Design and Validation of a Deep Learning Model for Renal Stone Detection and Segmentation on Kidney–Ureter–Bladder Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Structure of Renal Stones
- Calcium (Ca)-containing stones:
- Non-calcium-containing stones:
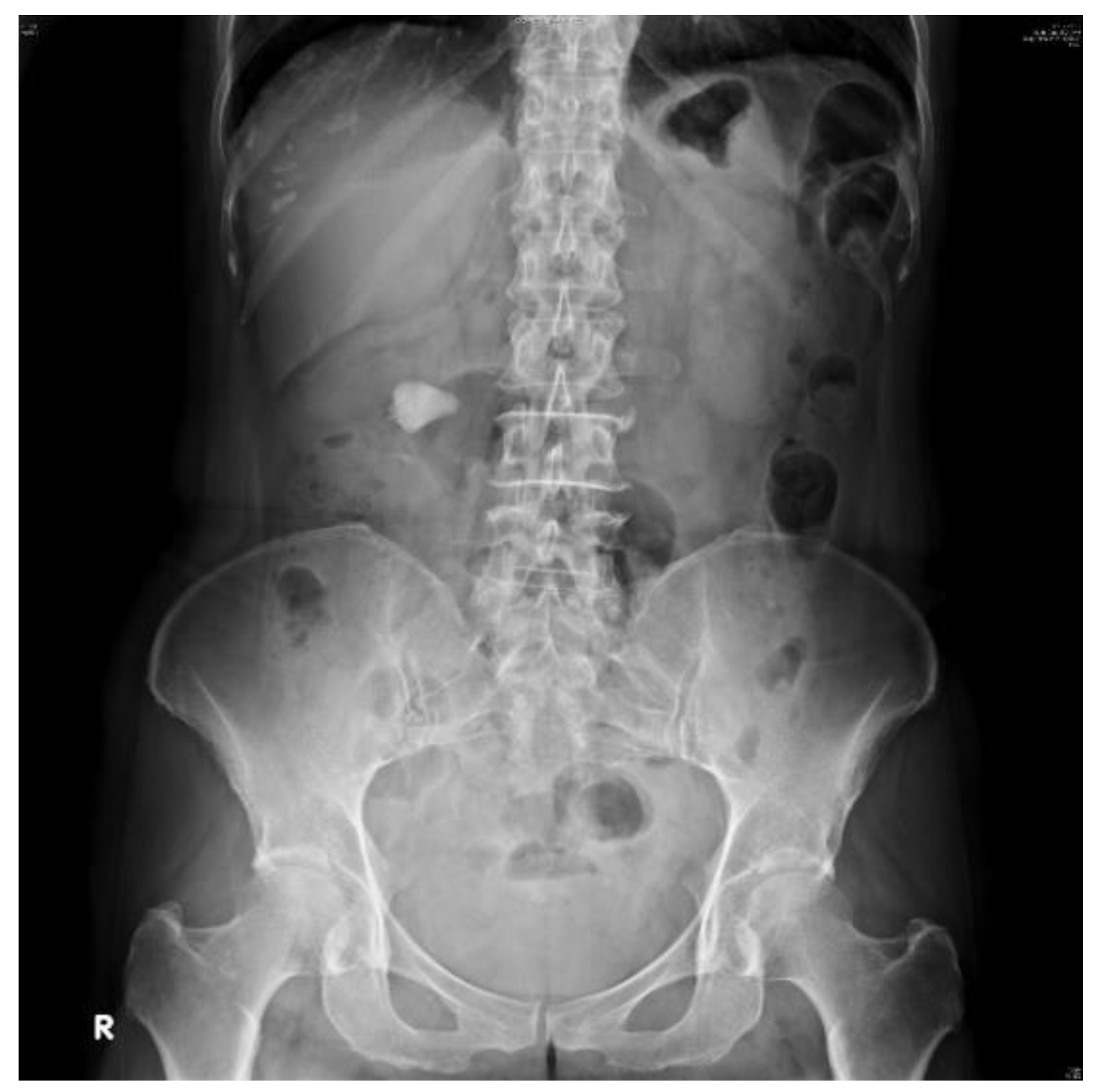
2.2. Datasets
2.3. Image Preprocessing
2.3.1. Contrast-Limited Adaptive Histogram Equalization
2.3.2. Image Mask
2.3.3. Image Cropping
2.4. Data Augmentation
2.5. Deep Learning Model
2.5.1. Residual Network
2.5.2. Inception-ResNetV2
2.5.3. U-Net
2.6. System Architecture
3. Results
3.1. Evaluation Metrics
3.2. Effect of Data Augmentation on the Training of the Classification Model
3.3. Subsystem 1—Classification Model for Medical Images
3.4. Subsystem 2—Segmentation Model for Medical Images
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Romero, V.; Akpinar, H.; Assimos, D.G. Kidney stones: A global picture of prevalence, incidence, and associated risk factors. Rev. Urol. 2010, 12, e86–e96. [Google Scholar] [PubMed]
- Chewcharat, A.; Curhan, G. Trends in the prevalence of kidney stones in the United States from 2007 to 2016. Urolithiasis 2020, 49, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Tundo, G.; Vollstedt, A.; Meeks, W.; Pais, V. Beyond Prevalence: Annual Cumulative Incidence of Kidney Stones in the United States. J. Urol. 2021, 205, 1704–1709. [Google Scholar] [CrossRef]
- Alghamdi, H.S.; Amoudi, G.; Elhag, S.; Saeedi, K.; Nasser, J. Deep Learning Approaches for Detecting COVID-19 From Chest X-Ray Images: A Survey. IEEE Access 2021, 9, 20235–20254. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Kaabouch, N.; Hu, W.C. A survey of medical imaging techniques used for breast cancer detection. In Proceedings of the IEEE International Conference on Electro-Information Technology, Rapid City, SD, USA, 9–11 May 2013; pp. 1–5. [Google Scholar] [CrossRef]
- Poovongsaroj, S.; Rattanachaisit, P.; Patcharatrakul, T.; Gonlachanvit, S.; Vateekul, P. AI-assisted diagnosis of DYSSYNER-GIC defecation using deep learning approach on abdominal radiography and symptom questionnaire. In Proceedings of the 2022 19th International Joint Conference on Computer Science and Software Engineering (JCSSE), Bangkok, Thailand, 8–10 June 2022. [Google Scholar]
- Ashour, A.S.; Dey, N.; Mohamed, W.S. Abdominal imaging in clinical applications: Computer aided diagnosis approaches. Med. Imaging Clin. Appl. 2016, 651, 3–17. [Google Scholar]
- Heidenreich, A.; Desgrandschamps, F.; Terrier, F. Modern Approach of Diagnosis and Management of Acute Flank Pain: Review of All Imaging Modalities. Eur. Urol. 2002, 41, 351–362. [Google Scholar] [CrossRef]
- Niall, O.; Russell, J.; MacGregor, R.; Duncan, H.; Mullins, J. A Comparison of Noncontrast Computerized Tomography with Excretory Urography in the Assessment of Acute Flank Pain. J. Urol. 1999, 161, 534–537. [Google Scholar] [CrossRef]
- Wang, J.-H.; Shen, S.-H.; Huang, S.-S.; Chang, C.-Y. Prospective Comparison of Unenhanced Spiral Computed Tomography and Intravenous Urography in the Evaluation of Acute Renal Colic. J. Chin. Med. Assoc. 2008, 71, 30–36. [Google Scholar] [CrossRef]
- Brisbane, W.; Bailey, M.R.; Sorensen, M.D. An overview of kidney stone imaging techniques. Nat. Rev. Urol. 2016, 13, 654–662. [Google Scholar] [CrossRef]
- Li, L.; Wei, M.; Liu, B.; Atchaneeyasakul, K.; Zhou, F.; Pan, Z.; Kumar, S.A.; Zhang, J.Y.; Pu, Y.; Liebeskind, D.S.; et al. Deep Learning for Hemorrhagic Lesion Detection and Segmentation on Brain CT Images. IEEE J. Biomed. Health Inform. 2020, 25, 1646–1659. [Google Scholar] [CrossRef]
- Huang, L.; Han, R.; Ai, T.; Yu, P.; Kang, H.; Tao, Q.; Xia, L. Serial Quantitative Chest CT Assessment of COVID-19: A Deep Learning Approach. Radiol. Cardiothorac. Imaging 2020, 2, e200075. [Google Scholar] [CrossRef] [PubMed]
- Mandeville, J.A.; Gnessin, E.; Lingeman, J.E. Imaging evaluation in the patient with kidney stone disease. Semin. Nephrol. 2011, 31, 254–258. [Google Scholar] [CrossRef]
- Fulgham, P.F.; Assimos, D.G.; Pearle, M.S.; Preminger, G.M. Clinical Effectiveness Protocols for Imaging in the Management of Ureteral Calculous Disease: AUA Technology Assessment. J. Urol. 2013, 189, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K.; Aoyama, T.; Koyama, S.; Kawaura, C. Comparative evaluation of organ and effective doses for paediatric patients with those for adults in chest and abdominal CT Examinations. Br. J. Radiol. 2007, 80, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Smith-Bindman, R.; Moghadassi, M.; Wilson, N.; Nelson, T.R.; Boone, J.M.; Cagnon, C.H.; Gould, R.; Hall, D.J.; Krishnam, M.; Lamba, R.; et al. Radiation doses in consecutive CT examinations from five University of California Medical Centers. Radiology 2015, 277, 134–141. [Google Scholar] [CrossRef]
- Metaxas, V.I.; Messaris, G.A.; Lekatou, A.N.; Petsas, T.G.; Panayiotakis, G.S. Patient doses in common diagnostic X-ray examinations. Radiat. Prot. Dosim. 2018, 184, 12–27. [Google Scholar] [CrossRef]
- Brenner, D.J.; Hall, E.J. Computed Tomography—An Increasing Source of Radiation Exposure. N. Engl. J. Med. 2007, 357, 2277–2284. [Google Scholar] [CrossRef]
- Sagara, Y.; Hara, A.K.; Pavlicek, W.; Silva, A.C.; Paden, R.G.; Wu, Q. Abdominal CT: Comparison of Low-Dose CT With Adaptive Statistical Iterative Reconstruction and Routine-Dose CT With Filtered Back Projection in 53 Patients. Am. J. Roentgenol. 2010, 195, 713–719. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Sarvamangala, D.R.; Kulkarni, R.V. Convolutional neural networks in medical image understanding: A survey. Evol. Intell. 2021, 15, 1–22. [Google Scholar] [CrossRef]
- Fu, Y.; Lei, Y.; Wang, T.; Curran, W.J.; Liu, T.; Yang, X. Deep learning in medical image registration: A review. Phys. Med. Biol. 2020, 65, 20TR01. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.-P.; Samala, R.K.; Hadjiiski, L.M.; Zhou, C. Deep learning in medical image analysis. Adv. Exp. Med. Biol. 2020, 1213, 3–21. [Google Scholar] [PubMed]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical Review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.P.; Hadjiiski, L.M.; Samala, R.K. Computer-aided diagnosis in the era of deep learning. Med. Phys. 2020, 47, e218–e227. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.H.; Hadjiiski, L.; Samala, R.K.; Chan, H.-P.; Caoili, E.M.; Cohan, R.H. Urinary bladder segmentation in CT urography using deep-learning convolutional neural network and level sets. Med. Phys. 2016, 43, 1882–1896. [Google Scholar] [CrossRef]
- Längkvist, M.; Jendeberg, J.; Thunberg, P.; Loutfi, A.; Lidén, M. Computer aided detection of ureteral stones in thin slice computed tomography volumes using Convolutional Neural Networks. Comput. Biol. Med. 2018, 97, 153–160. [Google Scholar] [CrossRef]
- Fitri, L.A.; Haryanto, F.; Arimura, H.; YunHao, C.; Ninomiya, K.; Nakano, R.; Haekal, M.; Warty, Y.; Fauzi, U. Automated classification of urinary stones based on microcomputed tomography images using convolutional neural network. Phys. Medica 2020, 78, 201–208. [Google Scholar] [CrossRef]
- Kobayashi, M.; Ishioka, J.; Matsuoka, Y.; Fukuda, Y.; Kohno, Y.; Kawano, K.; Morimoto, S.; Muta, R.; Fujiwara, M.; Kawamura, N.; et al. Computer-aided diagnosis with a convolutional neural network algorithm for automated detection of urinary tract stones on plain X-ray. BMC Urol. 2021, 21, 102. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Huang, Z.-H.; Huang, K.-W. Deep Learning Model for Computer-Aided Diagnosis of Urolithiasis Detection from Kidney–Ureter–Bladder Images. Bioengineering 2022, 9, 811. [Google Scholar] [CrossRef]
- Pearle, M.S.; Lotan, Y. Urinary lithiasis. In Campbell-Walsh Urology; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Zimmerman, J.; Pizer, S.; Staab, E.; Perry, J.; McCartney, W.; Brenton, B. An evaluation of the effectiveness of adaptive histogram equalization for contrast enhancement. IEEE Trans. Med. Imaging 1988, 7, 304–312. [Google Scholar] [CrossRef]
- Pizer, S.M.; Amburn, E.P.; Austin, J.D.; Cromartie, R.; Geselowitz, A.; Greer, T.; ter Haar Romeny, B.; Zimmerman, J.B.; Zuiderveld, K. Adaptive histogram equalization and its variations. Comput. Vis. Graph. Image Process. 1987, 39, 355–368. [Google Scholar] [CrossRef]
- Zuiderveld, K. Contrast limited adaptive histogram equalization. Graph. Gems 1994, 474–485. [Google Scholar]
- Lucknavalai, K.; Schulze, J.P. Real-Time Contrast Enhancement for 3D Medical Images Using Histogram Equalization. In Proceedings of the Advances in Visual Computing: 15th International Symposium, ISVC 2020, San Diego, CA, USA, 5–7 October 2020; Volume 12509, pp. 224–235. [Google Scholar] [CrossRef]
- Hayati, M.; Muchtar, K.; Roslidar; Maulina, N.; Syamsuddin, I.; Elwirehardja, G.N.; Pardamean, B. Impact of CLAHE-based image enhancement for diabetic retinopathy classification through deep learning. Procedia Comput. Sci. 2023, 216, 57–66. [Google Scholar] [CrossRef]
- Dinh, P.; Giang, N.L. A new medical image enhancement algorithm using adaptive parameters. Int. J. Imaging Syst. Technol. 2022, 32, 2198–2218. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Shen, W.; Xu, W.; Zhang, H.; Sun, Z.; Ma, J.; Ma, X.; Zhou, S.; Guo, S.; Wang, Y. Automatic segmentation of the femur and tibia bones from X-ray images based on pure dilated residual U-Net. Inverse Probl. Imaging 2021, 15, 1333. [Google Scholar] [CrossRef]
- Fawzi, A.; Samulowitz, H.; Turaga, D.; Frossard, P. Adaptive data augmentation for Image Classification. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016. [Google Scholar]
- Perez, L.; Wang, J. The effectiveness of data augmentation in image classification using deep learning. Convolutional Neural Netw. Vis. Recognit. 2017, 11, 1–8. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Nanni, L.; Paci, M.; Brahnam, S.; Lumini, A. Comparison of Different Image Data Augmentation Approaches. J. Imaging 2021, 7, 254. [Google Scholar] [CrossRef]
- Islam, Z.; Islam, M.; Asraf, A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform. Med. Unlocked 2020, 20, 100412. [Google Scholar] [CrossRef]
- Pathan, S.; Siddalingaswamy, P.; Ali, T. Automated Detection of Covid-19 from Chest X-ray scans using an optimized CNN architecture. Appl. Soft Comput. 2021, 104, 107238. [Google Scholar] [CrossRef]
- Gazda, M.; Plavka, J.; Gazda, J.; Drotar, P. Self-supervised deep convolutional neural network for chest X-ray classification. IEEE Access 2021, 9, 151972–151982. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, X.; Wang, Y.; Lei, X.; Teo, S.K.; Sim, J.Z.; Ting, Y.; Zhen, L.; Zhou, J.T.; Liu, Y.; et al. Deep supervised do-main adaptation for pneumonia diagnosis from chest X-ray images. IEEE J. Biomed. Health Inform. 2022, 26, 1080–1090. [Google Scholar] [CrossRef] [PubMed]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. Gan-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, J.; Liu, Y.; Fu, H.; Hu, Y.; Cheng, J.; Qi, H.; Wu, Y.; Zhang, J.; Zhao, Y. Structure and Illumination Constrained GAN for Medical Image Enhancement. IEEE Trans. Med. Imaging 2021, 40, 3955–3967. [Google Scholar] [CrossRef]
- Wright, L. Ranger—A Synergistic Optimizer. 2019. Available online: https://github.com/lessw2020/Ranger-Deep-Learning-Optimizer (accessed on 28 February 2023).
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-V4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, K.; Wan, Z.; Xie, S.; Lv, Z. Popular deep learning algorithms for disease prediction: A review. Clust. Comput. 2022, 26, 1231–1251. [Google Scholar] [CrossRef]
- Dakhli, R.; Barhoumi, W. A skin lesion classification method based on expanding the surrounding lesion-shaped border for an end-to-end inception-ResNet-V2 classifier. Signal Image Video Process. 2023, 5, 1–9. [Google Scholar] [CrossRef]
- Bozkurt, F. Skin lesion classification on dermatoscopic images using effective data augmentation and pre-trained deep learning approach. Multimed. Tools Appl. 2022, 82, 18985–19003. [Google Scholar] [CrossRef]
- Miserlis, D.; Munian, Y.; Cardona, L.M.F.; Teixeira, P.G.; DuBose, J.J.; Davies, M.G.; Alamaniotis, M.; Bohannon, W.; Koutakis, P. Benchmarking efficientnetb7, InceptionResNetV2, inceptionv3, and xception artificial neural networks ap-plications for aortic pathologies analysis. J. Vasc. Surg. 2023, 77, E345. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted Res-UNet for high-quality retina vessel segmentation. In Proceedings of the 2018 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Johansen, D.; de Lange, T.; Johansen, H.D.; Halvorsen, P.; Riegler, M.A. A comprehensive study on colorectal polyp segmentation with resunet++, conditional random field and test-time augmentation. IEEE J. Biomed. Health Inform. 2021, 25, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-C.; Yang, Y.; Wang, J.; Xu, W.; Yuille, A.L. Attention to scale: Scale-aware Semantic Image segmentation. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Y.; Wang, O.; Zhang, R.; Owens, A.; Efros, A.A. CNN-generated images are surprisingly easy to spot… for now. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Liu, L.; Jiang, H.; He, P.; Chen, W.; Liu, X.; Gao, J.; Han, J. On the variance of the adaptive learning rate and beyond. arXiv preprint 2019, arXiv:1908.03265. [Google Scholar]
- Zhang, M.J.; Ba, L.J.; Hinton, G.E. Lookahead optimizer: K steps forward, 1 step back. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2019; Volume 32. [Google Scholar]
- Dai, J.C.; Johnson, B.A. Artificial intelligence in endourology: Emerging technology for individualized care. Curr. Opin. Urol. 2022, 32, 379–392. [Google Scholar] [CrossRef]
- Parakh, A.; Lee, H.; Lee, J.H.; Eisner, B.H.; Sahani, D.V.; Do, S. Urinary stone detection on CT images using deep convolutional neural networks: Evaluation of model performance and generalization. Radiol. Artif. Intell. 2019, 1, e180066. [Google Scholar] [CrossRef]
- Chen, C.; Liu, M.-Y.; Tuzel, O.; Xiao, J. R-CNN for small object detection. In Proceedings of the Asian Conference on Computer Vision, Taipei, Taiwan, 20–24 November 2016; pp. 214–230. [Google Scholar]


























| CPU | Graphics Card | Memory |
|---|---|---|
| Intel Core i7-8700 @ 3.19 GHz | Nvidia GeForce RTX3070 8 G | 32 G |
| Epochs | 50 |
|---|---|
| Batch size | 16 |
| Learning rate | 0.001 |
| Loss function | Binary cross-entropy |
| Optimization algorithm | Ranger |
| Predicted Label | |||
|---|---|---|---|
| Have Stone | NO Stone | ||
| True label | Have stone | 182 | 3 |
| No stone | 1 | 184 | |
| Accuracy | Sensitivity | Specificity | Precision | F1-Score | |
|---|---|---|---|---|---|
| Testing dataset | 0.989 | 0.995 | 0.984 | 0.984 | 0.989 |
| Predicted Label | |||
|---|---|---|---|
| Have Stone | No Stone | ||
| True label | Have stone | 184 | 0 |
| No stone | 1 | 185 | |
| Accuracy | Sensitivity | Specificity | Precision | F1-Score | |
|---|---|---|---|---|---|
| Testing dataset | 0.997 | 1.000 | 0.995 | 0.995 | 0.997 |
| ResNet50 [31] | Inception-ResNetV2 | |
|---|---|---|
| Accuracy | 0.989 | 0.997 |
| Sensitivity | 0.995 | 1.000 |
| Specificity | 0.984 | 0.995 |
| Precision | 0.984 | 0.995 |
| F1-score | 0.989 | 0.997 |
| Epochs | 100 |
|---|---|
| Batch size | 8 |
| Learning rate | 0.0001 |
| Loss function | Focal loss + Jaccard loss |
| Optimization algorithm | Ranger |
| Bce_dice_loss | Bce_jaccard_loss | Binary_focal_dice_loss | Binary_focal_jaccard_loss | |
|---|---|---|---|---|
| TP | 270,382 | 267,678 | 259,717 | 268,203 |
| FP | 72,813 | 75,517 | 83,478 | 74,992 |
| TN | 1,480,475 | 1,480,115 | 1,481,879 | 1,482,633 |
| FN | 26,330 | 26,690 | 24,926 | 27,751 |
| Bce_dice_loss | Bce_jaccard_loss | Binary_focal_dice_loss | Binary_focal_jaccard_loss | |
|---|---|---|---|---|
| TP | 268,540 | 270,597 | 256,915 | 266,816 |
| FP | 74,655 | 72,598 | 86,280 | 76,379 |
| TN | 1,479,054 | 1,472,688 | 1,487,191 | 1,480,639 |
| FN | 27,751 | 34,117 | 19,614 | 26,166 |
| Bce_dice_loss | Bce_jaccard_loss | Binary_focal_dice_loss | Binary_focal_jaccard_loss | |||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | |
| Accuracy | 0.946 | 0.946 | 0.945 | 0.945 | 0.941 | 0.941 | 0.946 | 0.946 |
| Sensitivity | 0.953 | 0.911 | 0.951 | 0.909 | 0.947 | 0.912 | 0.952 | 0.917 |
| Precision | 0.983 | 0.788 | 0.982 | 0.780 | 0.983 | 0.757 | 0.984 | 0.781 |
| F1-score | 0.968 | 0.845 | 0.967 | 0.840 | 0.965 | 0.827 | 0.968 | 0.844 |
| IoU | 0.937 | 0.732 | 0.935 | 0.724 | 0.932 | 0.706 | 0.937 | 0.730 |
| MIoU | 0.834 | 0.834 | 0.830 | 0.830 | 0.819 | 0.819 | 0.834 | 0.834 |
| FWIoU | 0.904 | 0.904 | 0.902 | 0.902 | 0.897 | 0.897 | 0.905 | 0.905 |
| Bce_dice_loss | Bce_jaccard_loss | Binary_focal_dice_loss | Binary_focal_jaccard_loss | |||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | Negative | Positive | |
| Accuracy | 0.945 | 0.945 | 0.942 | 0.942 | 0.943 | 0.943 | 0.945 | 0.945 |
| Sensitivity | 0.952 | 0.906 | 0.953 | 0.888 | 0.945 | 0.929 | 0.951 | 0.911 |
| Precision | 0.982 | 0.782 | 0.977 | 0.788 | 0.987 | 0.749 | 0.983 | 0.777 |
| F1-score | 0.967 | 0.840 | 0.965 | 0.836 | 0.966 | 0.829 | 0.967 | 0.839 |
| IoU | 0.935 | 0.724 | 0.932 | 0.717 | 0.934 | 0.708 | 0.935 | 0.722 |
| MIoU | 0.830 | 0.830 | 0.825 | 0.825 | 0.821 | 0.821 | 0.829 | 0.829 |
| FWIoU | 0.901 | 0.901 | 0.897 | 0.897 | 0.900 | 0.900 | 0.902 | 0.902 |
| ResNet34′s Bce_dice_loss | ResNet34′s Binary_focal_jaccard_loss | ResNet50′s Bce_dice_loss | ResNet50′s Binary_focal_jaccard_loss | |
|---|---|---|---|---|
| Accuracy | 0.946 | 0.946 | 0.945 | 0.945 |
| Sensitivity | 0.953 | 0.952 | 0.952 | 0.951 |
| Precision | 0.983 | 0.984 | 0.982 | 0.983 |
| F1-score | 0.968 | 0.968 | 0.967 | 0.967 |
| IoU | 0.937 | 0.937 | 0.935 | 0.935 |
| MIoU | 0.834 | 0.834 | 0.830 | 0.829 |
| FWIoU | 0.904 | 0.905 | 0.901 | 0.902 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.-H.; Liu, Y.-Y.; Wu, W.-J.; Huang, K.-W. Design and Validation of a Deep Learning Model for Renal Stone Detection and Segmentation on Kidney–Ureter–Bladder Images. Bioengineering 2023, 10, 970. https://doi.org/10.3390/bioengineering10080970
Huang Z-H, Liu Y-Y, Wu W-J, Huang K-W. Design and Validation of a Deep Learning Model for Renal Stone Detection and Segmentation on Kidney–Ureter–Bladder Images. Bioengineering. 2023; 10(8):970. https://doi.org/10.3390/bioengineering10080970
Chicago/Turabian StyleHuang, Zih-Hao, Yi-Yang Liu, Wei-Juei Wu, and Ko-Wei Huang. 2023. "Design and Validation of a Deep Learning Model for Renal Stone Detection and Segmentation on Kidney–Ureter–Bladder Images" Bioengineering 10, no. 8: 970. https://doi.org/10.3390/bioengineering10080970
APA StyleHuang, Z.-H., Liu, Y.-Y., Wu, W.-J., & Huang, K.-W. (2023). Design and Validation of a Deep Learning Model for Renal Stone Detection and Segmentation on Kidney–Ureter–Bladder Images. Bioengineering, 10(8), 970. https://doi.org/10.3390/bioengineering10080970







