.; Doench, J.G.; Garraway, L.A.; Struhl, K. Alternative to the Soft-Agar Assay That Permits High-Throughput Drug and Genetic Screens for Cellular Transformation. Proc. Natl. Acad. Sci. USA 2015, 112, 5708–5713. [Google Scholar] [CrossRef]
- Kusakawa, S.; Yasuda, S.; Kuroda, T.; Kawamata, S.; Sato, Y. Ultra-Sensitive Detection of Tumorigenic Cellular Impurities in Human Cell-Processed Therapeutic Products by Digital Analysis of Soft Agar Colony Formation. Sci. Rep. 2016, 5, 17892. [Google Scholar] [CrossRef]
- Lemmens, M.; Fischer, B.; Zogg, M.; Rodrigues, L.; Kerr, G.; del Rio-Espinola, A.; Schaeffer, F.; Maddalo, D.; Dubost, V.; Piaia, A.; et al. Evaluation of Two in Vitro Assays for Tumorigenicity Assessment of CRISPR-Cas9 Genome-Edited Cells. Mol. Ther.-Methods Clin. Dev. 2021, 23, 241–253. [Google Scholar] [CrossRef]
- Sedeeq, M.; Maklad, A.; Gueven, N.; Azimi, I. Development of a High-Throughput Agar Colony Formation Assay to Identify Drug Candidates against Medulloblastoma. Pharmaceuticals 2020, 13, 368. [Google Scholar] [CrossRef]
- Abe-Fukasawa, N.; Otsuka, K.; Aihara, A.; Itasaki, N.; Nishino, T. Novel 3D Liquid Cell Culture Method for Anchorage-Independent Cell Growth, Cell Imaging and Automated Drug Screening. Sci. Rep. 2018, 8, 3627. [Google Scholar] [CrossRef]
- Barkholt, L.; Flory, E.; Jekerle, V.; Lucas-Samuel, S.; Ahnert, P.; Bisset, L.; Büscher, D.; Fibbe, W.; Foussat, A.; Kwa, M.; et al. Risk of Tumorigenicity in Mesenchymal Stromal Cell–Based Therapies—Bridging Scientific Observations and Regulatory Viewpoints. Cytotherapy 2013, 15, 753–759. [Google Scholar] [CrossRef]
- Sato, Y.; Bando, H.; Di Piazza, M.; Gowing, G.; Herberts, C.; Jackman, S.; Leoni, G.; Libertini, S.; MacLachlan, T.; McBlane, J.W.; et al. Tumorigenicity Assessment of Cell Therapy Products: The Need for Global Consensus and Points to Consider. Cytotherapy 2019, 21, 1095–1111. [Google Scholar] [CrossRef] [PubMed]
- Bialecka, M.; Montilla-Rojo, J.; Roelen, B.A.J.; Gillis, A.J.; Looijenga, L.H.J.; Salvatori, D.C.F. Humanised Mice and Immunodeficient Mice (NSG) Are Equally Sensitive for Prediction of Stem Cell Malignancy in the Teratoma Assay. Int. J. Mol. Sci. 2022, 23, 4680. [Google Scholar] [CrossRef] [PubMed]
- The International Stem Cell Initiative. Assessment of Established Techniques to Determine Developmental and Malignant Potential of Human Pluripotent Stem Cells. Nat. Commun. 2018, 9, 1925. [Google Scholar] [CrossRef] [PubMed]
- Tristan, C.A.; Ormanoglu, P.; Slamecka, J.; Malley, C.; Chu, P.-H.; Jovanovic, V.M.; Gedik, Y.; Jethmalani, Y.; Bonney, C.; Barnaeva, E.; et al. Robotic High-Throughput Biomanufacturing and Functional Differentiation of Human Pluripotent Stem Cells. Stem Cell Rep. 2021, 16, 3076–3092. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, M.; Nakajima, K.; Kino-oka, M. Approach of Resource Expenditure Estimation toward Mechanization in the Manufacturing of Cell-Based Products. Regen. Ther. 2022, 20, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Park, E.; Yoo, H.S.; Park, J.; Jung, Y.M.; Park, J.H. Recent Advances in Monitoring Stem Cell Status and Differentiation Using Nano-Biosensing Technologies. Nanomaterials 2022, 12, 2934. [Google Scholar] [CrossRef]
- Desa, D.E.; Qian, T.; Skala, M.C. Label-Free Optical Imaging and Sensing for Quality Control of Stem Cell Manufacturing. Curr. Opin. Biomed. Eng. 2023, 25, 100435. [Google Scholar] [CrossRef]
- Zaman, W.S.W.K.; Karman, S.B.; Ramlan, E.I.; Tukimin, S.N.B.; Ahmad, M.Y.B. Machine Learning in Stem Cells Research: Application for Biosafety and Bioefficacy Assessment. IEEE Access 2021, 9, 25926–25945. [Google Scholar] [CrossRef]
- Srinivasan, M.; Thangaraj, S.R.; Ramasubramanian, K.; Thangaraj, P.P.; Ramasubramanian, K.V. Exploring the Current Trends of Artificial Intelligence in Stem Cell Therapy: A Systematic Review. Cureus 2021, 13, e20083. [Google Scholar] [CrossRef]
- Kusumoto, D.; Yuasa, S.; Fukuda, K. Induced Pluripotent Stem Cell-Based Drug Screening by Use of Artificial Intelligence. Pharmaceuticals 2022, 15, 562. [Google Scholar] [CrossRef]

| Marker | Approach of Discovery | LLOD | References |
|---|---|---|---|
| LIN28 or LIN28A | RT-PCR on a panel of 7 common iPSC markers | 0.001% | [70] |
| ESRG, CAMKV, IDO1, CNMD, LIDT1, LCK, VRTN, ZSCAN10 | Bulk RNA-sequencing | 0.001–0.1% | [72] |
| MIR302CHG | Bulk RNA-sequencing | 0.0001% | [73] |
| microRNA 300 and 500 families | miRNA microarray | 0.0003% | [74] |
| Marker | Approach of Discovery | LLOD | References |
|---|---|---|---|
| SSEA-4 | Flow cytometry on a panel of 11 common iPSC markers | Not reported | [57,96] |
| TRA-1-60 | Flow cytometry on a panel of 11 common iPSC markers | <0.0005% | [57] |
| EpCAM (CD326) | Flow cytometry on a panel of 11 common iPSC markers | Not reported | [57,96] |
| Tetramethylrhodamine methyl ester perchlorate (TMRM) | Rationale analysis | <0.1% | [50] |
| Animal Model | PCR | Cytometry | Soft Agar | |
|---|---|---|---|---|
| Principle | In vivo cell outgrowth | mRNA expression | Protein expression | In vitro cell outgrowth |
| Sensitivity | N/A | 0.001% | 0.0005% | 0.0001% |
| Assay time | 1–7 months | 3 h | 1.5–2 h | 2–6 weeks |
| Bias | Low | High (due to cDNA synthesis) | Low | Medium (may discriminate suspended cells) |
| Major limitations | Long assay time Non-scalable Labor-intensive | Subjective cutoff Marker-mediated | Marker-mediated Non-standard equipment | Long assay time Labor-intensive |
| References | [55,65] | [70] | [57] | [96,99] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z. Assessing Tumorigenicity in Stem Cell-Derived Therapeutic Products: A Critical Step in Safeguarding Regenerative Medicine. Bioengineering 2023, 10, 857. https://doi.org/10.3390/bioengineering10070857
Wang Z. Assessing Tumorigenicity in Stem Cell-Derived Therapeutic Products: A Critical Step in Safeguarding Regenerative Medicine. Bioengineering. 2023; 10(7):857. https://doi.org/10.3390/bioengineering10070857
Chicago/Turabian StyleWang, Zongjie. 2023. "Assessing Tumorigenicity in Stem Cell-Derived Therapeutic Products: A Critical Step in Safeguarding Regenerative Medicine" Bioengineering 10, no. 7: 857. https://doi.org/10.3390/bioengineering10070857
APA StyleWang, Z. (2023). Assessing Tumorigenicity in Stem Cell-Derived Therapeutic Products: A Critical Step in Safeguarding Regenerative Medicine. Bioengineering, 10(7), 857. https://doi.org/10.3390/bioengineering10070857







