Elucidating Multimodal Imaging Patterns in Accelerated Brain Aging: Heterogeneity through a Discriminant Analysis Approach Using the UK Biobank Dataset
Abstract
:1. Introduction
2. Materials and Methods
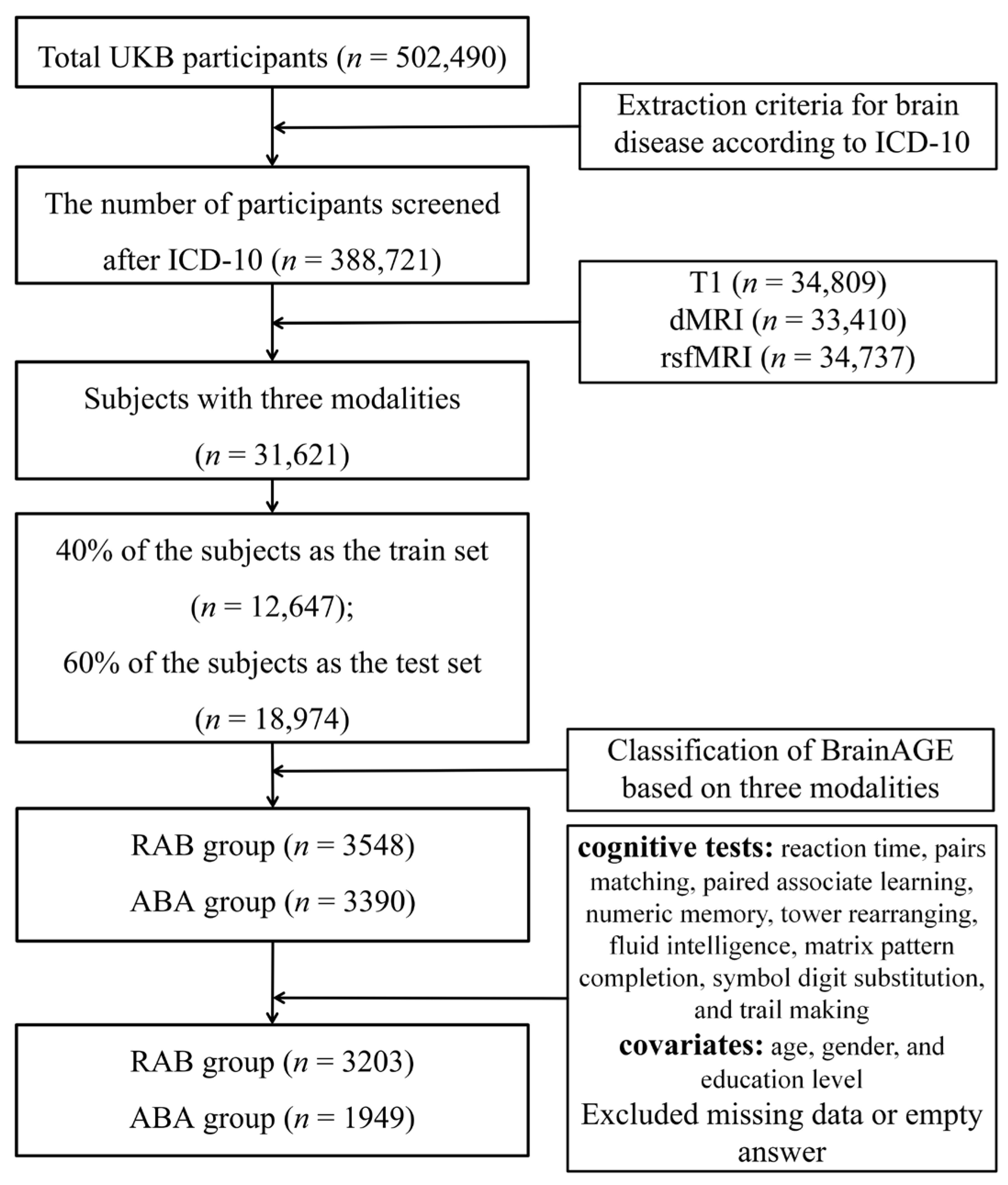
2.1. Participants
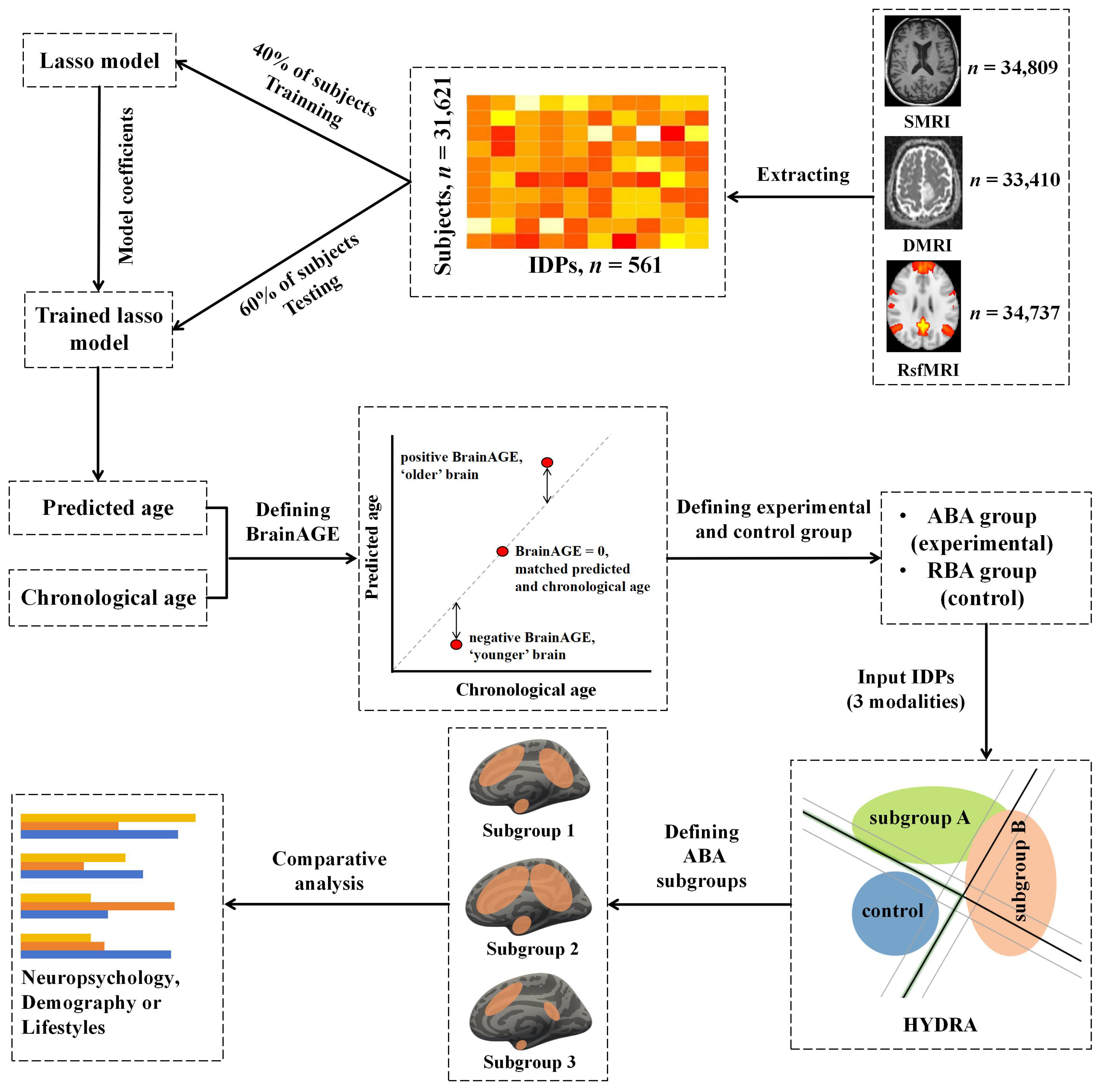
2.2. Imaging-Derived Phenotypes (IDPs)
2.3. Brain-Age Prediction Model
2.4. Non-Imaging Derived Phenotypes (Non-IDPs)
2.5. Neuropsychological Tests
2.6. Identification of ABA Subgroups Using HYDRA
2.7. Statistical Analysis
3. Results
3.1. Brain-Age Prediction
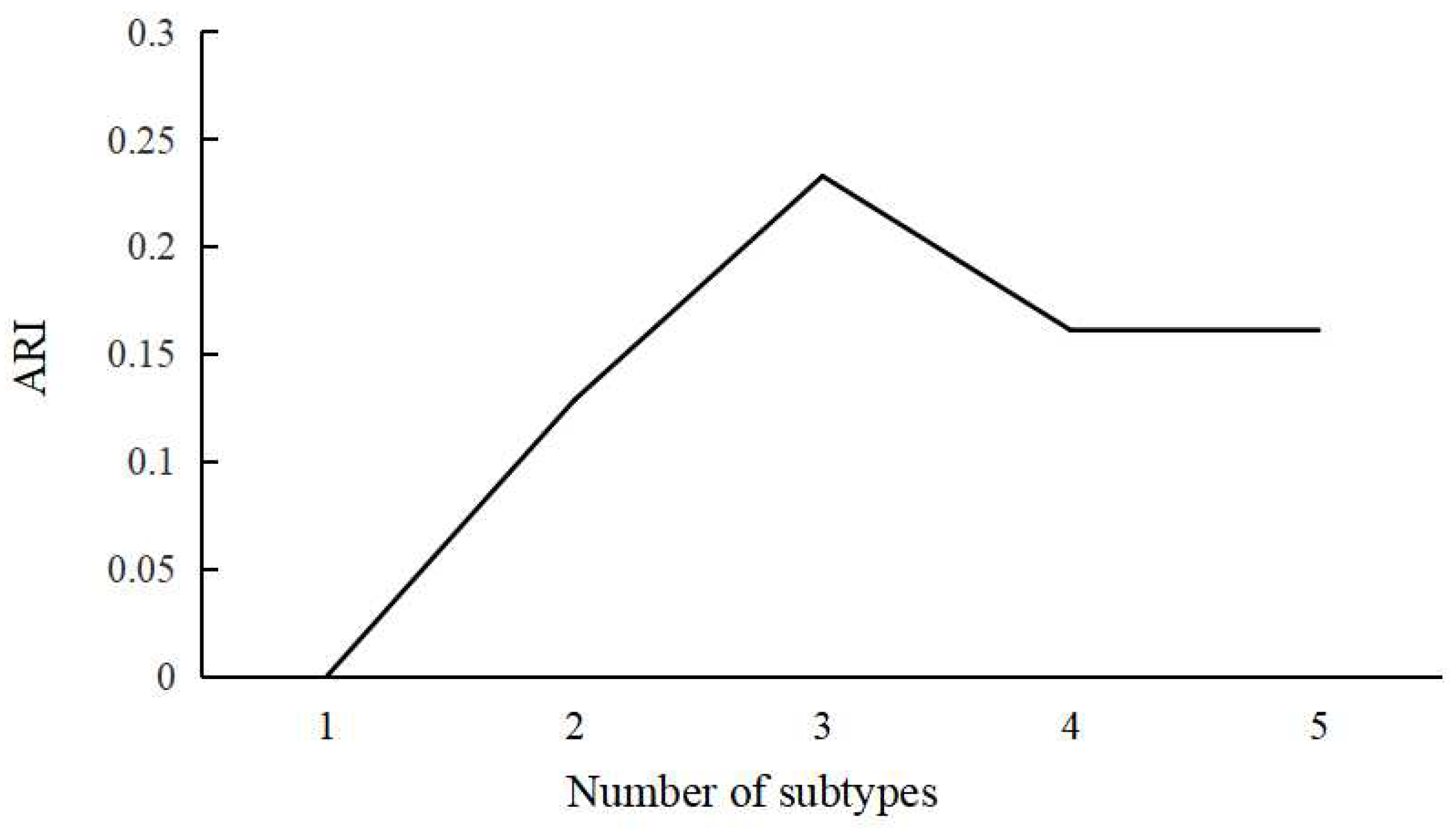
3.2. Definition of ABA Subgroups
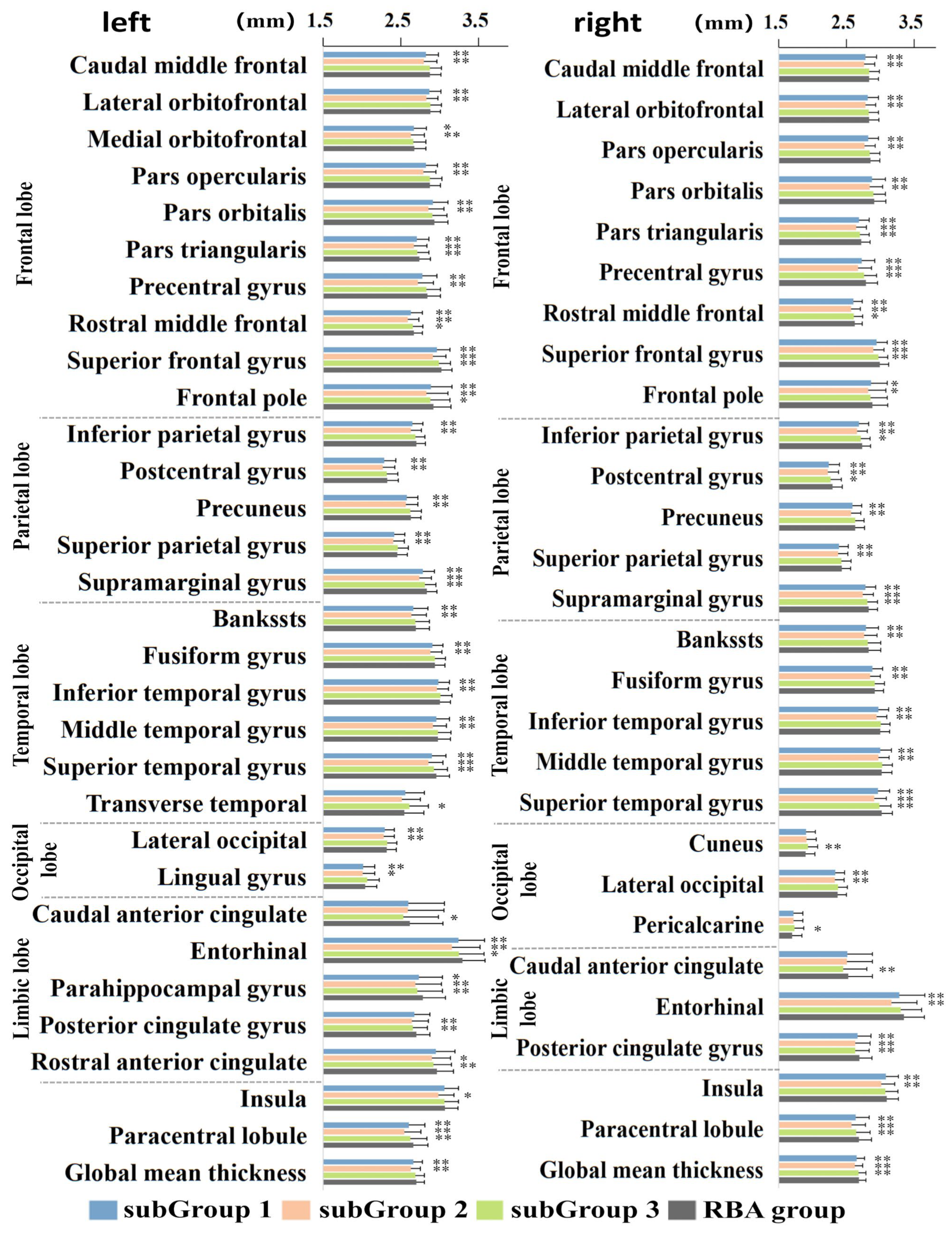
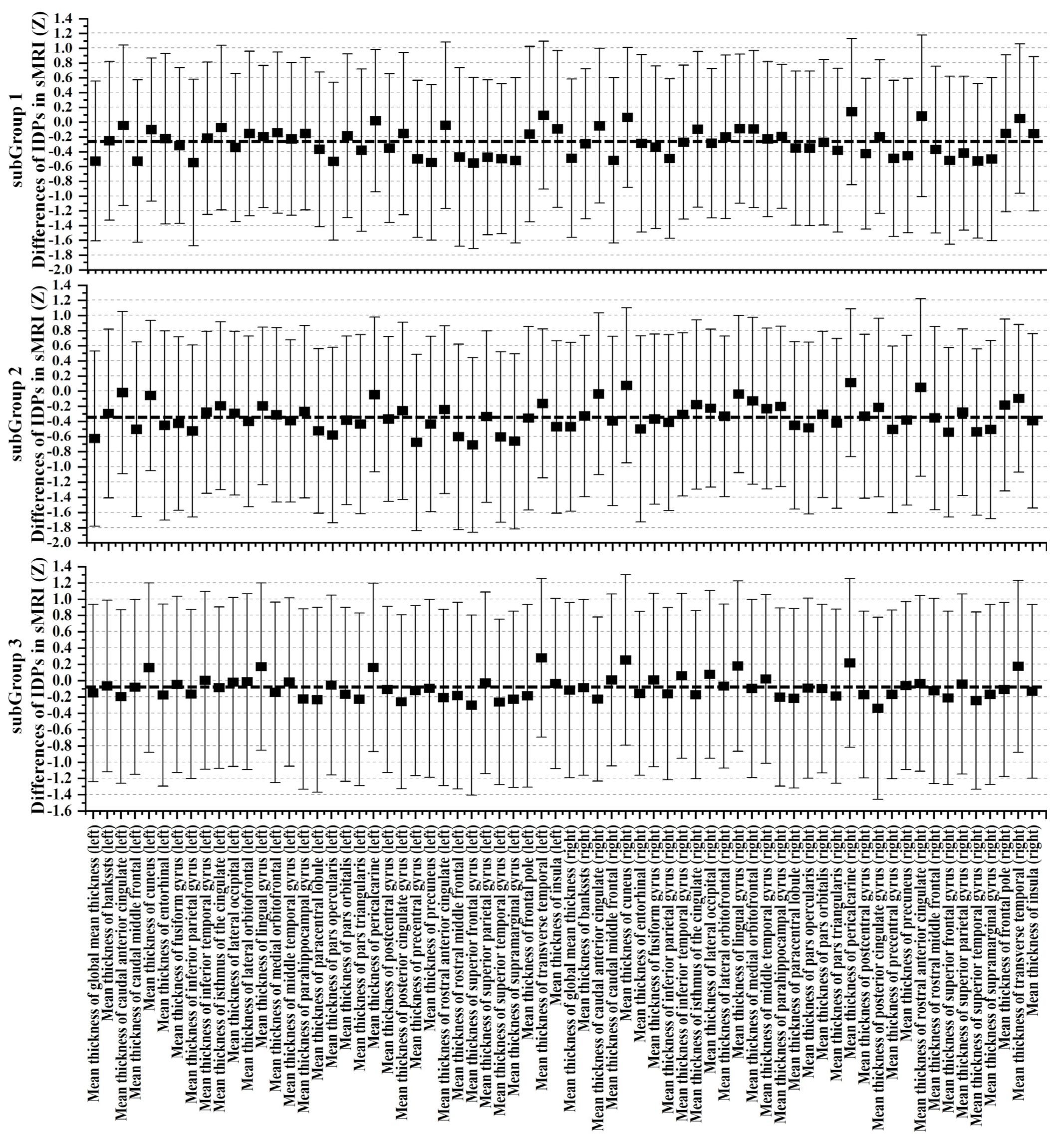

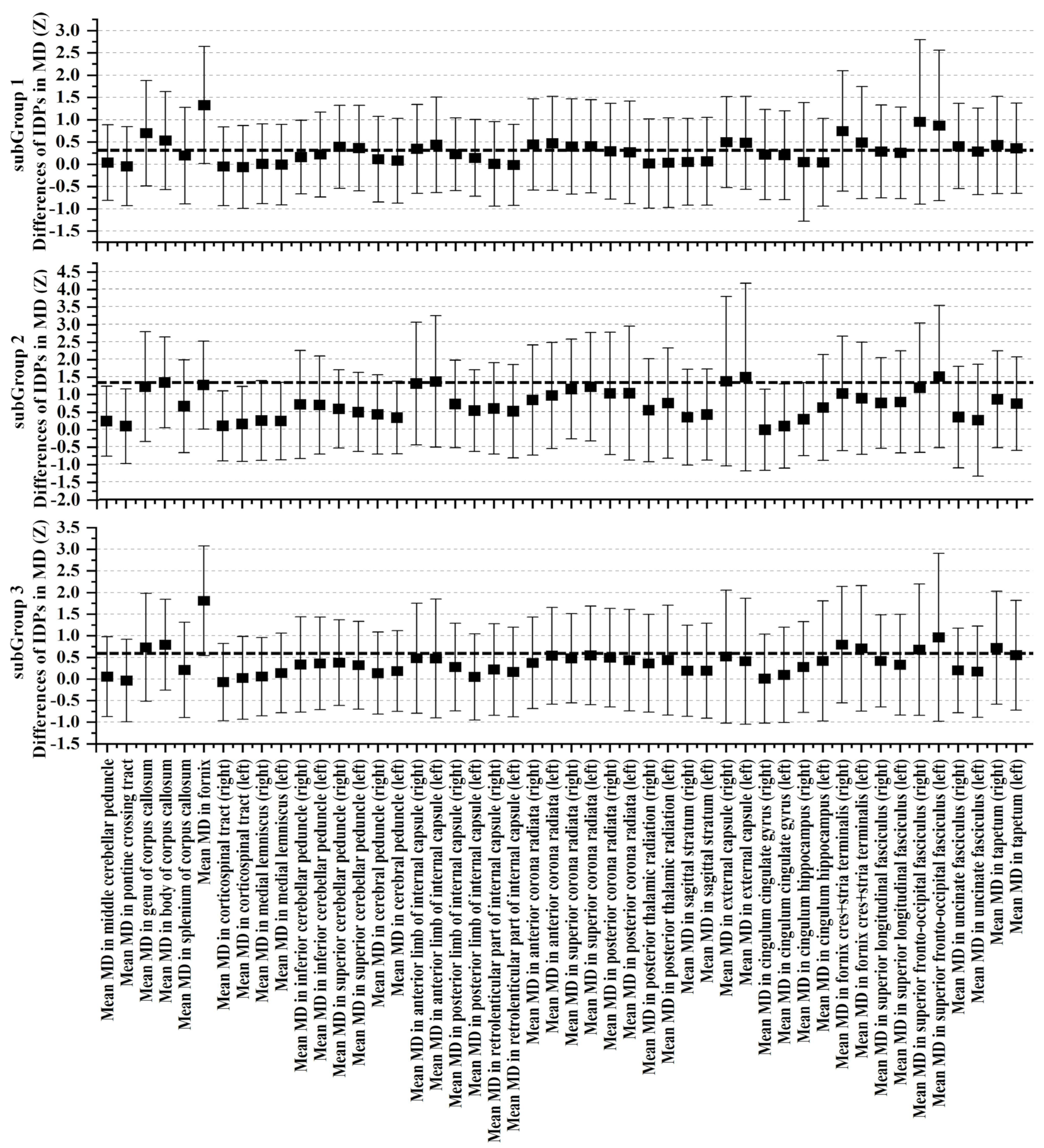
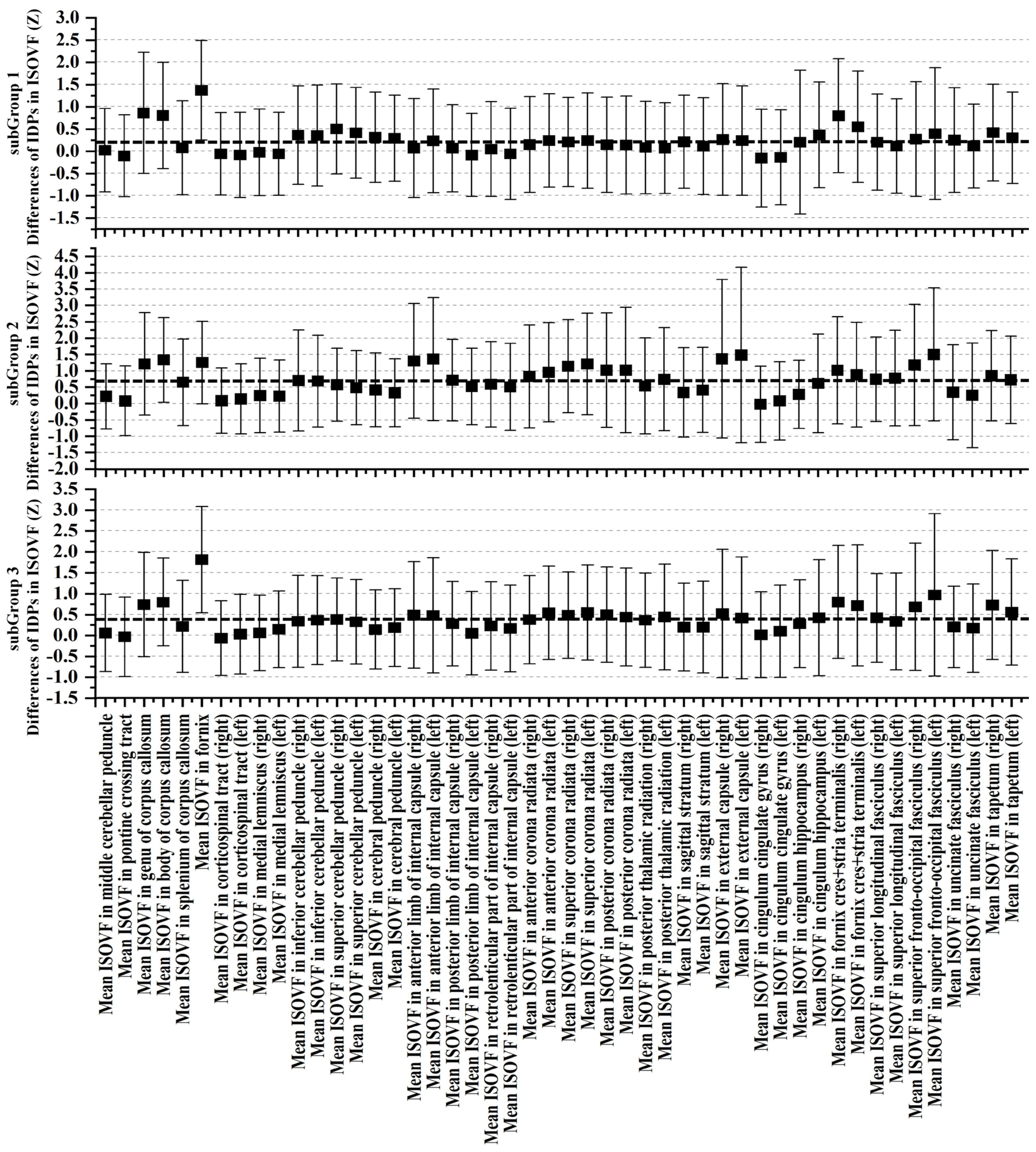

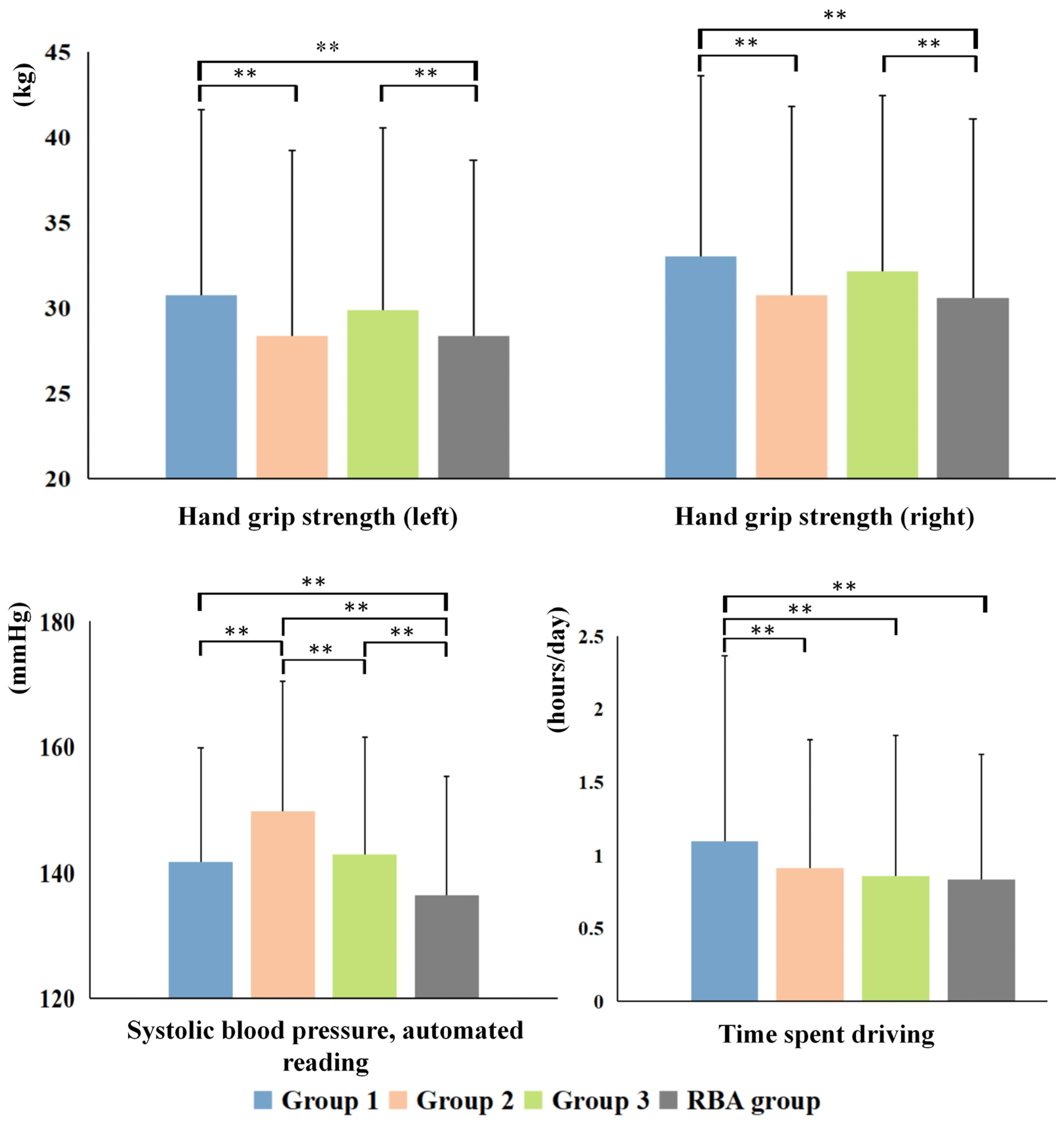
3.3. Cognitive and Non-IDPs Characteristics between Matched Subtypes
4. Discussion
4.1. Complex Landscape of ABA
4.2. ABA Subtype and Cognitive Reserve
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salat, D.H.; Buckner, R.L.; Snyder, A.Z.; Greve, D.N.; Desikan, R.S.; Busa, E.; Morris, J.C.; Dale, A.M.; Fischl, B. Thinning of the cerebral cortex in aging. Cereb. Cortex 2004, 14, 721–730. [Google Scholar] [CrossRef]
- Vernooij, M.W.; de Groot, M.; van der Lugt, A.; Ikram, M.A.; Krestin, G.P.; Hofman, A.; Niessen, W.J.; Breteler, M.M. White matter atrophy and lesion formation explain the loss of structural integrity of white matter in aging. Neuroimage 2008, 43, 470–477. [Google Scholar] [CrossRef]
- Salami, A.; Pudas, S.; Nyberg, L. Elevated hippocampal resting-state connectivity underlies deficient neurocognitive function in aging. Proc. Natl. Acad. Sci. USA 2014, 111, 17654–17659. [Google Scholar] [CrossRef]
- Planche, V.; Coupé, P.; Helmer, C.; Le Goff, M.; Amieva, H.; Tison, F.; Dartigues, J.F.; Catheline, G. Evolution of brain atrophy subtypes during aging predicts long-term cognitive decline and future Alzheimer’s clinical syndrome. Neurobiol. Aging 2019, 79, 22–29. [Google Scholar] [CrossRef]
- Lee, A.J.; Ma, Y.; Yu, L.; Dawe, R.J.; McCabe, C.; Arfanakis, K.; Mayeux, R.; Bennett, D.A.; Klein, H.U.; De Jager, P.L. Multi-region brain transcriptomes uncover two subtypes of aging individuals with differences in Alzheimer risk and the impact of APOEε4. bioRxiv 2023. preprint. [Google Scholar] [CrossRef]
- Mather, K.A. Genetic and Environmental Factors in Ageing and Age-Related Disease. Genes 2022, 13, 396. [Google Scholar] [CrossRef] [PubMed]
- Pang, S.Y.; Ho, P.W.; Liu, H.F.; Leung, C.T.; Li, L.; Chang, E.E.S.; Ramsden, D.B.; Ho, S.L. The interplay of aging, genetics and environmental factors in the pathogenesis of Parkinson’s disease. Transl. Neurodegener. 2019, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Xiong, M.; Jin, Y.; Kang, W.; Wu, S.; Sun, S.; Fu, Z. Quantifying Brain and Cognitive Maintenance as Key Indicators for Sustainable Cognitive Aging: Insights from the UK Biobank. Sustainability 2023, 15, 9620. [Google Scholar] [CrossRef]
- Liu, L.; Sun, S.; Kang, W.; Wu, S.; Lin, L. A review of neuroimaging-based data-driven approach for Alzheimer’s disease heterogeneity analysis. Rev. Neurosci. 2024, in press. [CrossRef] [PubMed]
- Abellaneda-Pérez, K.; Cattaneo, G.; Cabello-Toscano, M.; Solana-Sánchez, J.; Mulet-Pons, L.; Vaqué-Alcázar, L.; Perellón-Alfonso, R.; Solé-Padullés, C.; Bargalló, N.; Tormos, J.M.; et al. Purpose in life promotes resilience to age-related brain burden in middle-aged adults. Alzheimers Res. Ther. 2023, 15, 49. [Google Scholar] [CrossRef] [PubMed]
- Montine, T.J.; Cholerton, B.A.; Corrada, M.M.; Edland, S.D.; Flanagan, M.E.; Hemmy, L.S.; Kawas, C.H.; White, L.R. Concepts for brain aging: Resistance, resilience, reserve, and compensation. Alzheimers Res. Ther. 2019, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Peeters, G.; Kok, A.; de Bruin, S.R.; van Campen, C.; Graff, M.; Nieuwboer, M.; Huisman, M.; van Munster, B.; van der Zee, E.A.; Kas, M.J.; et al. Supporting Resilience of Older Adults with Cognitive Decline Requires a Multi-Level System Approach. Gerontology 2023, 69, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Kozic, D.B.; Thurnher, M.M.; Boban, J.; Sundgren, P.C. Editorial: Accelerated Brain Aging: Different Diseases-Different Imaging Patterns. Front. Neurol. 2022, 13, 889538. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, P.R.; Singla, A.; Vaidya, V.A. Early Adversity and Accelerated Brain Aging: A Mini-Review. Front. Mol. Neurosci. 2022, 15, 822917. [Google Scholar] [CrossRef]
- Darrow, S.M.; Verhoeven, J.E.; Révész, D.; Lindqvist, D.; Penninx, B.W.; Delucchi, K.L.; Wolkowitz, O.M.; Mathews, C.A. The Association Between Psychiatric Disorders and Telomere Length: A Meta-Analysis Involving 14,827 Persons. Psychosom. Med. 2016, 78, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Franke, K.; Ziegler, G.; Klöppel, S.; Gaser, C. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: Exploring the influence of various parameters. Neuroimage 2010, 50, 883–892. [Google Scholar] [CrossRef]
- Weihs, A.; Frenzel, S.; Wittfeld, K.; Obst, A.; Stubbe, B.; Habes, M.; Szentkirályi, A.; Berger, K.; Fietze, I.; Penzel, T.; et al. Associations between sleep apnea and advanced brain aging in a large-scale population study. Sleep 2021, 44, zsaa204. [Google Scholar] [CrossRef] [PubMed]
- Klaus, F.; Nguyen, T.T.; Thomas, M.L.; Liou, S.C.; Soontornniyomkij, B.; Mitchell, K.; Daly, R.; Sutherland, A.N.; Jeste, D.V.; Eyler, L.T. Peripheral inflammation levels associated with degree of advanced brain aging in schizophrenia. Front. Psychiatry 2022, 13, 966439. [Google Scholar] [CrossRef]
- Habes, M.; Janowitz, D.; Erus, G.; Toledo, J.B.; Resnick, S.M.; Doshi, J.; Van der Auwera, S.; Wittfeld, K.; Hegenscheid, K.; Hosten, N.; et al. Advanced brain aging: Relationship with epidemiologic and genetic risk factors, and overlap with Alzheimer disease atrophy patterns. Transl. Psychiatry 2016, 6, e775. [Google Scholar] [CrossRef]
- Han, L.K.M.; Schnack, H.G.; Brouwer, R.M.; Veltman, D.J.; van der Wee, N.J.A.; van Tol, M.J.; Aghajani, M.; Penninx, B. Contributing factors to advanced brain aging in depression and anxiety disorders. Transl. Psychiatry 2021, 11, 402. [Google Scholar] [CrossRef]
- Busby, N.; Wilmskoetter, J.; Gleichgerrcht, E.; Rorden, C.; Roth, R.; Newman-Norlund, R.; Hillis, A.E.; Keller, S.S.; de Bezenac, C.; Kristinsson, S.; et al. Advanced Brain Age and Chronic Poststroke Aphasia Severity. Neurology 2023, 100, e1166–e1176. [Google Scholar] [CrossRef]
- Chen, C.L.; Kuo, M.C.; Wu, W.C.; Hsu, Y.C.; Wu, R.M.; Tseng, W.I. Advanced brain aging in multiple system atrophy compared to Parkinson’s disease. NeuroImage Clin. 2022, 34, 102997. [Google Scholar] [CrossRef] [PubMed]
- Whitwell, J.L.; Petersen, R.C.; Negash, S.; Weigand, S.D.; Kantarci, K.; Ivnik, R.J.; Knopman, D.S.; Boeve, B.F.; Smith, G.E.; Jack, C.R., Jr. Patterns of atrophy differ among specific subtypes of mild cognitive impairment. Arch. Neurol. 2007, 64, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Wrigglesworth, J.; Ryan, J.; Ward, P.G.D.; Woods, R.L.; Storey, E.; Egan, G.F.; Murray, A.; Espinoza, S.E.; Shah, R.C.; Trevaks, R.E.; et al. Health-related heterogeneity in brain aging and associations with longitudinal change in cognitive function. Front. Aging Neurosci. 2022, 14, 1063721. [Google Scholar] [CrossRef]
- Eavani, H.; Habes, M.; Satterthwaite, T.D.; An, Y.; Hsieh, M.K.; Honnorat, N.; Erus, G.; Doshi, J.; Ferrucci, L.; Beason-Held, L.L.; et al. Heterogeneity of structural and functional imaging patterns of advanced brain aging revealed via machine learning methods. Neurobiol. Aging 2018, 71, 41–50. [Google Scholar] [CrossRef]
- Varol, E.; Sotiras, A.; Davatzikos, C. HYDRA: Revealing heterogeneity of imaging and genetic patterns through a multiple max-margin discriminative analysis framework. Neuroimage 2017, 145, 346–364. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, H.; Li, J.; Fan, Y.; Jülich, S.T.; Lei, X. Subtypes of insomnia revealed by the heterogeneity of neuroanatomical patterns: A structural MRI study. Biol. Psychol. 2023, 180, 108591. [Google Scholar] [CrossRef]
- Liu, H.; Zheng, R.; Zhang, Y.; Zhang, B.; Hou, H.; Cheng, J.; Han, S. Two distinct neuroanatomical subtypes of migraine without aura revealed by heterogeneity through discriminative analysis. Brain Imaging Behav. 2023, 17, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Xu, Y.; Guo, H.R.; Fang, K.; Wei, Y.; Liu, L.; Cheng, J.; Zhang, Y.; Cheng, J. Two distinct subtypes of obsessive compulsive disorder revealed by heterogeneity through discriminative analysis. Hum. Brain Mapp. 2022, 43, 3037–3046. [Google Scholar] [CrossRef]
- Liu, G.; Shi, L.; Qiu, J.; Lu, W. Two neuroanatomical subtypes of males with autism spectrum disorder revealed using semi-supervised machine learning. Mol. Autism 2022, 13, 9. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Miller, K.L.; Alfaro-Almagro, F.; Bangerter, N.K.; Thomas, D.L.; Yacoub, E.; Xu, J.; Bartsch, A.J.; Jbabdi, S.; Sotiropoulos, S.N.; Andersson, J.L.; et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 2016, 19, 1523–1536. [Google Scholar] [CrossRef]
- Chung, B.S.; Park, J.S. Automatic segmentation of true color sectioned images using FMRIB Software Library: First trial in brain, gray matter, and white matter. Clin. Anat. 2020, 33, 1197–1203. [Google Scholar] [CrossRef]
- Beckmann, C.F.; Smith, S.M. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging 2004, 23, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.H. Multimodality neuroimaging brain-age in UK biobank: Relationship to biomedical, lifestyle, and cognitive factors. Neurobiol. Aging 2020, 92, 34–42. [Google Scholar] [CrossRef]
- Xiong, M.; Lin, L.; Jin, Y.; Kang, W.; Wu, S.; Sun, S. Comparison of Machine Learning Models for Brain Age Prediction Using Six Imaging Modalities on Middle-Aged and Older Adults. Sensors 2023, 23, 3622. [Google Scholar] [CrossRef]
- Franke, K.; Gaser, C. Ten Years of BrainAGE as a Neuroimaging Biomarker of Brain Aging: What Insights Have We Gained? Front. Neurol. 2019, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Anatürk, M.; Kaufmann, T.; Cole, J.H.; Suri, S.; Griffanti, L.; Zsoldos, E.; Filippini, N.; Singh-Manoux, A.; Kivimäki, M.; Westlye, L.T.; et al. Prediction of brain age and cognitive age: Quantifying brain and cognitive maintenance in aging. Hum. Brain Mapp. 2021, 42, 1626–1640. [Google Scholar] [CrossRef]
- Smith, S.M.; Vidaurre, D.; Alfaro-Almagro, F.; Nichols, T.E.; Miller, K.L. Estimation of brain age delta from brain imaging. Neuroimage 2019, 200, 528–539. [Google Scholar] [CrossRef]
- Fawns-Ritchie, C.; Deary, I.J. Reliability and validity of the UK Biobank cognitive tests. PLoS ONE 2020, 15, e0231627. [Google Scholar] [CrossRef] [PubMed]
- Amin, V.; Fletcher, J.M.; Sun, Z.; Lu, Q. Higher educational attainment is associated with longer telomeres in midlife: Evidence from sibling comparisons in the UK Biobank. SSM-Popul. Health 2022, 17, 101018. [Google Scholar] [CrossRef]
- Xu, X.; Lin, L.; Wu, S.; Sun, S. Exploring Successful Cognitive Aging: Insights Regarding Brain Structure, Function, and Demographics. Brain Sci. 2023, 13, 1651. [Google Scholar] [CrossRef] [PubMed]
- Franke, K.; Gaser, C.; Initiative, F.S.D.N. Longitudinal Changes in Individual BrainAGE in Healthy Aging, Mild Cognitive Impairment, and Alzheimer’s Disease. Geriatr. Psychiatry 2012, 25, 235–245. [Google Scholar] [CrossRef]
- Ziegler, G.; Ridgway, G.R.; Dahnke, R.; Gaser, C. Individualized Gaussian process-based prediction and detection of local and global gray matter abnormalities in elderly subjects. Neuroimage 2014, 97, 333–348. [Google Scholar] [CrossRef]
- Gaser, C.; Franke, K.; Klöppel, S.; Koutsouleris, N.; Sauer, H. BrainAGE in Mild Cognitive Impaired Patients: Predicting the Conversion to Alzheimer’s Disease. PLoS ONE 2013, 8, e67346. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.H.; Leech, R.; Sharp, D.J. Prediction of brain age suggests accelerated atrophy after traumatic brain injury. Ann. Neurol. 2015, 77, 571–581. [Google Scholar] [CrossRef]
- Cole, J.H.; Underwood, J.; Caan, M.W.; De Francesco, D.; van Zoest, R.A.; Leech, R.; Wit, F.W.; Portegies, P.; Geurtsen, G.J.; Schmand, B.A.; et al. Increased brain-predicted aging in treated HIV disease. Neurology 2017, 88, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Almeida, Y.; Fillingim, R.B.; Riley, J.L., 3rd; Woods, A.J.; Porges, E.; Cohen, R.; Cole, J. Chronic pain is associated with a brain aging biomarker in community-dwelling older adults. Pain 2019, 160, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Franke, K.; Gaser, C.; Manor, B.; Novak, V. Advanced BrainAGE in older adults with type 2 diabetes mellitus. Front. Aging Neurosci. 2013, 5, 90. [Google Scholar] [CrossRef]
- Cole, J.H.; Ritchie, S.J.; Bastin, M.E.; Valdés Hernández, M.C.; Muñoz Maniega, S.; Royle, N.; Corley, J.; Pattie, A.; Harris, S.E.; Zhang, Q.; et al. Brain age predicts mortality. Mol. Psychiatry 2018, 23, 1385–1392. [Google Scholar] [CrossRef]
- Steffener, J.; Habeck, C.; O’Shea, D.; Razlighi, Q.; Bherer, L.; Stern, Y. Differences between chronological and brain age are related to education and self-reported physical activity. Neurobiol. Aging 2016, 40, 138–144. [Google Scholar] [CrossRef]
- Stern, Y. What is cognitive reserve? Theory and research application of the reserve concept. J. Int. Neuropsychol. Soc. 2002, 8, 448–460. [Google Scholar] [CrossRef]
- Corbo, I.; Marselli, G.; Di Ciero, V.; Casagrande, M. The Protective Role of Cognitive Reserve in Mild Cognitive Impairment: A Systematic Review. J. Clin. Med. 2023, 12, 1759. [Google Scholar] [CrossRef] [PubMed]
- Panico, F.; Sagliano, L.; Magliacano, A.; Santangelo, G.; Trojano, L. The relationship between cognitive reserve and cognition in healthy adults: A systematic review. Curr. Psychol. 2022, 42, 24751–24763. [Google Scholar] [CrossRef]
- Nogueira, J.; Gerardo, B.; Santana, I.; Simões, M.R.; Freitas, S. The Assessment of Cognitive Reserve: A Systematic Review of the Most Used Quantitative Measurement Methods of Cognitive Reserve for Aging. Front. Psychol. 2022, 13, 847186. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Lin, L.; Xiong, M.; Sun, S.; Wu, S.C. Moderating effects of cognitive reserve on the relationship between brain structure and cognitive abilities in middle-aged and older adults. Neurobiol. Aging 2023, 128, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Jin, Y.; Xiong, M.; Wu, S.; Sun, S. The Protective Power of Cognitive Reserve: Examining White Matter Integrity and Cognitive Function in the Aging Brain for Sustainable Cognitive Health. Sustainability 2023, 15, 11336. [Google Scholar] [CrossRef]
- Stern, Y.; Barnes, C.A.; Grady, C.; Jones, R.N.; Raz, N. Brain reserve, cognitive reserve, compensation, and maintenance: Operationalization, validity, and mechanisms of cognitive resilience. Neurobiol. Aging 2019, 83, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Stern, Y.; Habeck, C.; Moeller, J.; Scarmeas, N.; Anderson, K.E.; Hilton, H.J.; Flynn, J.; Sackeim, H.; van Heertum, R. Brain networks associated with cognitive reserve in healthy young and old adults. Cereb. Cortex 2005, 15, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Wu, F.; Kong, L.; Tang, Y.; Zhou, Q.; Chang, M.; Zhou, Y.; Jiang, X.; Li, S.; Wang, F. Disrupted Structural and Functional Connectivity in Prefrontal-Hippocampus Circuitry in First-Episode Medication-Naïve Adolescent Depression. PLoS ONE 2016, 11, e0148345. [Google Scholar] [CrossRef]
- Toccaceli, V.; Brescianini, S.; Fagnani, C.; Gigantesco, A.; D’Abramo, F.; Stazi, M.A. What Potential Donors in Research Biobanking Want to Know: A Large Population Study of the Italian Twin Registry. Biopreserv. Biobank. 2016, 14, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A. The Need for Research Infrastructures: A Narrative Review of Large-Scale Research Infrastructures in Biobanking. Biopreserv. Biobank. 2017, 15, 375–383. [Google Scholar] [CrossRef] [PubMed]

| Testing | Description | Cognitive Domain | UKB ID |
|---|---|---|---|
| Pairs matching | Number of incorrect matches made in round | Visual declarative memory | 399 |
| Numeric memory | Maximum number of digits remembered correctly | Working memory | 4282 |
| Fluid intelligence | Fluid intelligence score assessment | Verbal and numerical reasoning | 20016 |
| Paired associate learning | Number of correctly associated word pairs | Verbal declarative memory | 20197 |
| Matrix pattern completion | Number of correctly solved puzzles | Non-verbal reasoning | 6373 |
| Reaction time | Mean time taken to correctly identify matches | Processing speed | 20023 |
| Symbol digit substitution | Number of correct symbol digit matches made | Processing speed | 23324 |
| Tower rearranging | Number of correctly solved puzzles | Executive function | 21004 |
| Trail-making | Duration to complete alphanumeric path | Executive function | 6350 |
| Characteristics | RBA Group | SubGroup 1 | SubGroup 2 | SubGroup 3 | p-Values |
|---|---|---|---|---|---|
| n | 3203 | 783 | 561 | 605 | |
| Age (years) | 64.62 | 61.57 | 66.78 | 63.40 | <0.0001 a,b,c |
| Education (years) | 16.17 | 15.46 | 15.54 | 15.75 | 0.527 |
| Women, n (%) | 1884 (58.8%) | 326 (41.6%) | 259 (46.2%) | 302 (49.9%) | 0.008 c |
| Cognitive Function Test | UKB ID | RBA Group | SubGroup 1 | SubGroup 2 | SubGroup 3 | p-Values |
|---|---|---|---|---|---|---|
| Pairs matching | 399 | 3.577 | 3.664 | 3.814 | 3.540 | 0.307 |
| Numeric memory | 4282 | 6.819 | 6.554 | 6.452 | 6.688 | 0.089 |
| Fluid intelligence | 20016 | 6.820 | 6.307 | 6.435 | 6.927 | <0.001 b,c |
| Paired associate learning | 20197 | 7.234 | 6.670 | 6.445 | 6.854 | 0.097 |
| Matrix pattern completion | 6373 | 8.227 | 7.756 | 7.745 | 8.088 | 0.036 b,c |
| Reaction time | 20023 | 2.764 | 2.764 | 2.784 | 2.769 | <0.001 a,c |
| Symbol digit substitution | 23324 | 19.634 | 18.654 | 17.633 | 18.832 | 0.003 a,c |
| Tower rearranging | 21004 | 10.041 | 9.807 | 9.580 | 9.958 | 0.246 |
| Trail-making | 6350 | 2.711 | 2.733 | 2.765 | 2.718 | <0.001 a,c |
| Group | sMRI | rsfMRI | dMRI |
|---|---|---|---|
| SubGroup 1 | 6.55 ± 4.51 | 11.51 ± 8.07 | 6.19 ± 4.55 |
| SubGroup 2 | 7.85 ± 5.94 | 10.73 ± 7.79 | 7.19 ± 5.52 |
| SubGroup 3 | 5.59 ± 4.39 | 11.41 ± 8.51 | 6.40 ± 4.64 |
| RBA group | −6.19 ± 4.56 | −10.85 ± 8.14 | −5.91 ± 4.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Lin, L.; Sun, S.; Wu, S. Elucidating Multimodal Imaging Patterns in Accelerated Brain Aging: Heterogeneity through a Discriminant Analysis Approach Using the UK Biobank Dataset. Bioengineering 2024, 11, 124. https://doi.org/10.3390/bioengineering11020124
Liu L, Lin L, Sun S, Wu S. Elucidating Multimodal Imaging Patterns in Accelerated Brain Aging: Heterogeneity through a Discriminant Analysis Approach Using the UK Biobank Dataset. Bioengineering. 2024; 11(2):124. https://doi.org/10.3390/bioengineering11020124
Chicago/Turabian StyleLiu, Lingyu, Lan Lin, Shen Sun, and Shuicai Wu. 2024. "Elucidating Multimodal Imaging Patterns in Accelerated Brain Aging: Heterogeneity through a Discriminant Analysis Approach Using the UK Biobank Dataset" Bioengineering 11, no. 2: 124. https://doi.org/10.3390/bioengineering11020124








