Functional and Molecular Analysis of Human Osteoarthritic Chondrocytes Treated with Bone Marrow-Derived MSC-EVs
Abstract
:1. Introduction
2. Materials and Methods
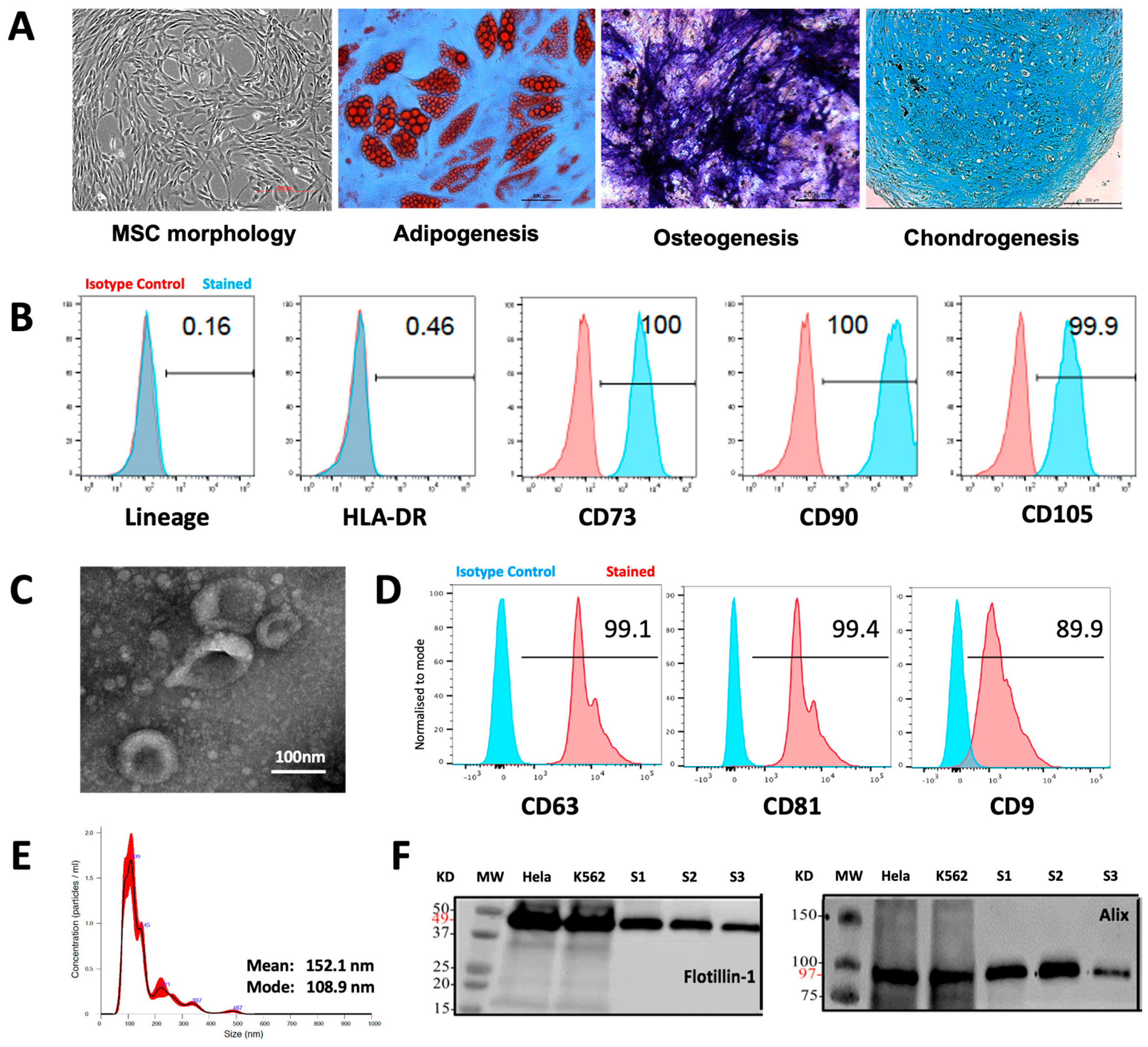
2.1. MSC Generation and Characterization
2.2. MSC-EV Isolation
2.3. MSC-EV Characterization
2.4. Chondrocyte Isolation and Culture
2.5. MSC-EV Uptake by Chondrocytes
2.6. Viability Assay
2.7. Scratch and Migration Assays
2.8. Secreted Protein, Cytokine, and Chemokine Assessment
2.9. Spheroid Culture of Chondrocytes
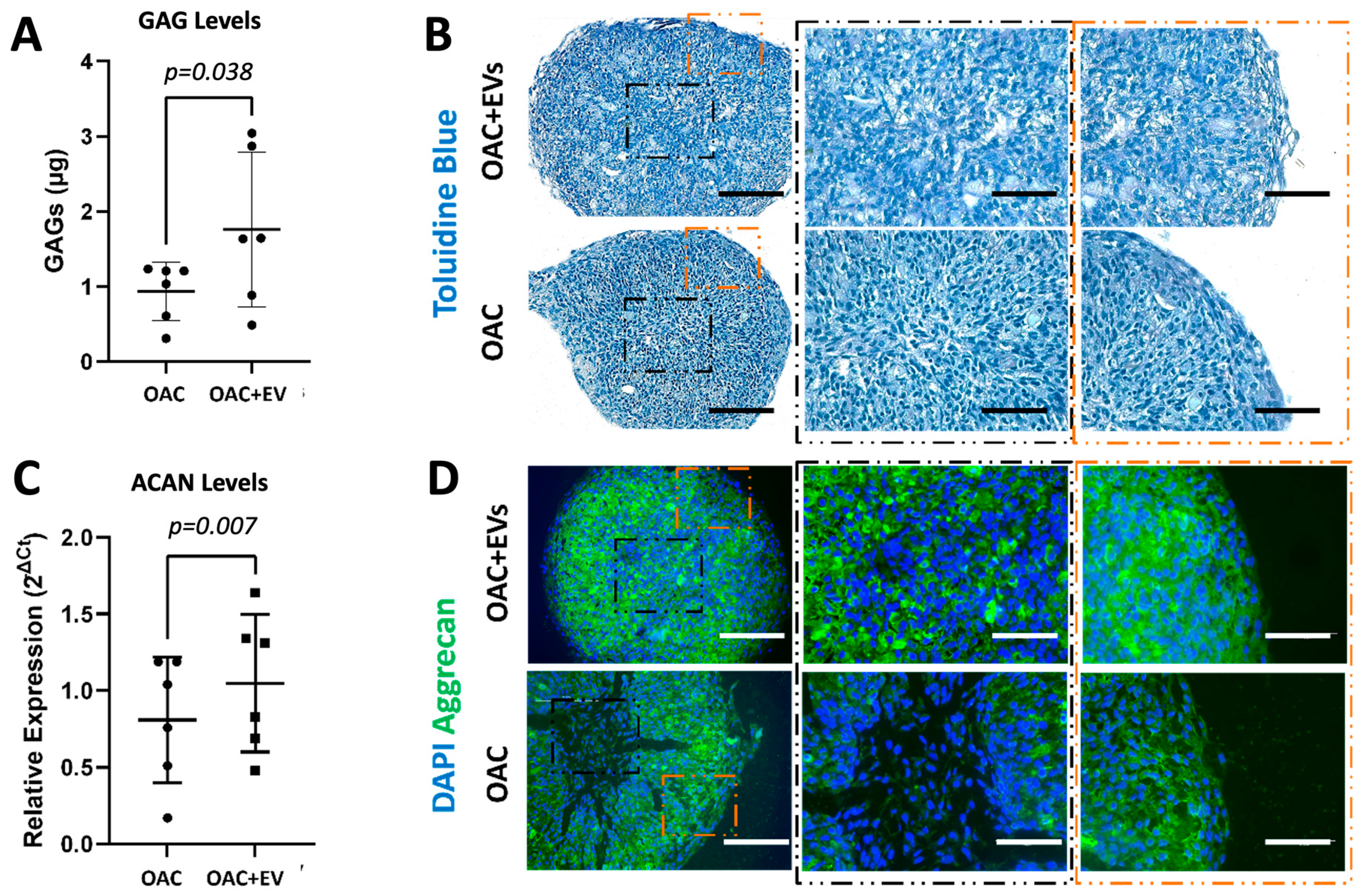
2.10. Sulphated GAG Quantification
2.11. Histology and Immunohistochemistry
2.12. RNA Isolation and Quantification
2.13. Quantitative Real-Time PCR
2.14. NanoString
2.15. RNA-Seq
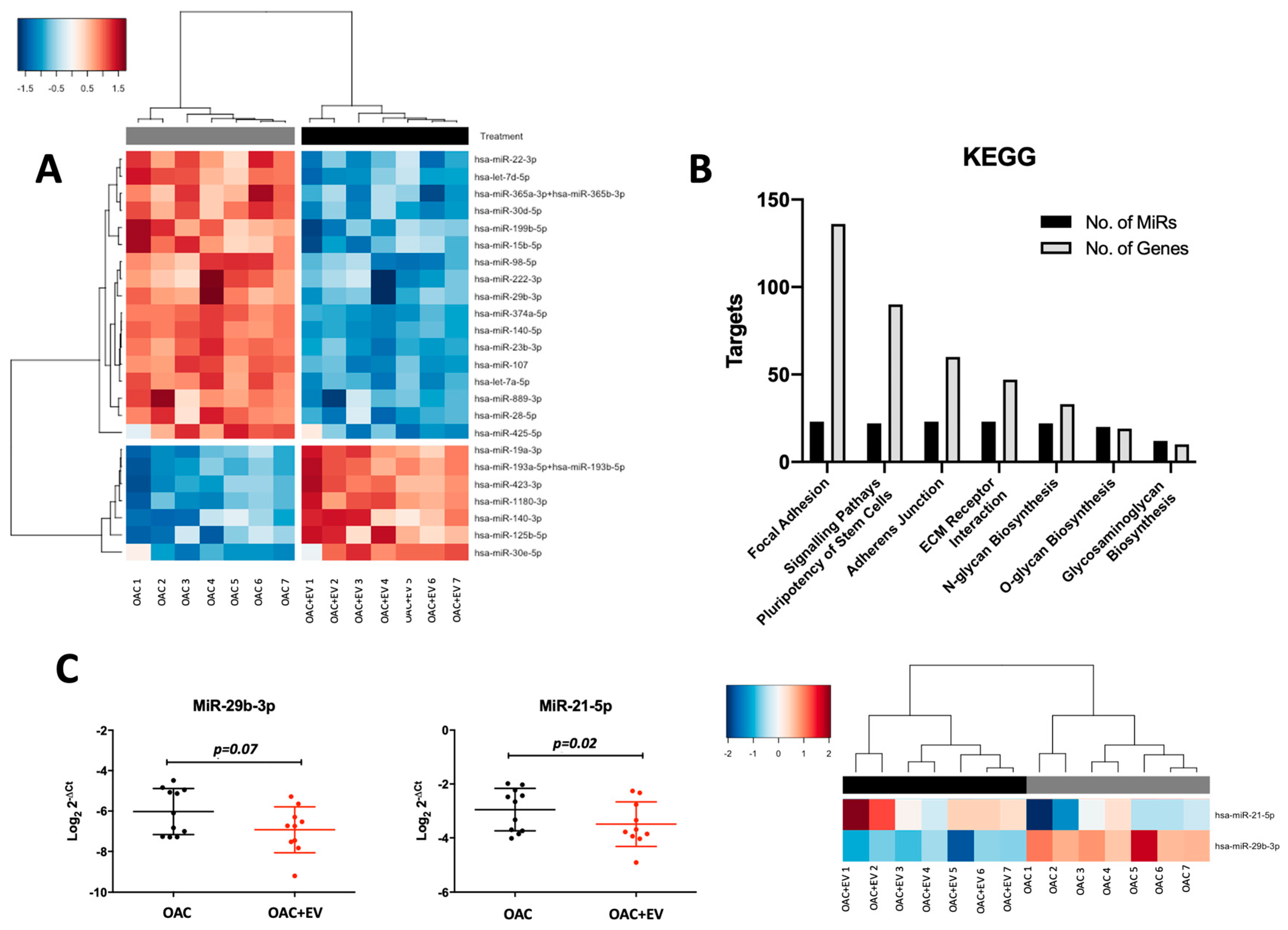
2.16. Pathway and Gene Enrichment Analysis
2.17. Statistical Analysis
3. Results
3.1. Characteristics of MSCs and MSC-EVs
3.2. MSC-EV-Treated OACs Display Increased Viability, Proliferation, and Migration
3.3. MSC-EV Switch OAC Cytokine, Catabolic Protein, Gene, and MicroRNA Expression to Favor Cartilage Repair
3.4. MSC-EV-Treated OACs Display a Pattern of Gene Expression and sGAG Production in Favor of Chondrogenesis in a 3D Spheroid Model
3.5. MSC-EVs Reverse the Pathological Impact of IL-1β on OAC Chondrogenic Gene Expression and ECM Component Production
3.6. MSC-EVs Change the Global Gene Expression Profiles of Treated OACs
3.7. MSC-EVs Affect the MicroRNA Expression Profiles of Treated OACs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- ter Huurne, M.; Schelbergen, R.; Blattes, R.; Blom, A.; de Munter, W.; Grevers, L.C.; Jeanson, J.; Noël, D.; Casteilla, L.; Jorgensen, C.; et al. Antiinflammatory and chondroprotective effects of intraarticular injection of adipose-derived stem cells in experimental osteoarthritis. Arthritis Rheum. 2012, 64, 3604–3613. [Google Scholar] [CrossRef] [PubMed]
- Quinn, C.; Flake, A.W. In vivo Differentiation Potential of Mesenchymal Stem Cells: Prenatal and Postnatal Model Systems. Transfus. Med. Hemother. 2008, 35, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Keating, A. Mesenchymal stromal cells: New directions. Cell Stem Cell 2012, 10, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chuah, S.J.; Lai, R.C.; Hui, J.H.P.; Lim, S.K.; Toh, W.S. MSC exosomes mediate cartilage repair by enhancing proliferation, attenuating apoptosis and modulating immune reactivity. Biomaterials 2018, 156, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Vonk, L.A.; van Dooremalen, S.F.J.; Liv, N.; Klumperman, J.; Coffer, P.J.; Saris, D.B.F.; Lorenowicz, M.J. Mesenchymal Stromal/stem Cell-derived Extracellular Vesicles Promote Human Cartilage Regeneration In Vitro. Theranostics 2018, 8, 906–920. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chu, W.C.; Lai, R.C.; Lim, S.K.; Hui, J.H.; Toh, W.S. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthr. Cartil. 2016, 24, 2135–2140. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Xiao, K.; Xiang, S.; Li, Z.; Weng, X. Emerging Role of Exosomes in the Joint Diseases. Cell. Physiol. Biochem. 2018, 47, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.-C.; Yuan, T.; Zhang, Y.-L.; Yin, W.-J.; Guo, S.-C.; Zhang, C.-Q. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics 2017, 7, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Zhang, Z.; Hu, S.; Zhang, Z.; Chang, Z.; Huang, Z.; Liao, W.; Kang, Y. Exosomes derived from miR-92a-3p-overexpressing human mesenchymal stem cells enhance chondrogenesis and suppress cartilage degradation via targeting WNT5A. Stem Cell Res. Ther. 2018, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Warmink, K.; Rios, J.L.; Varderidou-Minasian, S.; Torres-Torrillas, M.; van Valkengoed, D.R.; Versteeg, S.; Eijkelkamp, N.; Weinans, H.; Korthagen, N.M.; Lorenowicz, M.J. Mesenchymal stem/stromal cells-derived extracellular vesicles as a potentially more beneficial therapeutic strategy than MSC-based treatment in a mild metabolic osteoarthritis model. Stem Cell Res. Ther. 2023, 14, 137. [Google Scholar] [CrossRef]
- Chen, P.; Zheng, L.; Wang, Y.; Tao, M.; Xie, Z.; Xia, C.; Gu, C.; Chen, J.; Qiu, P.; Mei, S.; et al. Desktop-stereolithography 3D printing of a radially oriented extracellular matrix/mesenchymal stem cell exosome bioink for osteochondral defect regeneration. Theranostics 2019, 9, 2439–2459. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zou, R.; Wang, Z.; Wen, C.; Zhang, F.; Lin, F. Exosomal KLF3-AS1 from hMSCs promoted cartilage repair and chondrocyte proliferation in osteoarthritis. Biochem. J. 2018, 475, 3629–3638. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, S.; Ruiz, M.; Toupet, K.; Jorgensen, C.; Noël, D. Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Sci. Rep. 2017, 7, 16214. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, D.; Liu, Z.; Zhou, F.; Dai, J.; Wu, B.; Zhou, J.; Heng, B.C.; Zou, X.H.; Ouyang, H.; et al. Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res. Ther. 2017, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Tofiño-Vian, M.; Guillén, M.I.; Pérez del Caz, M.D.; Silvestre, A.; Alcaraz, M.J. Microvesicles from Human Adipose Tissue-Derived Mesenchymal Stem Cells as a New Protective Strategy in Osteoarthritic Chondrocytes. Cell. Physiol. Biochem. 2018, 47, 11–25. [Google Scholar] [CrossRef]
- Reis, M.; Mavin, E.; Nicholson, L.; Green, K.; Dickinson, A.M.; Wang, X.N. Mesenchymal Stromal Cell-Derived Extracellular Vesicles Attenuate Dendritic Cell Maturation and Function. Front. Immunol. 2018, 9, 2538. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. In Current Protocols in Cell Biology; Wiley: Hoboken, NJ, USA, 2006; Chapter 3, Unit 3.22. [Google Scholar] [CrossRef]
- Young, D.A.; Lakey, R.L.; Pennington, C.J.; Jones, D.; Kevorkian, L.; Edwards, D.R.; Cawston, T.E.; Clark, I.M. Histone deacetylase inhibitors modulate metalloproteinase gene expression in chondrocytes and block cartilage resorption. Arthritis Res. Ther. 2005, 7, R503. [Google Scholar] [CrossRef] [PubMed]
- Qing, C.; Wei-ding, C.; Wei-min, F. Co-culture of chondrocytes and bone marrow mesenchymal stem cells in vitro enhances the expression of cartilaginous extracellular matrix components. Braz. J. Med. Biol. Res. 2011, 44, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Scalzone, A.; Cerqueni, G.; Wang, X.N.; Dalgarno, K.; Mattioli-Belmonte, M.; Ferreira-Duarte, A.M.; Gentile, P. A cytokine-induced spheroid-based in vitro model for studying osteoarthritis pathogenesis. Front. Bioeng. Biotechnol. 2023, 11, 1167623. [Google Scholar] [CrossRef] [PubMed]
- Sanjurjo-Rodríguez, C.; Crossland, R.E.; Reis, M.; Pandit, H.; Wang, X.-N.; Jones, E. Characterization and miRNA Profiling of Extracellular Vesicles from Human Osteoarthritic Subchondral Bone Multipotential Stromal Cells (MSCs). Stem Cells Int. 2021, 2021, 7232773. [Google Scholar] [CrossRef] [PubMed]
- Crossland, R.E.; Albiero, A.; Sanjurjo-Rodríguez, C.; Reis, M.; Resteu, A.; Anderson, A.E.; Dickinson, A.M.; Pratt, A.G.; Birch, M.; McCaskie, A.W.; et al. MicroRNA profiling of low concentration extracellular vesicle RNA utilizing NanoString nCounter technology. J. Extracell. Biol. 2023, 2, e72. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T. svaseq: Removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014, 42, e161. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]
- Soul, J.; Hardingham, T.E.; Boot-Handford, R.P.; Schwartz, J.-M. SkeletalVis: An exploration and meta-analysis data portal of cross-species skeletal transcriptomics data. Bioinformatics 2019, 35, 2283–2290. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.B.; Shon, O.J.; Seo, M.S.; Choi, Y.; Park, W.T.; Lee, G.W. Mesenchymal Stem Cell-Derived Exosomes and Their Therapeutic Potential for Osteoarthritis. Biology 2021, 10, 285. [Google Scholar] [CrossRef] [PubMed]
- Velot, É.; Madry, H.; Venkatesan, J.K.; Bianchi, A.; Cucchiarini, M. Is Extracellular Vesicle-Based Therapy the Next Answer for Cartilage Regeneration? Front. Bioeng. Biotechnol. 2021, 9, 645039. [Google Scholar] [CrossRef] [PubMed]
- Samvelyan, H.J.; Hughes, D.; Stevens, C.; Staines, K.A. Models of Osteoarthritis: Relevance and New Insights. Calcif. Tissue Int. 2021, 109, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Hu, J.; Zhao, J.; Liu, J.; Ouyang, W.; Yang, C.; Gong, N.; Du, L.; Khanal, A.; Chen, L. Side-by-side comparison of the biological characteristics of human umbilical cord and adipose tissue-derived mesenchymal stem cells. Biomed. Res. Int. 2013, 2013, 438243. [Google Scholar] [CrossRef] [PubMed]
- Kodama, J.; Wilkinson, K.J.; Otsuru, S. MSC-EV therapy for bone/cartilage diseases. Bone Rep. 2022, 17, 101636. [Google Scholar] [CrossRef]
- He, Y.; Li, Z.; Alexander, P.G.; Ocasio-Nieves, B.D.; Yocum, L.; Lin, H.; Tuan, R.S. Pathogenesis of Osteoarthritis: Risk Factors, Regulatory Pathways in Chondrocytes, and Experimental Models. Biology 2020, 9, 194. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-X.; Zhao, Z.-D.; Wang, Q.; Li, Z.-L.; Huang, Y.; Zhao, S.; Hu, W.; Liang, J.-W.; Li, P.-L.; Wang, H.; et al. Biological potential alterations of migratory chondrogenic progenitor cells during knee osteoarthritic progression. Arthritis Res. Ther. 2020, 22, 62. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.C.; Yeo, R.W.; Lim, S.K. Mesenchymal stem cell exosomes. Semin. Cell Dev. Biol. 2015, 40, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Hopper, N.; Henson, F.; Brooks, R.; Ali, E.; Rushton, N.; Wardale, J. Peripheral blood derived mononuclear cells enhance osteoarthritic human chondrocyte migration. Arthritis Res. Ther. 2015, 17, 199. [Google Scholar] [CrossRef] [PubMed]
- Seol, D.; McCabe, D.J.; Choe, H.; Zheng, H.; Yu, Y.; Jang, K.; Walter, M.W.; Lehman, A.D.; Ding, L.; Buckwalter, J.A.; et al. Chondrogenic progenitor cells respond to cartilage injury. Arthritis Rheum. 2012, 64, 3626–3637. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Y.; Zhao, B.; Niu, X.; Hu, B.; Li, Q.; Zhang, J.; Ding, J.; Chen, Y.; Wang, Y. Comparison of exosomes secreted by induced pluripotent stem cell-derived mesenchymal stem cells and synovial membrane-derived mesenchymal stem cells for the treatment of osteoarthritis. Stem Cell Res. Ther. 2017, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Stöckl, S.; Lukas, C.; Götz, J.; Herrmann, M.; Federlin, M.; Grässel, S. hBMSC-Derived Extracellular Vesicles Attenuate IL-1β-Induced Catabolic Effects on OA-Chondrocytes by Regulating Pro-inflammatory Signaling Pathways. Front. Bioeng. Biotechnol. 2020, 8, 603598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tian, X.; Qu, Z.; Hao, J.; Zhang, W. Hypoxia-Preconditioned Extracellular Vesicles from Mesenchymal Stem Cells Improve Cartilage Repair in Osteoarthritis. Membranes 2022, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Kawata, K.; Koga, H.; Tsuji, K.; Miyatake, K.; Nakagawa, Y.; Yokota, T.; Sekiya, I.; Katagiri, H. Extracellular vesicles derived from mesenchymal stromal cells mediate endogenous cell growth and migration via the CXCL5 and CXCL6/CXCR2 axes and repair menisci. Stem Cell Res. Ther. 2021, 12, 414. [Google Scholar] [CrossRef] [PubMed]
- Monaco, G.; El Haj, A.J.; Alini, M.; Stoddart, M.J. Ex Vivo Systems to Study Chondrogenic Differentiation and Cartilage Integration. J. Funct. Morphol. Kinesiol. 2021, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Dufour, A.; Zucker, S.; Sampson, N.S.; Kuscu, C.; Cao, J. Role of Matrix Metalloproteinase-9 Dimers in Cell Migration: Design of Inhibitory Peptides. J. Biol. Chem. 2010, 285, 35944–35956. [Google Scholar] [CrossRef] [PubMed]
- Molnar, V.; Matišić, V.; Kodvanj, I.; Bjelica, R.; Jeleč, Ž.; Hudetz, D.; Rod, E.; Čukelj, F.; Vrdoljak, T.; Vidović, D.; et al. Cytokines and Chemokines Involved in Osteoarthritis Pathogenesis. Int. J. Mol. Sci. 2021, 22, 9208. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, Y.; Li, Q. Therapeutic mechanisms of ibuprofen, prednisone and betamethasone in osteoarthritis. Mol. Med. Rep. 2017, 15, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Merz, D.; Liu, R.; Johnson, K.; Terkeltaub, R. IL-8/CXCL8 and Growth-Related Oncogene α/CXCL1 Induce Chondrocyte Hypertrophic Differentiation. J. Immunol. 2003, 171, 4406. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; de Andrés, M.C.; Hashimoto, K.; Itoi, E.; Oreffo, R.O. Epigenetic regulation of interleukin-8, an inflammatory chemokine, in osteoarthritis. Osteoarthr. Cartil. 2015, 23, 1946–1954. [Google Scholar] [CrossRef]
- García-Manrique, M.; Calvet, J.; Orellana, C.; Berenguer-Llergo, A.; Garcia-Cirera, S.; Llop, M.; Albiñana-Giménez, N.; Galisteo-Lencastre, C.; Gratacós, J. Synovial fluid but not plasma interleukin-8 is associated with clinical severity and inflammatory markers in knee osteoarthritis women with joint effusion. Sci. Rep. 2021, 11, 5258. [Google Scholar] [CrossRef]
- Ruan, G.; Xu, J.; Wang, K.; Zheng, S.; Wu, J.; Bian, F.; Chang, B.; Zhang, Y.; Meng, T.; Zhu, Z.; et al. Associations between serum IL-8 and knee symptoms, joint structures, and cartilage or bone biomarkers in patients with knee osteoarthritis. Clin. Rheumatol. 2019, 38, 3609–3617. [Google Scholar] [CrossRef]
- Gilbert, S.J.; Blain, E.J.; Mason, D.J. Interferon-gamma modulates articular chondrocyte and osteoblast metabolism through protein kinase R-independent and dependent mechanisms. Biochem. Biophys. Rep. 2022, 32, 101323. [Google Scholar] [CrossRef] [PubMed]
- Woods, J.M.; Amin, M.A.; Katschke, K.J., Jr.; Volin, M.V.; Ruth, J.H.; Connors, M.A.; Woodruff, D.C.; Kurata, H.; Arai, K.; Haines, G.K., 3rd; et al. Interleukin-13 gene therapy reduces inflammation, vascularization, and bony destruction in rat adjuvant-induced arthritis. Hum. Gene Ther. 2002, 13, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Nabbe, K.C.; van Lent, P.L.; Holthuysen, A.E.; Sloëtjes, A.W.; Koch, A.E.; Radstake, T.R.; van den Berg, W.B. Local IL-13 gene transfer prior to immune-complex arthritis inhibits chondrocyte death and matrix-metalloproteinase-mediated cartilage matrix degradation despite enhanced joint inflammation. Arthritis Res. Ther. 2005, 7, R392–R401. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kuang, L.; Chen, C.; Yang, J.; Zeng, W.N.; Li, T.; Chen, H.; Huang, S.; Fu, Z.; Li, J.; et al. miR-100-5p-abundant exosomes derived from infrapatellar fat pad MSCs protect articular cartilage and ameliorate gait abnormalities via inhibition of mTOR in osteoarthritis. Biomaterials 2019, 206, 87–100. [Google Scholar] [CrossRef]
- Zhang, S.; Teo, K.Y.W.; Chuah, S.J.; Lai, R.C.; Lim, S.K.; Toh, W.S. MSC exosomes alleviate temporomandibular joint osteoarthritis by attenuating inflammation and restoring matrix homeostasis. Biomaterials 2019, 200, 35–47. [Google Scholar] [CrossRef]
- Wong, K.L.; Zhang, S.; Wang, M.; Ren, X.; Afizah, H.; Lai, R.C.; Lim, S.K.; Lee, E.H.; Hui, J.H.P.; Toh, W.S. Intra-Articular Injections of Mesenchymal Stem Cell Exosomes and Hyaluronic Acid Improve Structural and Mechanical Properties of Repaired Cartilage in a Rabbit Model. Arthroscopy 2020, 36, 2215–2228.e2212. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Y.; Li, Y.; Niu, X.; Zhao, B.; Wang, Y.; Bao, C.; Xie, Z.; Lin, Q.; Zhu, L. Integration of stem cell-derived exosomes with in situ hydrogel glue as a promising tissue patch for articular cartilage regeneration. Nanoscale 2017, 9, 4430–4438. [Google Scholar] [CrossRef]
- Wang, B.; Liu, W.; Li, J.J.; Chai, S.; Xing, D.; Yu, H.; Zhang, Y.; Yan, W.; Xu, Z.; Zhao, B.; et al. A low dose cell therapy system for treating osteoarthritis: In vivo study and in vitro mechanistic investigations. Bioact. Mater. 2022, 7, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Zhang, L.; Meisgen, F.; Harada, M.; Heilborn, J.; Homey, B.; Grandér, D.; Ståhle, M.; Sonkoly, E.; Pivarcsi, A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 2012, 287, 29899–29908. [Google Scholar] [CrossRef]
- Killock, D. miR-125b—A new target in cartilage destruction? Nat. Rev. Rheumatol. 2013, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, T.; Sakai, T.; Yonezawa, T.; Hiraiwa, H.; Hamada, T.; Nakashima, M.; Ono, Y.; Ishizuka, S.; Nakahara, H.; Lotz, M.K.; et al. MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res. Ther. 2013, 15, R28. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, Z.; Rasheed, N.; Abdulmonem, W.A.; Khan, M.I. MicroRNA-125b-5p regulates IL-1β induced inflammatory genes via targeting TRAF6-mediated MAPKs and NF-κB signaling in human osteoarthritic chondrocytes. Sci. Rep. 2019, 9, 6882. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Tian, B.; Qu, X.; Liu, F.; Tang, T.; Qin, A.; Zhu, Z.; Dai, K. MicroRNAs play a role in chondrogenesis and osteoarthritis (Review). Int. J. Mol. Med. 2014, 34, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Wang, Y. RETRACTED: MicroRNA-19a promotes cell viability and migration of chondrocytes via up-regulating SOX9 through NF-κB pathway. Biomed. Pharmacother. 2018, 98, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, S.; Jin, P.; Shang, T.; Sun, R.; Lu, L.; Guo, K.; Liu, J.; Tong, Y.; Wang, J.; et al. Dual functions of microRNA-17 in maintaining cartilage homeostasis and protection against osteoarthritis. Nat. Commun. 2022, 13, 2447. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Liu, X.; Yang, Y.; Sun, Z.; Deng, S.; Jiang, Z.; Li, W.; Wu, F. NEAT1/miR-193a-3p/SOX5 axis regulates cartilage matrix degradation in human osteoarthritis. Cell Biol. Int. 2020, 44, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Gao, J.; Si, Y.; Zhao, D. Combination of circulating miR-19b-3p, miR-122-5p and miR-486-5p expressions correlates with risk and disease severity of knee osteoarthritis. Am. J. Transl. Res. 2017, 9, 2852–2864. [Google Scholar] [PubMed]
- Hu, J.; Zhao, W.; Huang, Y.; Wang, Z.; Jiang, T.; Wang, L. MiR-1180 from bone marrow MSCs promotes cell proliferation and glycolysis in ovarian cancer cells via SFRP1/Wnt pathway. Cancer Cell Int. 2019, 19, 66. [Google Scholar] [CrossRef] [PubMed]
- Ntoumou, E.; Tzetis, M.; Braoudaki, M.; Lambrou, G.; Poulou, M.; Malizos, K.; Stefanou, N.; Anastasopoulou, L.; Tsezou, A. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin. Epigenet. 2017, 9, 127. [Google Scholar] [CrossRef]
- Yin, C.M.; Suen, W.C.W.; Lin, S.; Wu, X.M.; Li, G.; Pan, X.H. Dysregulation of both miR-140-3p and miR-140-5p in synovial fluid correlate with osteoarthritis severity. Bone Jt. Res. 2017, 6, 612–618. [Google Scholar] [CrossRef] [PubMed]
- McAlinden, A.; Varghese, N.; Wirthlin, L.; Chang, L.-W. Differentially Expressed MicroRNAs in Chondrocytes from Distinct Regions of Developing Human Cartilage. PLoS ONE 2013, 8, e75012. [Google Scholar] [CrossRef]
- Wang, X.; Guo, Y.; Wang, C.; Yu, H.; Yu, X.; Yu, H. MicroRNA-142-3p Inhibits Chondrocyte Apoptosis and Inflammation in Osteoarthritis by Targeting HMGB1. Inflammation 2016, 39, 1718–1728. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Wei, P.; Song, Q.; Ye, Z.; Wang, Y.; Huang, L. MiR-140-3p Ameliorates the Progression of Osteoarthritis via Targeting CXCR4. Biol. Pharm. Bull. 2020, 43, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.-Y.; Wang, Y.; An, J.; Mao, C.-M.; Hou, N.; Lv, Y.-X.; Wang, Y.-L.; Cui, F.; Huang, M.; Yang, X. Mutation analysis of the Smad3 gene in human osteoarthritis. Eur. J. Hum. Genet. 2003, 11, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Niu, Y.; Peng, Y.; Pan, X.; Wang, F. COL3A1, COL5A1 and COL6A2 serve as potential molecular biomarkers for osteoarthritis based on weighted gene co-expression network analysis bioinformatics analysis. Exp. Ther. Med. 2023, 26, 540. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zinkle, A.; Chen, L.; Mohammadi, M. Fibroblast growth factor signalling in osteoarthritis and cartilage repair. Nat. Rev. Rheumatol. 2020, 16, 547–564. [Google Scholar] [CrossRef]
- Almeria, C.; Kreß, S.; Weber, V.; Egger, D.; Kasper, C. Heterogeneity of mesenchymal stem cell-derived extracellular vesicles is highly impacted by the tissue/cell source and culture conditions. Cell Biosci. 2022, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Cavallo, C.; Merli, G.; Borzì, R.M.; Zini, N.; D’Adamo, S.; Guescini, M.; Grigolo, B.; Di Martino, A.; Santi, S.; Filardo, G. Small Extracellular Vesicles from adipose derived stromal cells significantly attenuate in vitro the NF-κB dependent inflammatory/catabolic environment of osteoarthritis. Sci. Rep. 2021, 11, 1053. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wong, K.L.; Ren, X.; Teo, K.Y.W.; Afizah, H.; Choo, A.B.H.; Lai, R.C.; Lim, S.K.; Hui, J.H.P.; Toh, W.S. Mesenchymal Stem Cell Exosomes Promote Functional Osteochondral Repair in a Clinically Relevant Porcine Model. Am. J. Sports Med. 2022, 50, 788–800. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.H.; Kim, H.K.; Jung, G.Y.; Jung, Y.J.; Lee, K.S.; Yun, Y.E.; Han, J.; Lee, J.; Kim, W.S.; Choi, J.S.; et al. Small extracellular vesicles from human adipose-derived stem cells attenuate cartilage degeneration. J. Extracell. Vesicles 2020, 9, 1735249. [Google Scholar] [CrossRef] [PubMed]




| Direction | Gene Ensemble | Gene Name | Accession | Description | PMID | Species | Tissue |
|---|---|---|---|---|---|---|---|
| Up | ENSG00000156966 | B3GNT7 | GSE97118 | Differentiation of Ezh2-deficient immature mouse chondrocytes | 29280310 | Mouse | Cartilage |
| ENSG00000155980 | KIF5A | E-MTAB-6266_B | Human knee OA subgroups and non-OA cartilage | 29273646 | Human | Cartilage | |
| GSE45233 | Human injured meniscus with age and chondrosis | 24692131 | Human | Meniscus | |||
| ENSG00000277377 | SERPINA1 | GSE55457 | RA, OA, and control synovial membrane | 24690414 | Human | Synovium | |
| E-MTAB-1123 | Mineralizing osteoblast-specific androgen receptor knockout | 22525354 | Mouse | Bone | |||
| GSE45233 | Human injured meniscus with age and chondrosis | 24692131 | Human | Meniscus | |||
| E-GEOD-1919 | Human OA or RA synovium treated with drug regimes | 20858714 | Human | Synovium | |||
| GSE55584 | RA and OA synovial membrane | 24690414 | Human | Synovium | |||
| ENSG00000224212 | TAP1 | GSE27492 | Synovial fluid cells from a mouse model of autoantibody-mediated arthritis timecourse | 20506316 | Mouse | Synovium | |
| GSE55457 | RA, OA, and control synovial membrane | 24690414 | Human | Synovium | |||
| E-GEOD-1919 | Human OA or RA synovium treated with drug regimes | 20858714 | Human | Synovium | |||
| GSE112413 | Constitutively active FGF8 signaling in embryonic skulls | 29752281 | Mouse | Embryo | |||
| E-GEOD-19664 | OA chondrocytes and MSCs during chondrogenic differentiation | 20883804 | Human | Cartilage | |||
| E-GEOD-1919 | Human OA or RA synovium treated with drug regimes | 20858714 | Human | Synovium | |||
| GSE55584 | RA and OA synovial membrane | 24690414 | Human | Synovium | |||
| ENSG00000235715 | PSMB8 | GSE27492 | Synovial fluid cells from a mouse model of autoantibody-mediated arthritis time course | 20506316 | Mouse | Synovium | |
| E-GEOD-1919 | Human OA or RA synovium treated with drug regimes | 20858714 | Human | Synovium | |||
| Down | ENSG00000137821 | LRRC49 | E-GEOD-19664 | Osteoarthritic chondrocytes and MSCs during chondrogenic differentiation | 20883804 | Human | Cartilage |
| ENSG00000122507 | BBS9 | E-MTAB-6417 | Periosteum and bone marrow from fractured bones | 29472541 | Mouse | Bone |
| MicroRNA | FC | Gene | FC | Source | Relationship | Pathway | |
|---|---|---|---|---|---|---|---|
| miR-98-5p | ↓ | CCND1 | ↑ | IEF | ↓ | ↑ | Cyclins and cell cycle regulation, IL-8 signaling |
| miR-98-5p | ↓ | COL5A1 | ↑ | EF | ↓ | ↑ | GP6 signaling |
| miR-98-5p | ↓ | PRRC2A | ↑ | TarBase | ↓ | ↑ | |
| miR-98-5p | ↓ | SCYL1 | ↑ | TarBase | ↓ | ↑ | |
| miR-125b-5p | ↑ | OSBPL9 | ↓ | IEF | ↑ | ↓ | |
| miR-140-5p | ↓ | SMAD3 | ↑ | miRecords | ↓ | ↑ | Human embryonic stem cell pluripotency, osteoarthritis pathway, IL-2 expression |
| miR-15b-5p | ↓ | CCND1 | ↑ | IEF | ↓ | ↑ | Cyclins and cell cycle regulation, IL-8 signaling |
| miR-15b-5p | ↓ | FGFR1 | ↑ | IEF | ↓ | ↑ | Human embryonic stem cell pluripotency, IL-15 production, osteoarthritis pathway |
| miR-23b-3p | ↓ | SMAD3 | ↑ | miRecords | ↓ | ↑ | Human embryonic Stem cell pluripotency, osteoarthritis pathway, IL-2 expression |
| miR-29b-3p | ↓ | COL5A1 | ↑ | IEF | ↓ | ↑ | GP6 signaling |
| miR-30e-5p | ↑ | COL5A2 | ↓ | IEF | ↑ | ↓ | GP6 signaling |
| miR-30e-5p | ↑ | NCL | ↓ | TarBase | ↑ | ↓ | |
| miR-30e-5p | ↑ | PLOD2 | ↓ | IEF | ↑ | ↓ | |
| miR-30e-5p | ↑ | SLC38A1 | ↓ | TarBase | ↑ | ↓ | Glutamate receptor signaling |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scalzone, A.; Sanjurjo-Rodríguez, C.; Berlinguer-Palmini, R.; Dickinson, A.M.; Jones, E.; Wang, X.-N.; Crossland, R.E. Functional and Molecular Analysis of Human Osteoarthritic Chondrocytes Treated with Bone Marrow-Derived MSC-EVs. Bioengineering 2024, 11, 388. https://doi.org/10.3390/bioengineering11040388
Scalzone A, Sanjurjo-Rodríguez C, Berlinguer-Palmini R, Dickinson AM, Jones E, Wang X-N, Crossland RE. Functional and Molecular Analysis of Human Osteoarthritic Chondrocytes Treated with Bone Marrow-Derived MSC-EVs. Bioengineering. 2024; 11(4):388. https://doi.org/10.3390/bioengineering11040388
Chicago/Turabian StyleScalzone, Annachiara, Clara Sanjurjo-Rodríguez, Rolando Berlinguer-Palmini, Anne M. Dickinson, Elena Jones, Xiao-Nong Wang, and Rachel E. Crossland. 2024. "Functional and Molecular Analysis of Human Osteoarthritic Chondrocytes Treated with Bone Marrow-Derived MSC-EVs" Bioengineering 11, no. 4: 388. https://doi.org/10.3390/bioengineering11040388
APA StyleScalzone, A., Sanjurjo-Rodríguez, C., Berlinguer-Palmini, R., Dickinson, A. M., Jones, E., Wang, X.-N., & Crossland, R. E. (2024). Functional and Molecular Analysis of Human Osteoarthritic Chondrocytes Treated with Bone Marrow-Derived MSC-EVs. Bioengineering, 11(4), 388. https://doi.org/10.3390/bioengineering11040388








