Non-Contrasted CT Radiomics for SAH Prognosis Prediction
Abstract
:1. Introduction
2. Results
2.1. Clinical Features
2.2. Stable Feature Selection
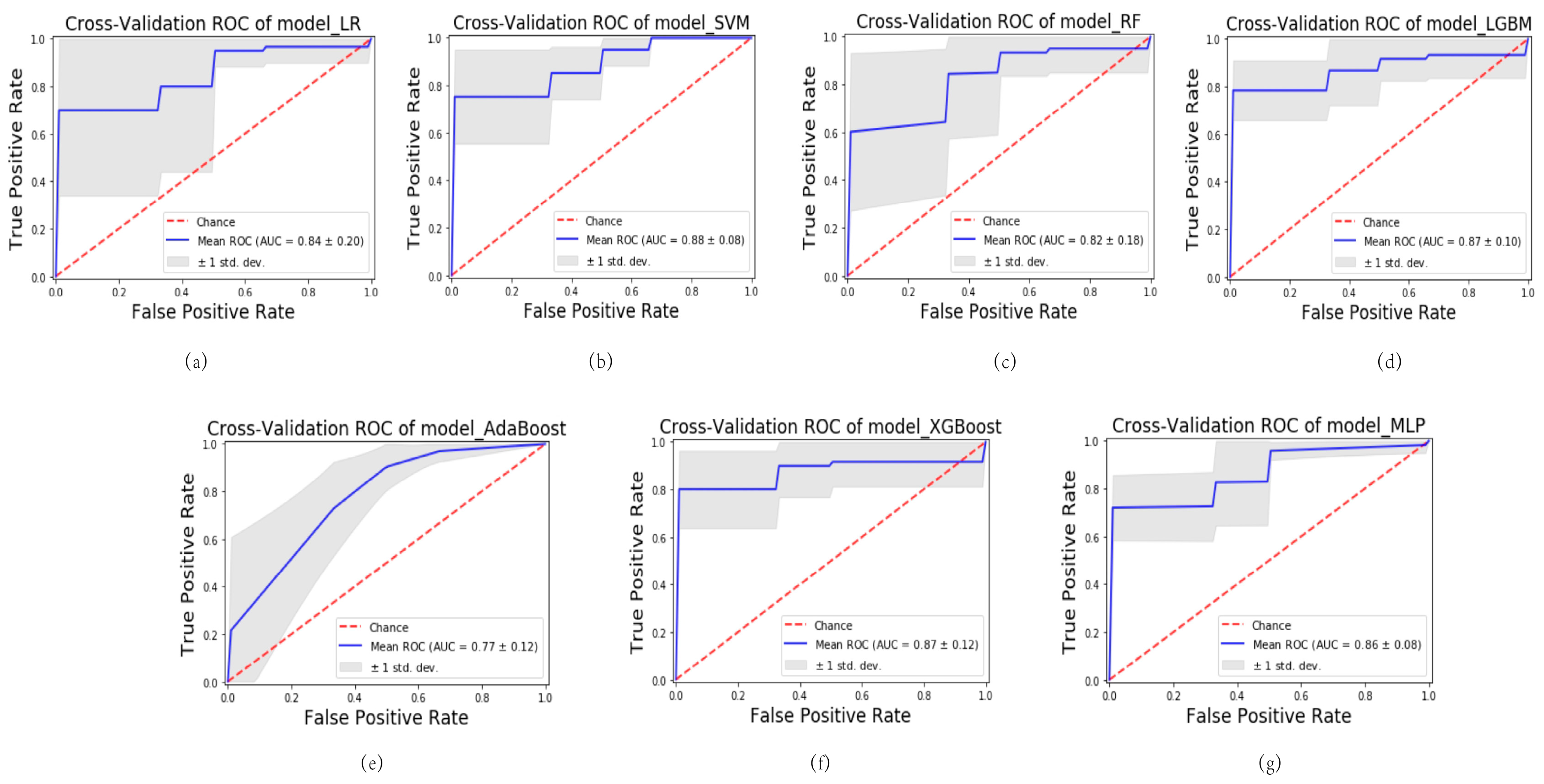
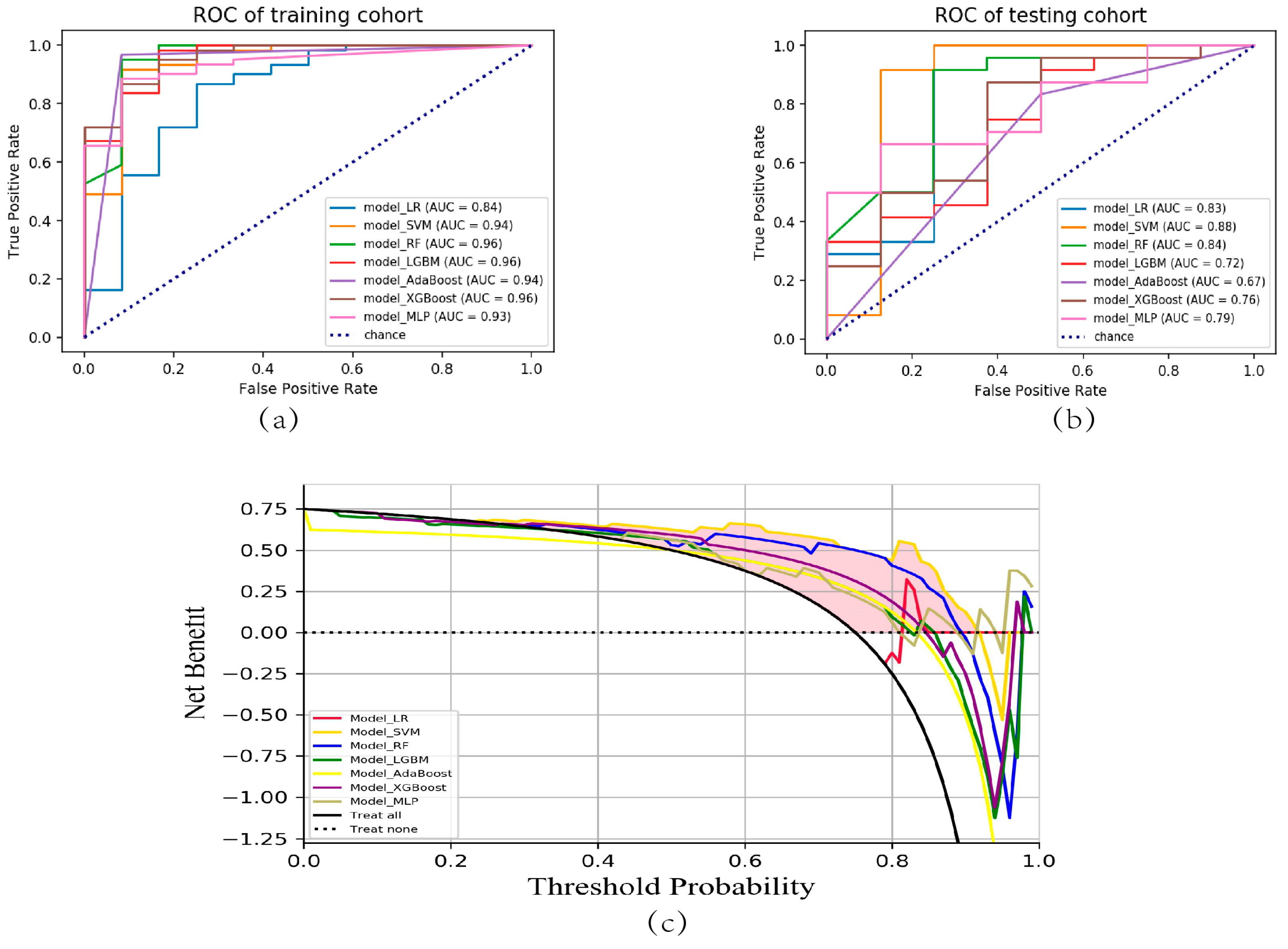
2.3. Predictive Model Construction and Evaluation
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Clinical and Imaging Data
4.3. Image Segmentation and Feature Extraction
4.4. Predictive Model Construction and Evaluation
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Claassen, J.; Park, S. Spontaneous subarachnoid haemorrhage. Lancet 2022, 400, 846–862. [Google Scholar] [CrossRef] [PubMed]
- Etminan, N.; Chang, H.-S.; Hackenberg, K.; de Rooij, N.K.; Vergouwen, M.D.I.; Rinkel, G.J.E.; Algra, A. Worldwide Incidence of Aneurysmal Subarachnoid Hemorrhage According to Region, Time Period, Blood Pressure, and Smoking Prevalence in the Population: A Systematic Review and Meta-analysis. JAMA Neurol. 2019, 76, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.J.; Sivilotti, M.L.A.; Émond, M.; Hohl, C.M.; Khan, M.; Lesiuk, H.; Abdulaziz, K.; Wells, G.A.; Stiell, I.G. Prospective Implementation of the Ottawa Subarachnoid Hemorrhage Rule and 6-Hour Computed Tomography Rule. Stroke 2020, 51, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, T.L.; Ueno, Y.; Dohan, A.; Chatterjee, A.; Vallières, M.; Winter-Reinhold, E.; Saif, S.; Levesque, I.R.; Zeng, X.Z.; Forghani, R.; et al. Development and Validation of Multiparametric MRI-based Radiomics Models for Preoperative Risk Stratification of Endometrial Cancer. Radiology 2022, 305, 375–386. [Google Scholar] [CrossRef]
- Patel, R.V.; Yao, S.; Huang, R.Y.; Bi, W.L. Application of radiomics to meningiomas: A systematic review. Neuro-Oncology 2023, 25, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.P.; O’Connor, J.P.B.; McShane, L.M.; Giger, M.L.; Lambin, P.; Kinahan, P.E.; Siegel, E.L.; Shankar, L.K. Criteria for the translation of radiomics into clinically useful tests. Nat. Rev. Clin. Oncol. 2023, 20, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Moon, G.; Choi, J.-R.; Lee, C.; Oh, Y.; Kim, K.H.; Kim, D. Machine learning-based design of meta-plasmonic biosensors with negative index metamaterials. Biosens. Bioelectron. 2020, 164, 112335. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Liu, Y.; Kang, B.; Wang, R.; Sun, M.-Y.; Wu, Q.; Meng, X.-F.; Lin, J.-P. Large-scale comparison of machine learning algorithms for target prediction of natural products. Brief. Bioinform. 2022, 23, bbac359. [Google Scholar] [CrossRef]
- Shamshirband, S.; Fathi, M.; Dehzangi, A.; Chronopoulos, A.T.; Alinejad-Rokny, H. A review on deep learning approaches in healthcare systems: Taxonomies, challenges, and open issues. J. Biomed. Inform. 2021, 113, 103627. [Google Scholar] [CrossRef]
- Eraslan, G.; Avsec, Ž.; Gagneur, J.; Theis, F.J. Deep learning: New computational modelling techniques for genomics. Nat. Rev. Genet. 2019, 20, 389–403. [Google Scholar] [CrossRef]
- Sung, C.; An, J.; Lee, S.; Park, J.; Lee, K.S.; Kim, I.-H.; Han, J.-Y.; Park, Y.H.; Kim, J.H.; Kang, E.J.; et al. Integrative analysis of risk factors for immune-related adverse events of checkpoint blockade therapy in cancer. Nat. Cancer 2023, 4, 844–859. [Google Scholar] [CrossRef] [PubMed]
- Doudesis, D.; Lee, K.K.; Boeddinghaus, J.; Bularga, A.; Ferry, A.V.; Tuck, C.; Lowry, M.T.H.; Lopez-Ayala, P.; Nestelberger, T.; Koechlin, L.; et al. Machine learning for diagnosis of myocardial infarction using cardiac troponin concentrations. Nat. Med. 2023, 29, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Christensen, S.; Ouyang, J.; Scalzo, F.; Liebeskind, D.S.; Lansberg, M.G.; Albers, G.W.; Zaharchuk, G. Predicting Hypoperfusion Lesion and Target Mismatch in Stroke from Diffusion-weighted MRI Using Deep Learning. Radiology 2023, 307, e220882. [Google Scholar] [CrossRef] [PubMed]
- Almandoz, J.E.D.; Schaefer, P.W.; Goldstein, J.N.; Rosand, J.; Lev, M.H.; González, R.G.; Romero, J.M. Practical scoring system for the identification of patients with intracerebral hemorrhage at highest risk of harboring an underlying vascular etiology: The Secondary Intracerebral Hemorrhage Score. Am. J. Neuroradiol. 2010, 31, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Jaja, B.N.R.; Saposnik, G.; Lingsma, H.F.; Macdonald, E.; Thorpe, K.E.; Mamdani, M.; Steyerberg, E.W.; Molyneux, A.; Manoel, A.L.d.O.; Schatlo, B.; et al. Development and validation of outcome prediction models for aneurysmal subarachnoid haemorrhage: The SAHIT multinational cohort study. BMJ 2018, 360, j5745. [Google Scholar] [CrossRef]
- Pace, A.; Mitchell, S.; Casselden, E.; Zolnourian, A.; Glazier, J.; Foulkes, L.; Bulters, D.; Galea, I. A subarachnoid haemorrhage-specific outcome tool. Brain 2018, 141, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Lindbohm, J.V.; Kaprio, J.; Jousilahti, P.; Salomaa, V.; Korja, M. Risk Factors of Sudden Death From Subarachnoid Hemorrhage. Stroke 2017, 48, 2399–2404. [Google Scholar] [CrossRef]
- Duan, C.; Liu, F.; Gao, S.; Zhao, J.; Niu, L.; Li, N.; Liu, S.; Wang, G.; Zhou, X.; Ren, Y.; et al. Comparison of Radiomic Models Based on Different Machine Learning Methods for Predicting Intracerebral Hemorrhage Expansion. Clin. Neuroradiol. 2022, 32, 215–223. [Google Scholar] [CrossRef]
- Song, Z.; Guo, D.; Tang, Z.; Liu, H.; Li, X.; Luo, S.; Yao, X.; Song, W.; Song, J.; Zhou, Z. Noncontrast Computed Tomography-Based Radiomics Analysis in Discriminating Early Hematoma Expansion after Spontaneous Intracerebral Hemorrhage. Korean J. Radiol. 2021, 22, 415–424. [Google Scholar] [CrossRef]
- Song, Z.; Tang, Z.; Liu, H.; Guo, D.; Cai, J.; Zhou, Z. A clinical-radiomics nomogram may provide a personalized 90-day functional outcome assessment for spontaneous intracerebral hemorrhage. Eur. Radiol. 2021, 31, 4949–4959. [Google Scholar] [CrossRef]
- Xia, X.; Ren, Q.; Cui, J.; Dong, H.; Huang, Z.; Jiang, Q.; Guan, S.; Huang, C.; Yin, J.; Xu, J.; et al. Radiomics for predicting revised hematoma expansion with the inclusion of intraventricular hemorrhage growth in patients with supratentorial spontaneous intraparenchymal hematomas. Ann. Transl. Med. 2022, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, F.; Li, H.; Wu, Y.; Fu, H.; Zhong, Q.; Chen, J.; Wang, X. Development and validation of a clinical-radiomics nomogram for predicting a poor outcome and 30-day mortality after a spontaneous intracerebral hemorrhage. Quant. Imaging Med. Surg. 2022, 12, 4900–4913. [Google Scholar] [CrossRef] [PubMed]
- Buscot, M.-J.; Chandra, R.V.; Maingard, J.; Nichols, L.; Blizzard, L.; Stirling, C.; Smith, K.; Lai, L.; Asadi, H.; Froelich, J.; et al. Association of Onset-to-Treatment Time with Discharge Destination, Mortality, and Complications among Patients with Aneurysmal Subarachnoid Hemorrhage. JAMA Netw. Open 2022, 5, e2144039. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Chen, J.; Zhang, Y.; Zhao, F.; Zhai, Y.; Xu, X.; Xue, L.; Zhao, Y.; Wang, H. Exploration of Risk Factors for Poor Prognosis of Non-Traumatic Non-Aneurysmal Subarachnoid Hemorrhage. Biomolecules 2022, 12, 948. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, D.; Zhang, Q.; Ma, Y.; Li, S.; Zhao, H.; Deng, J.; Yang, J.; Ren, J.; Xu, M.; et al. Development and Validation of a Clinical-Based Signature to Predict the 90-Day Functional Outcome for Spontaneous Intracerebral Hemorrhage. Front. Aging Neurosci. 2022, 14, 904085. [Google Scholar] [CrossRef] [PubMed]
- Saydjari, A.K.; Finkbeiner, D.P. Equivariant Wavelets: Fast Rotation and Translation Invariant Wavelet Scattering Transforms. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 1716–1731. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Lin, Z.; Deng, B.; Zhu, J.; Li, Q. Cascade Superpixel Regularized Gabor Feature Fusion for Hyperspectral Image Classification. IEEE Trans. Neural Netw. Learn. Syst. 2020, 31, 1638–1652. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Lau, M.C.; Haruki, K.; Väyrynen, J.P.; Gurjao, C.; Väyrynen, S.A.; Dias Costa, A.; Borowsky, J.; Fujiyoshi, K.; Arima, K.; et al. Bayesian risk prediction model for colorectal cancer mortality through integration of clinicopathologic and genomic data. npj Precis. Oncol. 2023, 7, 57. [Google Scholar] [CrossRef]
- Ferreiro, A.L.; Choi, J.; Ryou, J.; Newcomer, E.P.; Thompson, R.; Bollinger, R.M.; Hall-Moore, C.; Ndao, I.M.; Sax, L.; Benzinger, T.L.S.; et al. Gut microbiome composition may be an indicator of preclinical Alzheimer’s disease. Sci. Transl. Med. 2023, 15, eabo2984. [Google Scholar] [CrossRef]
- Yang, X.; Chen, D.; Sun, Q.; Wang, Y.; Xia, Y.; Yang, J.; Lin, C.; Dang, X.; Cen, Z.; Liang, D.; et al. A live-cell image-based machine learning strategy for reducing variability in PSC differentiation systems. Cell Discov. 2023, 9, 53. [Google Scholar] [CrossRef]
- Huang, X.; Liu, B.; Guo, S.; Guo, W.; Liao, K.; Hu, G.; Shi, W.; Kuss, M.; Duryee, M.J.; Anderson, D.R.; et al. SERS spectroscopy with machine learning to analyze human plasma derived sEVs for coronary artery disease diagnosis and prognosis. Bioeng. Transl. Med. 2023, 8, e10420. [Google Scholar] [CrossRef] [PubMed]
- Danieli, M.G.; Paladini, A.; Longhi, E.; Tonacci, A.; Gangemi, S. A machine learning analysis to evaluate the outcome measures in inflammatory myopathies. Autoimmun. Rev. 2023, 22, 103353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Al-Shaheri, F.N.; Alhamdani, M.S.S.; Bauer, A.S.; Hoheisel, J.D.; Schenk, M.; Hinz, U.; Goedecke, P.; Al-Halabi, K.; Büchler, M.W.; et al. Blood-Based Diagnosis and Risk Stratification of Patients with Pancreatic Intraductal Papillary Mucinous Neoplasm (IPMN). Clin. Cancer Res. 2023, 29, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ye, J.; Zhang, K.; Ding, L.; Granzier-Nakajima, T.; Ranasinghe, J.C.; Xue, Y.; Sharma, S.; Biase, I.; Terrones, M.; et al. Rapid Biomarker Screening of Alzheimer’s Disease by Interpretable Machine Learning and Graphene-Assisted Raman Spectroscopy. ACS Nano 2022, 16, 6426–6436. [Google Scholar] [CrossRef]



| Variable | Total n = 105 | p Value | ||
|---|---|---|---|---|
| Good Outcome (85) | Poor Outcome (20) | |||
| Sex | Male | 31 | 8 | 0.77 |
| Female | 54 | 12 | ||
| Age | <60 | 46 | 6 | 0.05 |
| ≥60 | 39 | 14 | ||
| Hypertension | Yes | 50 | 12 | 0.92 |
| No | 35 | 8 | ||
| Diabetes Mellitus | Yes | 11 | 5 | 0.18 |
| No | 74 | 15 | ||
| Smoking | Yes | 20 | 6 | 0.55 |
| No | 65 | 14 | ||
| Drinking | Yes | 22 | 4 | 0.58 |
| No | 63 | 16 | ||
| Model | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Model_LR | 0.75 | 0.56 | 0.75 | 0.64 |
| Model_SVM | 0.84 | 0.87 | 0.84 | 0.82 |
| Model_RF | 0.78 | 0.76 | 0.78 | 0.74 |
| Model_LGBM | 0.81 | 0.80 | 0.81 | 0.80 |
| Model_AdaBoost | 0.75 | 0.75 | 0.75 | 0.75 |
| Model_XGBoost | 0.84 | 0.84 | 0.84 | 0.83 |
| Model_MLP | 0.81 | 0.85 | 0.81 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, D.; Wang, J.; Qi, P.; Lu, J.; Wang, D. Non-Contrasted CT Radiomics for SAH Prognosis Prediction. Bioengineering 2023, 10, 967. https://doi.org/10.3390/bioengineering10080967
Shan D, Wang J, Qi P, Lu J, Wang D. Non-Contrasted CT Radiomics for SAH Prognosis Prediction. Bioengineering. 2023; 10(8):967. https://doi.org/10.3390/bioengineering10080967
Chicago/Turabian StyleShan, Dezhi, Junjie Wang, Peng Qi, Jun Lu, and Daming Wang. 2023. "Non-Contrasted CT Radiomics for SAH Prognosis Prediction" Bioengineering 10, no. 8: 967. https://doi.org/10.3390/bioengineering10080967
APA StyleShan, D., Wang, J., Qi, P., Lu, J., & Wang, D. (2023). Non-Contrasted CT Radiomics for SAH Prognosis Prediction. Bioengineering, 10(8), 967. https://doi.org/10.3390/bioengineering10080967






