Development of a 3D Vascular Network Visualization Platform for One-Dimensional Hemodynamic Simulation
Abstract
1. Introduction
2. Requirements
- Medical Image Input/Output and Processing: This involves the processing of medical images, primarily focusing on segmentation to extract vascular regions from the image.
- Centerline Extraction: The platform should be able to obtain centerlines from extracted vascular regions, encompassing two key aspects: accurately capturing the vasculature’s topological structure and computing geometric parameters of the vessels, such as radius and length.
- Centerline Denoising: With the extracted centerlines, the platform should be able to remove noise inherent in the imaging data, generating smooth numerical data for simulation.
- Simulation Results Integration: This includes reading the simulation outputs and mapping these results onto the corresponding centerlines, ensuring a coherent relationship between the simulated data and the anatomical structures.
- Simulation Results Visualization: This involves generating surface representations of vessel lumens and displaying the simulation results on these surfaces, enabling clear and detailed visualization of hemodynamic parameters.
3. 1D–0D Computational Model
4. Architecture
4.1. Medical Image Processing
4.2. Arterial Geometry Modeling
4.3. Simulation
- Flow Rate : This is the velocity of blood through the vasculature over time, quantifying the volumetric flow rate along the length of the vessel, where s represents the spatial coordinate along the vessel’s length, and t denotes time.
- Blood Pressure : This represents the pressure exerted by the blood against the vascular walls over time, providing insight into the forces that the heart must overcome to circulate blood and the potential stresses on vessel walls.
- Cross-sectional Area : This is the area of the blood vessel cross-section at different spatial locations and time points, reflecting changes in vessel diameter due to pressure dynamics and the mechanical properties of the vessel wall.
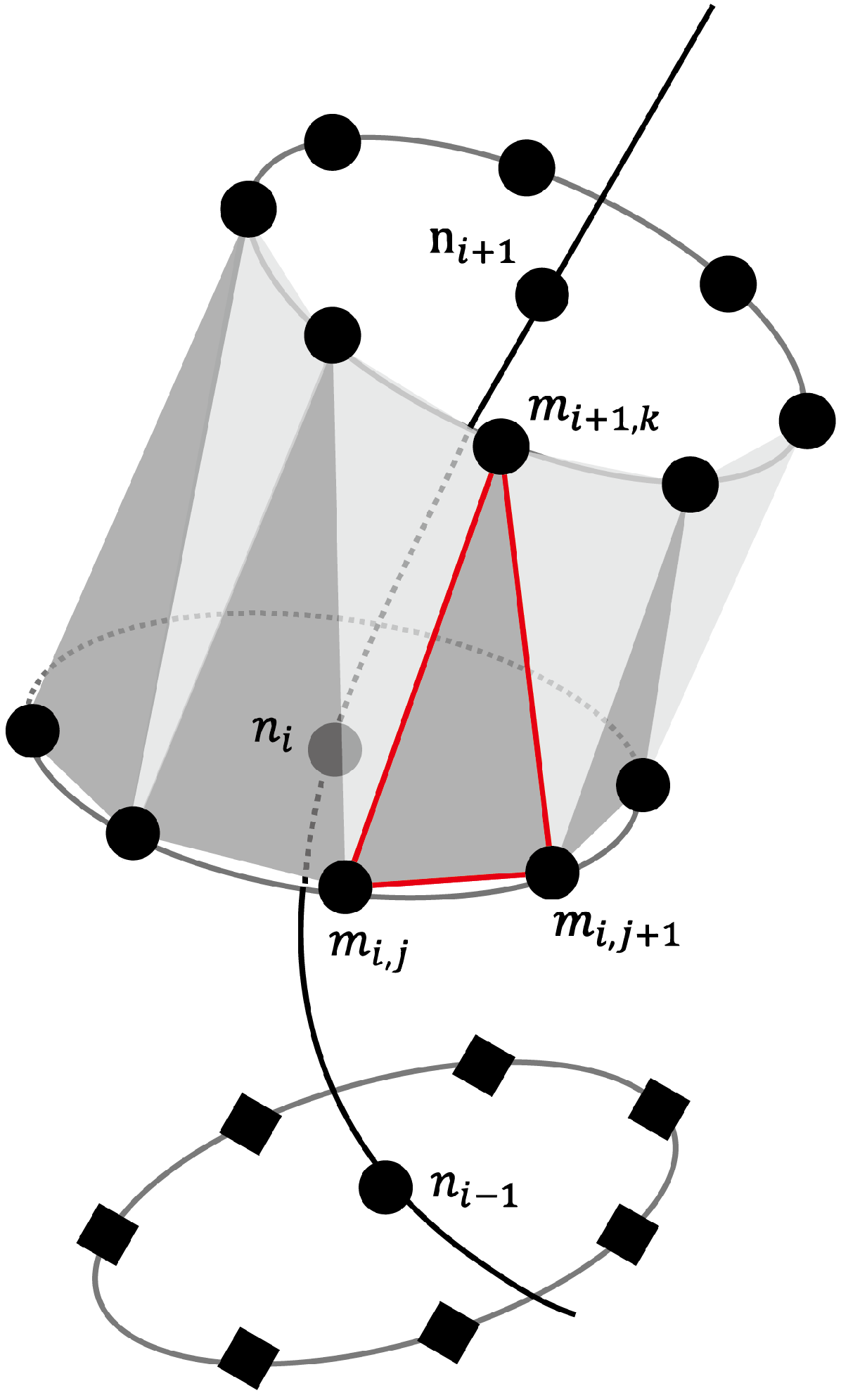
4.4. 3D Visualization
5. Results
5.1. Case CS: Post-Surgical Flow Rate Prediction
5.2. Case VRCA: Vascular Diameter Remodeling
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hughes, T.J.; Lubliner, J. On the one-dimensional theory of blood flow in the larger vessels. Math. Biosci. 1973, 18, 161–170. [Google Scholar] [CrossRef]
- Westerhof, N.; Bosman, F.; De Vries, C.J.; Noordergraaf, A. Analog studies of the human systemic arterial tree. J. Biomech. 1969, 2, 121–143. [Google Scholar] [CrossRef]
- Alastruey, J.P.K.H.; Parker, K.H.; Peiró, J.; Byrd, S.M.; Sherwin, S.J. Modelling the circle of Willis to assess the effects of anatomical variations and occlusions on cerebral flows. J. Biomech. 2007, 40, 1794–1805. [Google Scholar] [CrossRef]
- Avolio, A.P. Multi-branched model of the human arterial system. Med. Biol. Eng. Comput. 1980, 18, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Azer, K.; Peskin, C.S. A one-dimensional model of blood flow in arteries with friction and convection based on the Womersley velocity profile. Cardiovasc. Eng. 2007, 7, 51–73. [Google Scholar] [CrossRef] [PubMed]
- Fitchett, D.H. LV-arterial coupling: Interactive model to predict effect of wave reflections on LV energetics. Am. J. Physiol. Heart Circ. Physiol. 1991, 261, H1026–H1033. [Google Scholar] [CrossRef]
- Formaggia, L.; Lamponi, D.; Tuveri, M.; Veneziani, A. Numerical modeling of 1D arterial networks coupled with a lumped parameters description of the heart. Comput. Methods Biomech. Biomed. Eng. 2006, 9, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Olufsen, M.S.; Peskin, C.S.; Kim, W.Y.; Pedersen, E.M.; Nadim, A.; Larsen, J. Numerical simulation and experimental validation of blood flow in arteries with structured-tree outflow conditions. Ann. Biomed. Eng. 2000, 28, 1281–1299. [Google Scholar] [CrossRef] [PubMed]
- Sherwin, S.; Franke, V.; Peiró, J.; Parker, K. One-dimensional modelling of a vascular network in space-time variables. J. Eng. Math. 2003, 47, 217–250. [Google Scholar] [CrossRef]
- Stergiopulos, N.; Segers, P.; Westerhof, N. Use of pulse pressure method for estimating total arterial compliance in vivo. Am. J. Physiol. Heart Circ. Physiol. 1999, 276, H424–H428. [Google Scholar] [CrossRef]
- Stergiopulos, N.; Young, D.; Rogge, T. Computer simulation of arterial flow with applications to arterial and aortic stenoses. J. Biomech. 1992, 25, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Blanco, P.J.; Watanabe, S.M.; Passos, M.A.R.F.; Lemos, P.A.; Feijóo, R.A. An anatomically detailed arterial network model for one-dimensional computational hemodynamics. IEEE Trans. Biomed. Eng. 2014, 62, 736–753. [Google Scholar] [CrossRef] [PubMed]
- Larrabide, I.; Blanco, P.J.; Urquiza, S.; Dari, E.; Vénere, M.J.; e Silva, N.d.S.; Feijóo, R.A. HeMoLab—Hemodynamics Modelling Laboratory: An application for modelling the human cardiovascular system. Comput. Biol. Med. 2012, 42, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Young, D.F.; Tsai, F.Y. Flow characteristics in models of arterial stenoses—II. Unsteady flow. J. Biomech. 1973, 6, 547–559. [Google Scholar] [CrossRef]
- Liang, F.; Fukasaku, K.; Liu, H.; Takagi, S. A computational model study of the influence of the anatomy of the circle of Willis on cerebral hyperperfusion following carotid artery surgery. Biomed. Eng. Online 2011, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Takagi, S.; Himeno, R.; Liu, H. Multi-scale modeling of the human cardiovascular system with applications to aortic valvular and arterial stenoses. Med. Biol. Eng. Comput. 2009, 47, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fujiwara, N.; Kobayashi, M.; Yamada, S.; Liang, F.; Takagi, S.; Oshima, M. Development of a numerical method for patient-specific cerebral circulation using 1d–0d simulation of the entire cardiovascular system with SPECT data. Ann. Biomed. Eng. 2016, 44, 2351–2363. [Google Scholar] [CrossRef] [PubMed]
- Yuhn, C.; Oshima, M. Development of a Numerical Method for Assessment of Cerebrovascular Reserve Using 1D-0D Hemodynamic Simulation with Cerebral Autoregulation Model. In Proceedings of the SB3C2017 Summer Biomechanics, Bioengineering, and Biotransport Conference, Tucson, AZ, USA, 21–24 June 2017. [Google Scholar]
- Yuhn, C.; Hoshina, K.; Miyahara, K.; Oshima, M. Computational simulation of flow-induced arterial remodeling of the pancreaticoduodenal arcade associated with celiac artery stenosis. J. Biomech. 2019, 92, 146–154. [Google Scholar] [CrossRef]
- Yuhn, C.; Oshima, M.; Chen, Y.; Hayakawa, M.; Yamada, S. Uncertainty quantification in cerebral circulation simulations focusing on the collateral flow: Surrogate model approach with machine learning. PLoS Comput. Biol. 2022, 18, e1009996. [Google Scholar] [CrossRef]
- Grinberg, L.; Cheever, E.; Anor, T.; Madsen, J.R.; Karniadakis, G.E. Modeling blood flow circulation in intracranial arterial networks: A comparative 3D/1D simulation study. Ann. Biomed. Eng. 2011, 39, 297–309. [Google Scholar] [CrossRef]
- Moore, S.M.; Moorhead, K.T.; Chase, J.G.; David, T.; Fink, J. One-dimensional and three-dimensional models of cerebrovascular flow. J. Biomech. Eng. 2005, 127, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Alastruey, J.; Figueroa, C.A. A systematic comparison between 1 and D and 3-D hemodynamics in compliant arterial models. Int. J. Numer. Methods Biomed. Eng. 2014, 30, 204–231. [Google Scholar] [CrossRef] [PubMed]
- Olufsen, M.S. Modeling the Arterial System with Reference to an Anestesia Simulator. Ph.D. Thesis, Institut for Studiet af Matematik og Fysik, Roskilde Universitetscenter, Roskilde, Denmark, 1998. [Google Scholar]
- Kobayashi, M.; Hoshina, K.; Yamamoto, S.; Nemoto, Y.; Akai, T.; Shigematsu, K.; Watanabe, T.; Ohshima, M. Development of an image-based modeling system to investigate evolutional geometric changes of a stent graft in an abdominal aortic aneurysm. Circ. J. 2015, 79, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Kobayashi, M.; Oshima, M. Development of an interface for computer-assisted surgery using 1D-0D blood flow simulation. In Proceedings of the CMBE 2019—6th International Conference on Computational and Mathematical Biomedical Engineering, Sendai, Japan, 10–12 June 2019; Volume 1, p. 473. [Google Scholar]
- Kobayashi, M.; Hoshina, K.; Nemoto, Y.; Takagi, S.; Shojima, M.; Hayakawa, M.; Yamada, S.; Oshima, M. A penalized spline fitting method to optimize geometric parameters of arterial centerlines extracted from medical images. Comput. Med. Imaging Graph. 2020, 84, 101746. [Google Scholar] [CrossRef] [PubMed]
- Sakorafas, G.H.; Sarr, M.G.; Peros, G. Celiac artery stenosis: An underappreciated and unpleasant surprise in patients undergoing pancreaticoduodenectomy. J. Am. Coll. Surg. 2008, 206, 349–356. [Google Scholar] [CrossRef]
- Humphrey, J. Vascular adaptation and mechanical homeostasis at tissue, cellular, and sub-cellular levels. Cell Biochem. Biophys. 2008, 50, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, A.; Togawa, T. Adaptive regulation of wall shear stress to flow change in the canine carotid artery. Am. J. Physiol. Heart Circ. Physiol. 1980, 239, H14–H21. [Google Scholar] [CrossRef]
- Langille, B.L.; O’Donnell, F. Reductions in arterial diameter produced by chronic decreases in blood flow are endothelium-dependent. Science 1986, 231, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Zarins, C.K.; Zatina, M.A.; Giddens, D.P.; Ku, D.N.; Glagov, S. Shear stress regulation of artery lumen diameter in experimental atherogenesis. J. Vasc. Surg. 1987, 5, 413–420. [Google Scholar] [CrossRef]
- Ii, S.; Kitade, H.; Ishida, S.; Imai, Y.; Watanabe, Y.; Wada, S. Multiscale modeling of human cerebrovasculature: A hybrid approach using image-based geometry and a mathematical algorithm. PLoS Comput. Biol. 2020, 16, e1007943. [Google Scholar] [CrossRef]
- Reymond, P.; Merenda, F.; Perren, F.; Rufenacht, D.; Stergiopulos, N. Validation of a one-dimensional model of the systemic arterial tree. Am. J. Physiol. Heart Circ. Physiol. 2009, 297, H208–H222. [Google Scholar] [CrossRef] [PubMed]





| Aspect | 3D Models | ROMs | Machine Learning Methods |
|---|---|---|---|
| Complexity | High; detailed spatial resolution | Lower; simplifies geometry and dynamics | Varies; algorithm and data-dependent |
| Computational Cost | High; requires more computational resources | Lower; more efficient due to simplifications | High for training; data acquisition costly |
| Result Fidelity | High; captures detailed flow dynamics | Lower due to simplifications | Varies; dependent on training quality |
| Flexibility | Less flexible; hard to adapt to new cases | More flexible; easier to adapt | Adaptable, but may require retraining |
| Data Requirement | Geometry and flow specifics | Simplified vessel and flow characteristics | Extensive data for training |
| Application | Detailed analysis and research | Quick assessments, preliminary studies | Predictive modeling, pattern recognition |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Kobayashi, M.; Yuhn, C.; Oshima, M. Development of a 3D Vascular Network Visualization Platform for One-Dimensional Hemodynamic Simulation. Bioengineering 2024, 11, 313. https://doi.org/10.3390/bioengineering11040313
Chen Y, Kobayashi M, Yuhn C, Oshima M. Development of a 3D Vascular Network Visualization Platform for One-Dimensional Hemodynamic Simulation. Bioengineering. 2024; 11(4):313. https://doi.org/10.3390/bioengineering11040313
Chicago/Turabian StyleChen, Yan, Masaharu Kobayashi, Changyoung Yuhn, and Marie Oshima. 2024. "Development of a 3D Vascular Network Visualization Platform for One-Dimensional Hemodynamic Simulation" Bioengineering 11, no. 4: 313. https://doi.org/10.3390/bioengineering11040313
APA StyleChen, Y., Kobayashi, M., Yuhn, C., & Oshima, M. (2024). Development of a 3D Vascular Network Visualization Platform for One-Dimensional Hemodynamic Simulation. Bioengineering, 11(4), 313. https://doi.org/10.3390/bioengineering11040313






