The Role of Biophysical Factors in Organ Development: Insights from Current Organoid Models
Abstract
1. Introduction
2. Clinical Demand for Improved In Vitro Organoid Models
3. Principles of Biophysical Factors in Regulating Organ Development
3.1. Stiffness, Viscoelasticity, and Shear Forces in Organ Development, Homeostasis, and Pathology
3.2. Mechanotransductive Signaling Pathways: YAP/TAZ and the Intersection between Biophysical and Biochemical Pathways

3.3. Geometric Constraints: Size and Shape
4. Approaches to Use Biophysical Factors in Regulating Organoid Development
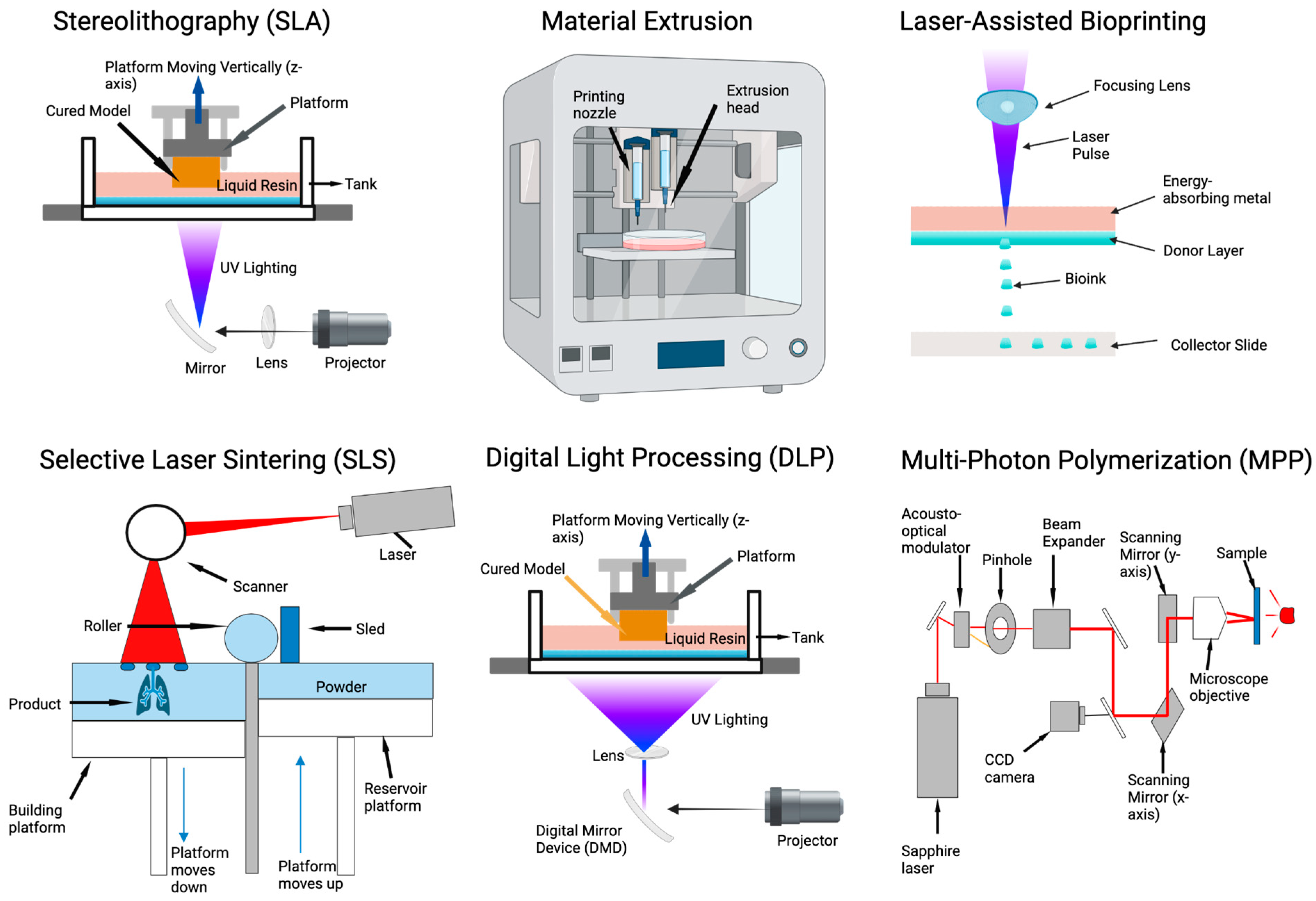
4.1. 3D Printing Approaches to Tissue Patterning and Modeling
4.2. Microfluidic Systems to Pattern Tissue Formation
4.3. Assembloids and Vascularized Organoids to Study Biophysical Components’ Organ Assembly and Angiogenesis
4.4. Biofunctionalization of the Microenvironment to Promote Organ Development
4.5. Dynamic Microenvironments: Towards Spatiotemporal Control of the Biophysical and Biochemical Niches
5. The Future of Organoid Clinical Applications
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Politis, M.D.; Bermejo-Sánchez, E.; Canfield, M.A.; Contiero, P.; Cragan, J.D.; Dastgiri, S.; de Walle, H.; Feldkamp, M.L.; Nance, A.; Groisman, B.; et al. Prevalence and Mortality among Children with Congenital Diaphragmatic Hernia: A Multi-Country Analysis. Ann. Epidemiol. 2021, 56, 61–69.e3. [Google Scholar] [CrossRef]
- Kunisaki, S.M.; Jiang, G.; Biancotti, J.C.; Ho, K.K.Y.; Dye, B.R.; Liu, A.P.; Spence, J.R. Human Induced Pluripotent Stem Cell-Derived Lung Organoids in an Ex Vivo Model of the Congenital Diaphragmatic Hernia Fetal Lung. Stem Cells Transl. Med. 2021, 10, 98–114. [Google Scholar] [CrossRef]
- Danzer, E.; Rintoul, N.E.; van Meurs, K.P.; Deprest, J. Prenatal Management of Congenital Diaphragmatic Hernia. Semin. Fetal. Neonatal Med. 2022, 27, 101406. [Google Scholar] [CrossRef]
- Antounians, L.; Catania, V.D.; Montalva, L.; Liu, B.D.; Hou, H.; Chan, C.; Matei, A.C.; Tzanetakis, A.; Li, B.; Figueira, R.L.; et al. Fetal Lung Underdevelopment Is Rescued by Administration of Amniotic Fluid Stem Cell Extracellular Vesicles in Rodents. Sci. Transl. Med. 2021, 13, eaax5941. [Google Scholar] [CrossRef]
- Vorselen, D.; Wang, Y.; de Jesus, M.M.; Shah, P.K.; Footer, M.J.; Huse, M.; Cai, W.; Theriot, J.A. Microparticle Traction Force Microscopy Reveals Subcellular Force Exertion Patterns in Immune Cell–Target Interactions. Nat. Commun. 2020, 11, 20. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Fong, E.L.S.; Balachander, G.M.; Chen, Z.; Soragni, A.; et al. Organoids. Nat. Rev. Methods Primer 2022, 2, 94. [Google Scholar] [CrossRef]
- Zhuang, C.; Gould, J.E.; Enninful, A.; Shao, S.; Mak, M. Biophysical and mechanobiological considerations for T-cell-based immunotherapy. Trends Pharmacol. Sci. 2023, 44, 366–378. [Google Scholar] [CrossRef]
- Najjari, A.; Mehdinavaz Aghdam, R.; Ebrahimi, S.A.S.; Suresh, K.S.; Krishnan, S.; Shanthi, C.; Ramalingam, M. Smart piezoelectric biomaterials for tissue engineering and regenerative medicine: A review. Biomed. Tech. 2022, 67, 71–88. [Google Scholar] [CrossRef]
- Zhu, R.; Sun, Z.; Li, C.; Ramakrishna, S.; Chiu, K.; He, L. Electrical stimulation affects neural stem cell fate and function in vitro. Exp. Neurol. 2019, 319, 112963. [Google Scholar] [CrossRef]
- Truebel, H.; Thurston, T. Danger in the Valley of Death: How the Transition from Preclinical Research to Clinical Trials Can Impact Valuations. Drug Discov. Today 2020, 25, 2089–2094. [Google Scholar] [CrossRef]
- Danopoulos, S.; Thornton, M.E.; Grubbs, B.H.; Frey, M.R.; Warburton, D.; Bellusci, S.; Al Alam, D. Discordant Roles for FGF Ligands in Lung Branching Morphogenesis between Human and Mouse. J. Pathol. 2019, 247, 254–265. [Google Scholar] [CrossRef]
- Singh, R.K.; Nasonkin, I.O. Limitations and Promise of Retinal Tissue From Human Pluripotent Stem Cells for Developing Therapies of Blindness. Front. Cell. Neurosci. 2020, 14, 179. [Google Scholar] [CrossRef]
- Gaston-Breton, R.; Maïza Letrou, A.; Hamoudi, R.; Stonestreet, B.S.; Mabondzo, A. Brain Organoids for Hypoxic-Ischemic Studies: From Bench to Bedside. Cell Mol. Life Sci. 2023, 80, 318. [Google Scholar] [CrossRef]
- Kreutzer, F.P.; Meinecke, A.; Schmidt, K.; Fiedler, J.; Thum, T. Alternative Strategies in Cardiac Preclinical Research and New Clinical Trial Formats. Cardiovasc. Res. 2022, 118, 746–762. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures in Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Volmert, B.; Kiselev, A.; Juhong, A.; Wang, F.; Riggs, A.; Kostina, A.; O’Hern, C.; Muniyandi, P.; Wasserman, A.; Huang, A.; et al. A patterned human primitive heart organoid model generated by pluripotent stem cell self-organization. Nat. Commun. 2023, 14, 8245. [Google Scholar] [CrossRef]
- Van Haute, L.; De Block, G.; Liebaers, I.; Sermon, K.; De Rycke, M. Generation of Lung Epithelial-like Tissue from Human Embryonic Stem Cells. Respir. Res. 2009, 10, 105. [Google Scholar] [CrossRef]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.A.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef]
- Zeng, Z.; Huang, B.; Parvez, R.K.; Li, Y.; Chen, J.; Vonk, A.C.; Thornton, M.E.; Patel, T.; Rutledge, E.A.; Kim, A.D.; et al. Generation of patterned kidney organoids that recapitulate the adult kidney collecting duct system from expandable ureteric bud progenitors. Nat. Commun. 2021, 12, 3641. [Google Scholar] [CrossRef]
- Yoshihara, E.; O’Connor, C.; Gasser, E.; Wei, Z.; Oh, T.G.; Tseng, T.W.; Wang, D.; Cayabyab, F.; Dai, Y.; Yu, R.T.; et al. Immune-Evasive Human Islet-Like Organoids Ameliorate Diabetes. Nature 2020, 586, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Baptista, L.S.; Mironov, V.; Koudan, E.; Amorim, É.A.; Pampolha, T.P.; Kasyanov, V.; Kovalev, A.; Senatov, F.; Granjeiro, J.M. Bioprinting Using Organ Building Blocks: Spheroids, Organoids, and Assembloids. Tissue Eng. Part A 2024. advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Ramezankhani, R.; Solhi, R.; Chai, Y.C.; Vosough, M.; Verfaillie, C. Organoid and Microfluidics-Based Platforms for Drug Screening in COVID-19. Drug Discov. Today 2022, 27, 1062–1076. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Cooper-White, J.; Janmey, P.A.; Mooney, D.J.; Shenoy, V.B. Effects of Extracellular Matrix Viscoelasticity on Cellular Behaviour. Nature 2020, 584, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Mierke, C.T. Viscoelasticity, Like Forces, Plays a Role in Mechanotransduction. Front. Cell Dev. Biol. 2022, 10, 789841. [Google Scholar] [CrossRef] [PubMed]
- Yavitt, F.M.; Kirkpatrick, B.E.; Blatchley, M.R.; Speckl, K.F.; Mohagheghian, E.; Moldovan, R.; Wang, N.; Dempsey, P.J.; Anseth, K.S. In Situ Modulation of Intestinal Organoid Epithelial Curvature through Photoinduced Viscoelasticity Directs Crypt Morphogenesis. Sci. Adv. 2023, 9, eadd5668. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-B.; Franze, K.; Seifert, G.; Steinhäuser, C.; Kirchhoff, F.; Wolburg, H.; Guck, J.; Janmey, P.; Wei, E.-Q.; Käs, J.; et al. Viscoelastic Properties of Individual Glial Cells and Neurons in the CNS. Proc. Natl. Acad. Sci. 2006, 103, 17759–17764. [Google Scholar] [CrossRef] [PubMed]
- Güney, T.G.; Herranz, A.M.; Mumby, S.; Dunlop, I.E.; Adcock, I.M. Epithelial-Stromal Cell Interactions and Extracellular Matrix Mechanics Drive the Formation of Airway-Mimetic Tubular Morphology in Lung Organoids. iScience 2021, 24, 103061. [Google Scholar] [CrossRef] [PubMed]
- Kuşoğlu, A.; Yangın, K.; Özkan, S.N.; Sarıca, S.; Örnek, D.; Solcan, N.; Karaoğlu, İ.C.; Kızılel, S.; Bulutay, P.; Fırat, P.; et al. Different Decellularization Methods in Bovine Lung Tissue Reveals Distinct Biochemical Composition, Stiffness, and Viscoelasticity in Reconstituted Hydrogels. ACS Appl. Bio. Mater. 2023, 6, 793–805. [Google Scholar] [CrossRef]
- Peng, Y.-H.; Hsiao, S.K.; Gupta, K.; Ruland, A.; Auernhammer, G.K.; Maitz, M.F.; Boye, S.; Lattner, J.; Gerri, C.; Honigmann, A.; et al. Dynamic Matrices with DNA-Encoded Viscoelasticity for Cell and Organoid Culture. Nat. Nanotechnol. 2023, 18, 1463–1473. [Google Scholar] [CrossRef]
- Rizwan, M.; Ling, C.; Guo, C.; Liu, T.; Jiang, J.-X.; Bear, C.E.; Ogawa, S.; Shoichet, M.S. Viscoelastic Notch Signaling Hydrogel Induces Liver Bile Duct Organoid Growth and Morphogenesis. Adv. Healthc. Mater. 2022, 11, e2200880. [Google Scholar] [CrossRef]
- Kleuskens, M.W.A.; Crispim, J.F.; van Doeselaar, M.; van Donkelaar, C.C.; Janssen, R.P.A.; Ito, K. Neo-Cartilage Formation Using Human Nondegenerate versus Osteoarthritic Chondrocyte-Derived Cartilage Organoids in a Viscoelastic Hydrogel. J. Orthop. Res. 2023, 41, 1902–1915. [Google Scholar] [CrossRef]
- Elosegui-Artola, A.; Gupta, A.; Najibi, A.J.; Seo, B.R.; Garry, R.; Tringides, C.M.; de Lázaro, I.; Darnell, M.; Gu, W.; Zhou, Q.; et al. Matrix Viscoelasticity Controls Spatiotemporal Tissue Organization. Nat. Mater. 2023, 22, 117–127. [Google Scholar] [CrossRef]
- Mountcastle, S.E.; Allen, P.; Mellors, B.O.L.; Lawless, B.M.; Cooke, M.E.; Lavecchia, C.E.; Fell, N.L.A.; Espino, D.M.; Jones, S.W.; Cox, S.C. Dynamic Viscoelastic Characterisation of Human Osteochondral Tissue: Understanding the Effect of the Cartilage-Bone Interface. BMC Musculoskelet. Disord. 2019, 20, 575. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Park, J.S.; Chu, J.S.; Tsou, A.D.; Diop, R.; Tang, Z.; Wang, A.; Li, S. The Effect of Matrix Stiffness on the Differentiation of Mesenchymal Stem Cells in Response to TGF-β. Biomaterials 2011, 32, 3921–3930. [Google Scholar] [CrossRef]
- Wells, R.G. The Role of Matrix Stiffness in Regulating Cell Behavior. Hepatology 2008, 47, 1394–1400. [Google Scholar] [CrossRef] [PubMed]
- Stowers, R.S.; Shcherbina, A.; Israeli, J.; Gruber, J.J.; Chang, J.; Nam, S.; Rabiee, A.; Teruel, M.N.; Snyder, M.P.; Kundaje, A.; et al. Matrix Stiffness Induces a Tumorigenic Phenotype in Mammary Epithelium through Changes in Chromatin Accessibility. Nat. Biomed. Eng. 2019, 3, 1009–1019. [Google Scholar] [CrossRef]
- Bao, M.; Chen, Y.; Liu, J.-T.; Bao, H.; Wang, W.-B.; Qi, Y.-X.; Lv, F. Extracellular Matrix Stiffness Controls VEGF165 Secretion and Neuroblastoma Angiogenesis via the YAP/RUNX2/SRSF1 Axis. Angiogenesis 2022, 25, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Nomdedeu-Sancho, G.; Gorkun, A.; Mahajan, N.; Willson, K.; Schaaf, C.R.; Votanopoulos, K.I.; Atala, A.; Soker, S. In Vitro Three-Dimensional (3D) Models for Melanoma Immunotherapy. Cancers 2023, 15, 5779. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Tan, W.J.; Pek, M.M.X.; Tan, M.-H.; Kurisawa, M. Mechanically and Chemically Defined Hydrogel Matrices for Patient-Derived Colorectal Tumor Organoid Culture. Biomaterials 2019, 219, 119400. [Google Scholar] [CrossRef] [PubMed]
- Mahvi, D.A.; Liu, R.; Grinstaff, M.W.; Colson, Y.L.; Raut, C.P. Local Cancer Recurrence: The Realities, Challenges, and Opportunities for New Therapies. CA Cancer J. Clin. 2018, 68, 488–505. [Google Scholar] [CrossRef]
- Even-Ram, S.; Artym, V.; Yamada, K.M. Matrix Control of Stem Cell Fate. Cell 2006, 126, 645–647. [Google Scholar] [CrossRef]
- Chaudhuri, O.; Gu, L.; Klumpers, D.; Darnell, M.; Bencherif, S.A.; Weaver, J.C.; Huebsch, N.; Lee, H.; Lippens, E.; Duda, G.N.; et al. Hydrogels with Tunable Stress Relaxation Regulate Stem Cell Fate and Activity. Nat. Mater. 2016, 15, 326–334. [Google Scholar] [CrossRef]
- Abbasi, F.; Ghanian, M.H.; Baharvand, H.; Vahidi, B.; Eslaminejad, M.B. Engineering Mesenchymal Stem Cell Spheroids by Incorporation of Mechanoregulator Microparticles. J. Mech. Behav. Biomed. Mater. 2018, 84, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Huang, H.; Ayers, D.C.; Song, J. Modulating Viscoelasticity, Stiffness, and Degradation of Synthetic Cellular Niches via Stoichiometric Tuning of Covalent versus Dynamic Noncovalent Cross-Linking. ACS Cent. Sci. 2018, 4, 971–981. [Google Scholar] [CrossRef]
- Singleton, R.C.; Pharr, G.M.; Nyman, J.S. Increased Tissue-Level Storage Modulus and Hardness with Age in Male Cortical Bone and Its Association with Decreased Fracture Toughness. Bone 2021, 148, 115949. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, S.; Paul, N.; Naguib, H.E. Standardized Static and Dynamic Evaluation of Myocardial Tissue Properties. Biomed. Mater. 2017, 12, 025013. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, A.; Salimi Bani, M.; Bahreinizad, H.; Karimi, A. Viscoelastic Properties of the Autologous Bypass Grafts: A Comparative Study among the Small Saphenous Vein and Internal Thoracic Artery. Artery Res. 2017, 19, 65–71. [Google Scholar] [CrossRef]
- Ohashi, T.; Ishii, Y.; Ishikawa, Y.; Matsumoto, T.; Sato, M. Experimental and Numerical Analyses of Local Mechanical Properties Measured by Atomic Force Microscopy for Sheared Endothelial Cells. Biomed. Mater. Eng. 2002, 12, 319–327. [Google Scholar]
- Pérez-Domínguez, S.; López-Alonso, J.; Lafont, F.; Radmacher, M. Comparison of Rheological Properties of Healthy versus Dupuytren Fibroblasts When Treated with a Cell Contraction Inhibitor by Atomic Force Microscope. Int. J. Mol. Sci. 2023, 24, 2043. [Google Scholar] [CrossRef] [PubMed]
- Budday, S.; Nay, R.; de Rooij, R.; Steinmann, P.; Wyrobek, T.; Ovaert, T.C.; Kuhl, E. Mechanical Properties of Gray and White Matter Brain Tissue by Indentation. J. Mech. Behav. Biomed. Mater. 2015, 46, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Holt, B.; Tripathi, A.; Morgan, J. Viscoelastic Response of Human Skin to Low Magnitude Physiologically Relevant Shear. J. Biomech. 2008, 41, 2689–2695. [Google Scholar] [CrossRef] [PubMed]
- Griffin, M.F.; Leung, B.C.; Premakumar, Y.; Szarko, M.; Butler, P.E. Comparison of the Mechanical Properties of Different Skin Sites for Auricular and Nasal Reconstruction. J. Otolaryngol.-Head Neck Surg. 2017, 46, 33. [Google Scholar] [CrossRef] [PubMed]
- Perni, S.; Prokopovich, P. Rheometer enabled study of cartilage frequency-dependent properties. Sci. Rep. 2020, 10, 20696. [Google Scholar] [CrossRef] [PubMed]
- Alkhouli, N.; Mansfield, J.; Green, E.; Bell, J.; Knight, B.; Liversedge, N.; Tham, J.C.; Welbourn, R.; Shore, A.C.; Kos, K.; et al. The Mechanical Properties of Human Adipose Tissues and Their Relationships to the Structure and Composition of the Extracellular Matrix. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E1427–E1435. [Google Scholar] [CrossRef] [PubMed]
- Juliar, B.A.; Strieder-Barboza, C.; Karmakar, M.; Flesher, C.G.; Baker, N.A.; Varban, O.A.; Lumeng, C.N.; Putnam, A.J.; O’Rourke, R.W. Viscoelastic Characterization of Diabetic and Non-Diabetic Human Adipose Tissue. Biorheology 2020, 57, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.W.; Moran, E.C.; Baptista, P.M.; Soker, S.; Sparks, J.L. Scale-Dependent Mechanical Properties of Native and Decellularized Liver Tissue. Biomech. Model. Mechanobiol. 2013, 12, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Khanafer, K.; Duprey, A.; Zainal, M.; Schlicht, M.; Williams, D.; Berguer, R. Determination of the Elastic Modulus of Ascending Thoracic Aortic Aneurysm at Different Ranges of Pressure Using Uniaxial Tensile Testing. J. Thorac. Cardiovasc. Surg. 2011, 142, 682–686. [Google Scholar] [CrossRef]
- Franchini, G.; Breslavsky, I.D.; Holzapfel, G.A.; Amabili, M. Viscoelastic Characterization of Human Descending Thoracic Aortas under Cyclic Load. Acta Biomater. 2021, 130, 291–307. [Google Scholar] [CrossRef]
- Nerger, B.A.; Sinha, S.; Lee, N.N.; Cheriyan, M.; Bertsch, P.; Johnson, C.P.; Mahadevan, L.; Bonventre, J.V.; Mooney, D.J. 3D Hydrogel Encapsulation Regulates Nephrogenesis in Kidney Organoids. Adv. Mater. Deerfield Beach Fla 2024, 36, e2308325. [Google Scholar] [CrossRef] [PubMed]
- Serluca, F.C.; Drummond, I.A.; Fishman, M.C. Endothelial Signaling in Kidney Morphogenesis: A Role for Hemodynamic Forces. Curr. Biol. 2002, 12, 492–497. [Google Scholar] [CrossRef]
- Natarajan, A.; Jose, P.A. Renal Modulation: The Renin-Angiotensin-Aldosterone System (RAAS). Nephrol. FluidElectrolyte Physiol. Neonatol. Quest. Controv. 2008, 47, 107–127. [Google Scholar] [CrossRef]
- Li, S.R.; Gulieva, R.E.; Helms, L.; Cruz, N.M.; Vincent, T.; Fu, H.; Himmelfarb, J.; Freedman, B.S. Glucose Absorption Drives Cystogenesis in a Human Organoid-on-Chip Model of Polycystic Kidney Disease. Nat. Commun. 2022, 13, 7918. [Google Scholar] [CrossRef] [PubMed]
- Castillo Bautista, C.M.; Sterneckert, J. Progress and Challenges in Directing the Differentiation of Human iPSCs into Spinal Motor Neurons. Front. Cell Dev. Biol. 2022, 10, 1089970. [Google Scholar] [CrossRef] [PubMed]
- Josephine Boder, E.; Banerjee, I.A. Alzheimer’s Disease: Current Perspectives and Advances in Physiological Modeling. Bioengineering 2021, 8, 211. [Google Scholar] [CrossRef]
- Hajal, C.; Offeddu, G.S.; Shin, Y.; Zhang, S.; Morozova, O.; Hickman, D.; Knutson, C.G.; Kamm, R.D. Engineered Human Blood–Brain Barrier Microfluidic Model for Vascular Permeability Analyses. Nat. Protoc. 2022, 17, 95–128. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.-J.; Park, J.-H.; Kang, J.H.; Sabaté Del Río, J.; Kong, S.-H.; Park, T.-E. Organoid-Based Human Stomach Micro-Physiological System to Recapitulate the Dynamic Mucosal Defense Mechanism. Adv. Sci. 2023, 10, e2300164. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in Mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.-Y.; Chinnaiyan, A.M.; et al. TEAD Mediates YAP-Dependent Gene Induction and Growth Control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef]
- Pan, D. The Hippo Signaling Pathway in Development and Cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.; Mayr, U.; Boni, A.; Lukonin, I.; Rempfler, M.; Challet Meylan, L.; Stadler, M.B.; Strnad, P.; Papasaikas, P.; Vischi, D.; et al. Self-Organization and Symmetry Breaking in Intestinal Organoid Development. Nature 2019, 569, 66–72. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo Pathway and Human Cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef]
- Edgar, L.T.; Underwood, C.J.; Guilkey, J.E.; Hoying, J.B.; Weiss, J.A. Extracellular Matrix Density Regulates the Rate of Neovessel Growth and Branching in Sprouting Angiogenesis. PLoS ONE 2014, 9, e85178. [Google Scholar] [CrossRef] [PubMed]
- Crosby, C.O.; Zoldan, J. Mimicking the Physical Cues of the ECM in Angiogenic Biomaterials. Regen. Biomater. 2019, 6, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Sousa-Ortega, A.; Vázquez-Marín, J.; Sanabria-Reinoso, E.; Corbacho, J.; Polvillo, R.; Campoy-López, A.; Buono, L.; Loosli, F.; Almuedo-Castillo, M.; Martínez-Morales, J.R. A Yap-Dependent Mechanoregulatory Program Sustains Cell Migration for Embryo Axis Assembly. Nat. Commun. 2023, 14, 2804. [Google Scholar] [CrossRef]
- Gjorevski, N.; Nikolaev, M.; Brown, T.E.; Brandenberg, N.; DelRio, F.W.; Yavitt, F.M.; Liberali, P.; Anseth, K.S.; Lutolf, M.P. Tissue Geometry Drives Deterministic Organoid Patterning. Science 2022, 375, eaaw9021. [Google Scholar] [CrossRef]
- Cordero-Espinoza, L.; Dowbaj, A.M.; Kohler, T.N.; Strauss, B.; Sarlidou, O.; Belenguer, G.; Pacini, C.; Martins, N.P.; Dobie, R.; Wilson-Kanamori, J.R.; et al. Dynamic Cell Contacts between Periportal Mesenchyme and Ductal Epithelium Act as a Rheostat for Liver Cell Proliferation. Cell Stem Cell 2021, 28, 1907–1921.e8. [Google Scholar] [CrossRef]
- Asaoka, Y.; Nishina, H.; Furutani-Seiki, M. YAP Is Essential for 3D Organogenesis Withstanding Gravity. Dev. Growth Differ. 2017, 59, 52–58. [Google Scholar] [CrossRef]
- Zhong, W.; Tian, K.; Zheng, X.; Li, L.; Zhang, W.; Wang, S.; Qin, J. Mesenchymal Stem Cell and Chondrocyte Fates in a Multishear Microdevice Are Regulated by Yes-Associated Protein. Stem Cells Dev. 2013, 22, 2083–2093. [Google Scholar] [CrossRef]
- Shorr, A.Z.; Sönmez, U.M.; Minden, J.S.; LeDuc, P.R. High-Throughput Mechanotransduction in Drosophila Embryos with Mesofluidics. Lab. Chip 2019, 19, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Batie, M.R.; Fan, Q.; Varisco, B.M. Mouse Lung Organoid Responses to Reduced, Increased, and Cyclic Stretch. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2022, 322, L162–L173. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Copland, I.; Post, M.; Yeger, H.; Cutz, E. Mechanical Stretch-Induced Serotonin Release from Pulmonary Neuroendocrine Cells: Implications for Lung Development. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006, 290, L185–L193. [Google Scholar] [CrossRef] [PubMed]
- Pantazi, D.; Kitsiouli, E.; Karkabounas, A.; Trangas, T.; Nakos, G.; Lekka, M.E. Dipalmitoyl-Phosphatidylcholine Biosynthesis Is Induced by Non-Injurious Mechanical Stretch in a Model of Alveolar Type II Cells. Lipids 2013, 48, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, Z.; Nayak, P.S.; Matthews, B.D.; Warburton, D.; Shi, W.; Sanchez-Esteban, J. Strain-Induced Differentiation of Fetal Type II Epithelial Cells Is Mediated via the Integrin A6β1-ADAM17/Tumor Necrosis Factor-α-Converting Enzyme (TACE) Signaling Pathway. J. Biol. Chem. 2013, 288, 25646–25657. [Google Scholar] [CrossRef] [PubMed]
- Ross, T.D.; Coon, B.G.; Yun, S.; Baeyens, N.; Tanaka, K.; Ouyang, M.; Schwartz, M.A. Integrins In Mechanotransduction. Curr. Opin. Cell Biol. 2013, 25, 613–618. [Google Scholar] [CrossRef]
- Qin, L.; He, T.; Chen, S.; Yang, D.; Yi, W.; Cao, H.; Xiao, G. Roles of Mechanosensitive Channel Piezo1/2 Proteins in Skeleton and Other Tissues. Bone Res. 2021, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Kuebler, W.M. Mechanotransduction by TRP Channels: General Concepts and Specific Role in the Vasculature. Cell Biochem. Biophys. 2010, 56, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Leckband, D.E.; de Rooij, J. Cadherin Adhesion and Mechanotransduction. Annu. Rev. Cell Dev. Biol. 2014, 30, 291–315. [Google Scholar] [CrossRef]
- Na, S.; Chowdhury, F.; Tay, B.; Ouyang, M.; Gregor, M.; Wang, Y.; Wiche, G.; Wang, N. Plectin Contributes to Mechanical Properties of Living Cells. Am. J. Physiol.-Cell Physiol. 2009, 296, C868–C877. [Google Scholar] [CrossRef]
- Goult, B.T.; Brown, N.H.; Schwartz, M.A. Talin in Mechanotransduction and Mechanomemory at a Glance. J. Cell Sci. 2021, 134, jcs258749. [Google Scholar] [CrossRef]
- Goldmann, W.H. Role of Vinculin in Cellular Mechanotransduction. Cell Biol. Int. 2016, 40, 241–256. [Google Scholar] [CrossRef]
- Razinia, Z.; Mäkelä, T.; Ylänne, J.; Calderwood, D.A. Filamins in Mechanosensing and Signaling. Annu. Rev. Biophys. 2012, 41, 227–246. [Google Scholar] [CrossRef]
- Ramirez, M.P.; Anderson, M.J.M.; Kelly, M.D.; Sundby, L.J.; Hagerty, A.R.; Wenthe, S.J.; Odde, D.J.; Ervasti, J.M.; Gordon, W.R. Dystrophin Missense Mutations Alter Focal Adhesion Tension and Mechanotransduction. Proc. Natl. Acad. Sci. USA 2022, 119, e2205536119. [Google Scholar] [CrossRef]
- André, A.A.M.; Spruijt, E. Liquid–Liquid Phase Separation in Crowded Environments. Int. J. Mol. Sci. 2020, 21, 5908. [Google Scholar] [CrossRef]
- Li, Y.; Chen, M.; Hu, J.; Sheng, R.; Lin, Q.; He, X.; Guo, M. Volumetric Compression Induces Intracellular Crowding to Control Intestinal Organoid Growth via Wnt/β-Catenin Signaling. Cell Stem Cell 2021, 28, 63–78.e7. [Google Scholar] [CrossRef]
- Schuth, S.; Le Blanc, S.; Krieger, T.G.; Jabs, J.; Schenk, M.; Giese, N.A.; Büchler, M.W.; Eils, R.; Conrad, C.; Strobel, O. Patient-Specific Modeling of Stroma-Mediated Chemoresistance of Pancreatic Cancer Using a Three-Dimensional Organoid-Fibroblast Co-Culture System. J. Exp. Clin. Cancer Res. CR 2022, 41, 312. [Google Scholar] [CrossRef] [PubMed]
- Kalukula, Y.; Stephens, A.D.; Lammerding, J.; Gabriele, S. Mechanics and Functional Consequences of Nuclear Deformations. Nat. Rev. Mol. Cell Biol. 2022, 23, 583–602. [Google Scholar] [CrossRef]
- van der Putten, C.; Buskermolen, A.B.C.; Werner, M.; Brouwer, H.F.M.; Bartels, P.A.A.; Dankers, P.Y.W.; Bouten, C.V.C.; Kurniawan, N.A. Protein Micropatterning in 2.5D: An Approach to Investigate Cellular Responses in Multi-Cue Environments. ACS Appl. Mater. Interfaces 2021, 13, 25589–25598. [Google Scholar] [CrossRef]
- Yavitt, F.M.; Kirkpatrick, B.E.; Blatchley, M.R.; Anseth, K.S. 4D Materials with Photoadaptable Properties Instruct and Enhance Intestinal Organoid Development. ACS Biomater. Sci. Eng. 2022, 8, 4634–4638. [Google Scholar] [CrossRef]
- Humenik, M.; Winkler, A.; Scheibel, T. Patterning of Protein-Based Materials. Biopolymers 2021, 112, e23412. [Google Scholar] [CrossRef] [PubMed]
- Warmflash, A.; Sorre, B.; Etoc, F.; Siggia, E.D.; Brivanlou, A.H. A Method to Recapitulate Early Embryonic Spatial Patterning in Human Embryonic Stem Cells. Nat. Methods 2014, 11, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.J.S.; Ang, Y.-S.; Fu, J.-D.; Rivas, R.N.; Mohamed, T.M.A.; Higgs, G.C.; Srivastava, D.; Pruitt, B.L. Contractility of Single Cardiomyocytes Differentiated from Pluripotent Stem Cells Depends on Physiological Shape and Substrate Stiffness. Proc. Natl. Acad. Sci. USA 2015, 112, 12705–12710. [Google Scholar] [CrossRef] [PubMed]
- Knight, G.T.; Lundin, B.F.; Iyer, N.; Ashton, L.M.; Sethares, W.A.; Willett, R.M.; Ashton, R.S. Engineering Induction of Singular Neural Rosette Emergence within hPSC-Derived Tissues. eLife 2018, 7, e37549. [Google Scholar] [CrossRef] [PubMed]
- Karzbrun, E.; Khankhel, A.H.; Megale, H.C.; Glasauer, S.M.K.; Wyle, Y.; Britton, G.; Warmflash, A.; Kosik, K.S.; Siggia, E.D.; Shraiman, B.I.; et al. Human Neural Tube Morphogenesis in Vitro by Geometric Constraints. Nature 2021, 599, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Harmansa, S.; Erlich, A.; Eloy, C.; Zurlo, G.; Lecuit, T. Growth Anisotropy of the Extracellular Matrix Shapes a Developing Organ. Nat. Commun. 2023, 14, 1220. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, T.; Lenne, P.-F. Cell Surface Mechanics and the Control of Cell Shape, Tissue Patterns and Morphogenesis. Nat. Rev. Mol. Cell Biol. 2007, 8, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Mammoto, T.; Ingber, D.E. Mechanical Control of Tissue and Organ Development. Dev. Camb. Engl. 2010, 137, 1407–1420. [Google Scholar] [CrossRef]
- Varner, V.D.; Gleghorn, J.P.; Miller, E.; Radisky, D.C.; Nelson, C.M. Mechanically Patterning the Embryonic Airway Epithelium. Proc. Natl. Acad. Sci. USA 2015, 112, 9230–9235. [Google Scholar] [CrossRef]
- Fedorchak, N.J.; Iyer, N.; Ashton, R.S. Bioengineering Tissue Morphogenesis and Function in Human Neural Organoids. Semin. Cell Dev. Biol. 2021, 111, 52–59. [Google Scholar] [CrossRef]
- Zahmatkesh, E.; Ghanian, M.H.; Zarkesh, I.; Farzaneh, Z.; Halvaei, M.; Heydari, Z.; Moeinvaziri, F.; Othman, A.; Ruoß, M.; Piryaei, A.; et al. Tissue-Specific Microparticles Improve Organoid Microenvironment for Efficient Maturation of Pluripotent Stem-Cell-Derived Hepatocytes. Cells 2021, 10, 1274. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-L.; Zhao, X.-Y.; Xiong, W.; Ji, L.-F.; Jia, M.-X.; Liu, Y.-Y.; Guo, H.-T.; Qu, F.; Cui, W.; Gu, Q.; et al. Cartilage Lacuna-Inspired Microcarriers Drive Hyaline Neocartilage Regeneration. Adv. Mater. 2023, 35, 2212114. [Google Scholar] [CrossRef]
- Mazalan, M.B.; Ramlan, M.A.B.; Shin, J.H.; Ohashi, T. Effect of Geometric Curvature on Collective Cell Migration in Tortuous Microchannel Devices. Micromachines 2020, 11, 659. [Google Scholar] [CrossRef]
- Alisafaei, F.; Jokhun, D.S.; Shivashankar, G.V.; Shenoy, V.B. Regulation of Nuclear Architecture, Mechanics, and Nucleocytoplasmic Shuttling of Epigenetic Factors by Cell Geometric Constraints. Proc. Natl. Acad. Sci. USA 2019, 116, 13200–13209. [Google Scholar] [CrossRef]
- Bao, M.; Xie, J.; Piruska, A.; Huck, W.T.S. 3D Microniches Reveal the Importance of Cell Size and Shape. Nat. Commun. 2017, 8, 1962. [Google Scholar] [CrossRef]
- Kress, S.J.P.; Richner, P.; Jayanti, S.V.; Galliker, P.; Kim, D.K.; Poulikakos, D.; Norris, D.J. Near-Field Light Design with Colloidal Quantum Dots for Photonics and Plasmonics. Nano Lett. 2014, 14, 5827–5833. [Google Scholar] [CrossRef]
- Arakawa, C.; Gunnarsson, C.; Howard, C.; Bernabeu, M.; Phong, K.; Yang, E.; DeForest, C.A.; Smith, J.D.; Zheng, Y. Biophysical and Biomolecular Interactions of Malaria-Infected Erythrocytes in Engineered Human Capillaries. Sci. Adv. 2020, 6, eaay7243. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.G.; Brunel, L.G.; Huang, M.S.; Liu, Y.; Cai, B.; Sinha, S.; Yang, F.; Pașca, S.P.; Shin, S.; Heilshorn, S.C. Spatially Controlled Construction of Assembloids Using Bioprinting. Nat. Commun. 2023, 14, 4346. [Google Scholar] [CrossRef]
- Huang, J.; Qin, Q.; Wang, J. A Review of Stereolithography: Processes and Systems. Processes 2020, 8, 1138. [Google Scholar] [CrossRef]
- Grigoryan, B.; Paulsen, S.J.; Corbett, D.C.; Sazer, D.W.; Fortin, C.L.; Stevens, K.R.; Miller, J.S.; Gounley, J.P.; Ta, A.H.; Johansson, F. Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 2019, 364, 458–464. [Google Scholar] [CrossRef]
- Kinstlinger, I.S.; Saxton, S.H.; Calderon, G.A.; Ruiz, K.V.; Yalacki, D.R.; Deme, P.R.; Rosenkrantz, J.E.; Louis-Rosenberg, J.D.; Johansson, F.; Janson, K.D.; et al. Generation of Model Tissues with Dendritic Vascular Networks via Sacrificial Laser-Sintered Carbohydrate Templates. Nat. Biomed. Eng. 2020, 4, 916–932. [Google Scholar] [CrossRef]
- Colucci, G.; Piano, M.; Lupone, F.; Baruffaldi, D.; Frascella, F.; Bondioli, F.; Messori, M. Printability Study by Selective Laser Sintering of Bio-Based Samples Obtained by Using PBS as Polymeric Matrix. Polym. Test. 2024, 131, 108327. [Google Scholar] [CrossRef]
- Ligon, S.C.; Liska, R.; Stampfl, J.; Gurr, M.; Mülhaupt, R. Polymers for 3D Printing and Customized Additive Manufacturing. Chem. Rev. 2017, 117, 10212–10290. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.N.; Strong, R.; Gold, S.A. A Review of Melt Extrusion Additive Manufacturing Processes: I. Process Design and Modeling. Rapid Prototyp. J. 2014, 20, 192–204. [Google Scholar] [CrossRef]
- Carneiro, O.S.; Silva, A.F.; Gomes, R. Fused Deposition Modeling with Polypropylene. Mater. Des. 2015, 83, 768–776. [Google Scholar] [CrossRef]
- Hong, D.; Moon, S.; Seo, J.B.; Kim, N. Development of a Patient-Specific Chest Computed Tomography Imaging Phantom with Realistic Lung Lesions Using Silicone Casting and Three-Dimensional Printing. Sci. Rep. 2023, 13, 3941. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Park, S.; Lee, C.C.; Ho, P.C.L.; Kwok, P.C.L.; Kang, L. A 3D Printed Human Upper Respiratory Tract Model for Particulate Deposition Profiling. Int. J. Pharm. 2021, 597, 120307. [Google Scholar] [CrossRef] [PubMed]
- Carberry, B.J.; Hergert, J.E.; Yavitt, F.M.; Hernandez, J.J.; Speckl, K.F.; Bowman, C.N.; McLeod, R.R.; Anseth, K.S. 3D Printing of Sacrificial Thioester Elastomers Using Digital Light Processing for Templating 3D Organoid Structures in Soft Biomatrices. Biofabrication 2021, 13, 044104. [Google Scholar] [CrossRef] [PubMed]
- Razavi Bazaz, S.; Kashaninejad, N.; Azadi, S.; Patel, K.; Asadnia, M.; Jin, D.; Ebrahimi Warkiani, M. Rapid Softlithography Using 3D-Printed Molds. Adv. Mater. Technol. 2019, 4, 1900425. [Google Scholar] [CrossRef]
- Xie, C.; Liang, R.; Ye, J.; Peng, Z.; Sun, H.; Zhu, Q.; Shen, X.; Hong, Y.; Wu, H.; Sun, W.; et al. High-Efficient Engineering of Osteo-Callus Organoids for Rapid Bone Regeneration within One Month. Biomaterials 2022, 288, 121741. [Google Scholar] [CrossRef]
- Roversi, K.; Orimi, H.E.; Erfanian, M.; Talbot, S.; Boutopoulos, C. LIST: A Newly Developed Laser-assisted Cell Bioprinting Technology. Bio. Protoc. 2022, 12, e4527. [Google Scholar] [CrossRef] [PubMed]
- Kawecki, F.; Clafshenkel, W.P.; Auger, F.A.; Bourget, J.-M.; Fradette, J.; Devillard, R. Self-Assembled Human Osseous Cell Sheets as Living Biopapers for the Laser-Assisted Bioprinting of Human Endothelial Cells. Biofabrication 2018, 10, 035006. [Google Scholar] [CrossRef]
- McLennan, H.J.; Blanch, A.J.; Wallace, S.J.; Ritter, L.J.; Heinrich, S.L.; Gardner, D.K.; Dunning, K.R.; Gauvin, M.J.; Love, A.K.; Thompson, J.G. Nano-Liter Perfusion Microfluidic Device Made Entirely by Two-Photon Polymerization for Dynamic Cell Culture with Easy Cell Recovery. Sci. Rep. 2023, 13, 562. [Google Scholar] [CrossRef]
- Rajasekar, S.; Lin, D.S.Y.; Abdul, L.; Liu, A.; Sotra, A.; Zhang, F.; Zhang, B. IFlowPlate-A Customized 384-Well Plate for the Culture of Perfusable Vascularized Colon Organoids. Adv. Mater. 2020, 32, e2002974. [Google Scholar] [CrossRef]
- Gijzen, L.; Yousef Yengej, F.A.; Schutgens, F.; Vormann, M.K.; Ammerlaan, C.M.E.; Nicolas, A.; Kurek, D.; Vulto, P.; Rookmaaker, M.B.; Lanz, H.L.; et al. Culture and Analysis of Kidney Tubuloids and Perfused Tubuloid Cells-on-a-Chip. Nat. Protoc. 2021, 16, 2023–2050. [Google Scholar] [CrossRef]
- Hosic, S.; Bindas, A.J.; Puzan, M.L.; Lake, W.; Soucy, J.R.; Zhou, F.; Koppes, R.A.; Breault, D.T.; Murthy, S.K.; Koppes, A.N. Rapid Prototyping of Multilayer Microphysiological Systems. ACS Biomater. Sci. Eng. 2021, 7, 2949–2963. [Google Scholar] [CrossRef]
- Manfrin, A.; Tabata, Y.; Paquet, E.R.; Vuaridel, A.R.; Rivest, F.R.; Naef, F.; Lutolf, M.P. Engineered Signaling Centers for the Spatially Controlled Patterning of Human Pluripotent Stem Cells. Nat. Methods 2019, 16, 640–648. [Google Scholar] [CrossRef]
- Shin, Y.; Choi, S.H.; Kim, E.; Bylykbashi, E.; Kim, J.A.; Chung, S.; Kim, D.Y.; Kamm, R.D.; Tanzi, R.E. Blood–Brain Barrier Dysfunction in a 3D In Vitro Model of Alzheimer’s Disease. Adv. Sci. 2019, 6, 1900962. [Google Scholar] [CrossRef]
- Rousset, N.; de Geus, M.; Chimisso, V.; Kaestli, A.J.; Hierlemann, A.; Lohasz, C. Controlling Bead and Cell Mobility in a Recirculating Hanging-Drop Network. Lab. Chip 2023, 23, 4834–4847. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kong, N.; Zhang, X.; Liu, Y.; Hu, P.; Mou, S.; Liljeström, P.; Shi, J.; Tan, W.; Kim, J.S.; et al. A Materials-Science Perspective on Tackling COVID-19. Nat. Rev. Mater. 2020, 5, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Kratz, S.R.A.; Höll, G.; Schuller, P.; Ertl, P.; Rothbauer, M. Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems. Biosensors 2019, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson-Bouchard, K.; Teles, D.; Yeager, K.; Tavakol, D.N.; Zhao, Y.; Chramiec, A.; Tagore, S.; Summers, M.; Stylianos, S.; Tamargo, M.; et al. A Multi-Organ Chip with Matured Tissue Niches Linked by Vascular Flow. Nat. Biomed. Eng. 2022, 6, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Wang, Y.; Xiong, Z.; Xue, Y.; Li, J.; Chen, M.; Zhou, K.; Xu, H.; Zhang, X.; Liu, J.; et al. Microvascularized Tumor Assembloids Model for Drug Delivery Evaluation in Colorectal Cancer-Derived Peritoneal Metastasis. Acta Biomater. 2023, 168, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Miura, Y.; Li, M.-Y.; Revah, O.; Yoon, S.-J.; Narazaki, G.; Pașca, S.P. Engineering Brain Assembloids to Interrogate Human Neural Circuits. Nat. Protoc. 2022, 17, 15–35. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, E.; Albanna, W.; Pasquini, G.; Ramani, A.; Josipovic, N.; Mariappan, A.; Schinzel, F.; Karch, C.M.; Bao, G.; Gottardo, M.; et al. Human Brain Organoids Assemble Functionally Integrated Bilateral Optic Vesicles. Cell Stem Cell 2021, 28, 1740–1757.e8. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, T.M.; Tryfonos, M.; Makwana, K.; Taylor, D.M.; Brosens, J.J.; Lucas, E.S. Endometrial Assembloids to Model Human Embryo Implantation In Vitro. Methods Mol. Biol. 2023, 7651, 495. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Erickson, S.; Nakayama, M.; Ihara, H.; Sugihara, K.; Nashimoto, Y.; Nishiyama, K.; Miura, T.; Yokokawa, R. Pericytes and Shear Stress Each Alter the Shape of a Self-Assembled Vascular Network. Lab Chip 2023, 23, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Dessalles, C.A.; Leclech, C.; Castagnino, A.; Barakat, A.I. Integration of Substrate- and Flow-Derived Stresses in Endothelial Cell Mechanobiology. Commun. Biol. 2021, 4, 764. [Google Scholar] [CrossRef] [PubMed]
- Lien, J.C.; Wang, Y.L. Cyclic Stretching-Induced Epithelial Cell Reorientation Is Driven by Microtubule-Modulated Transverse Extension During the Relaxation Phase. Sci. Rep. 2021, 11, 14803. [Google Scholar] [CrossRef]
- Mandrycky, C.; Hadland, B.; Zheng, Y. 3D Curvature-Instructed Endothelial Flow Response and Tissue Vascularization. Sci. Adv. 2020, 6, eabb3629. [Google Scholar] [CrossRef]
- Licata, J.P.; Schwab, K.H.; Har-El, Y.-E.; Gerstenhaber, J.A.; Lelkes, P.I. Bioreactor Technologies for Enhanced Organoid Culture. Int. J. Mol. Sci. 2023, 24, 11427. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Xie, W.; Wang, T.; Li, Y.; de Dieu Habimana, J.; Amissah, O.B.; Huang, J.; Chen, Y.; Ni, B.; Li, Z. HIF-1α promotes kidney organoid vascularization and applications in disease modeling. Stem Cell Res. Ther. 2023, 14, 336. [Google Scholar] [CrossRef] [PubMed]
- Basara, G.; Bahcecioglu, G.; Ozcebe, S.G.; Ellis, B.W.; Ronan, G.; Zorlutuna, P. Myocardial Infarction from a Tissue Engineering and Regenerative Medicine Point of View: A Comprehensive Review on Models and Treatments. Biophys. Rev. 2022, 3, 031305. [Google Scholar] [CrossRef] [PubMed]
- Strobel, H.A.; Moss, S.M.; Hoying, J.B. Vascularized Tissue Organoids. Bioengineering 2023, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.A.; Gonçalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An in Vivo Model of Functional and Vascularized Human Brain Organoids. Nat. Biotechnol. 2018, 36, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.K.; Tsuru, Y.; Hasegawa, K.; Kuwako, K.-I. Vascularization of Human Brain Organoids. Stem Cells 2021, 39, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Cortez, A.R.; Poling, H.M.; Brown, N.E.; Singh, A.; Mahe, M.M.; Helmrath, M.A. Transplantation of Human Intestinal Organoids into the Mouse Mesentery: A More Physiologic and Anatomic Engraftment Site. Surgery 2018, 164, 643–650. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, C.W.; Ritsma, L.; Avramut, M.C.; Wiersma, L.E.; van den Berg, B.M.; Leuning, D.G.; Lievers, E.; Koning, M.; Vanslambrouck, J.M.; Koster, A.J.; et al. Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-Vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo. Stem Cell Rep. 2018, 10, 751–765. [Google Scholar] [CrossRef]
- Watson, C.L.; Mahe, M.M.; Múnera, J.; Howell, J.C.; Sundaram, N.; Poling, H.M.; Schweitzer, J.I.; Vallance, J.E.; Mayhew, C.N.; Sun, Y.; et al. An in Vivo Model of Human Small Intestine Using Pluripotent Stem Cells. Nat. Med. 2014, 20, 1310–1314. [Google Scholar] [CrossRef]
- Mohiuddin, M.M.; Singh, A.K.; Scobie, L.; Goerlich, C.E.; Grazioli, A.; Saharia, K.; Crossan, C.; Burke, A.; Drachenberg, C.; Oguz, C.; et al. Graft Dysfunction in Compassionate Use of Genetically Engineered Pig-to-Human Cardiac Xenotransplantation: A Case Report. Lancet 2023, 402, 397–410. [Google Scholar] [CrossRef]
- Hynes, R.O. The Extracellular Matrix: Not Just Pretty Fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The Extracellular Matrix at a Glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.-P.; Stehle, T.; Zhang, R.; Joachimiak, A.; Frech, M.; Goodman, S.L.; Arnaout, M.A. Crystal Structure of the Extracellular Segment of Integrin Alpha Vbeta3 in Complex with an Arg-Gly-Asp Ligand. Science 2002, 296, 151–155. [Google Scholar] [CrossRef]
- Shimaoka, M.; Xiao, T.; Liu, J.-H.; Yang, Y.; Dong, Y.; Jun, C.-D.; McCormack, A.; Zhang, R.; Joachimiak, A.; Takagi, J.; et al. Structures of the Alpha L I Domain and Its Complex with ICAM-1 Reveal a Shape-Shifting Pathway for Integrin Regulation. Cell 2003, 112, 99–111. [Google Scholar] [CrossRef]
- Emsley, J.; Knight, C.G.; Farndale, R.W.; Barnes, M.J.; Liddington, R.C. Structural Basis of Collagen Recognition by Integrin Alpha2beta1. Cell 2000, 101, 47–56. [Google Scholar] [CrossRef]
- Humphries, J.D.; Byron, A.; Humphries, M.J. Integrin Ligands. J. Cell Sci. 2006, 119, 3901–3903. [Google Scholar] [CrossRef] [PubMed]
- Arimori, T.; Miyazaki, N.; Mihara, E.; Takizawa, M.; Taniguchi, Y.; Cabañas, C.; Sekiguchi, K.; Takagi, J. Structural Mechanism of Laminin Recognition by Integrin. Nat. Commun. 2021, 12, 4012. [Google Scholar] [CrossRef]
- Nagase, H.; Visse, R.; Murphy, G. Structure and Function of Matrix Metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef]
- Sternlicht, M.D.; Werb, Z. How Matrix Metalloproteinases Regulate Cell Behavior. Annu. Rev. Cell Dev. Biol. 2001, 17, 463–516. [Google Scholar] [CrossRef]
- Balion, Z.; Cėpla, V.; Svirskiene, N.; Svirskis, G.; Druceikaitė, K.; Inokaitis, H.; Rusteikaitė, J.; Masilionis, I.; Stankevičienė, G.; Jelinskas, T.; et al. Cerebellar Cells Self-Assemble into Functional Organoids on Synthetic, Chemically Crosslinked ECM-Mimicking Peptide Hydrogels. Biomolecules 2020, 10, 754. [Google Scholar] [CrossRef]
- Hernandez-Gordillo, V.; Kassis, T.; Lampejo, A.; Choi, G.; Gamboa, M.E.; Gnecco, J.S.; Brown, A.; Breault, D.T.; Carrier, R.; Griffith, L.G. Fully Synthetic Matrices for in Vitro Culture of Primary Human Intestinal Enteroids and Endometrial Organoids. Biomaterials 2020, 254, 120125. [Google Scholar] [CrossRef] [PubMed]
- Nason-Tomaszewski, C.E.; Thomas, E.E.; Matera, D.L.; Baker, B.M.; Shikanov, A. Extracellular Matrix-Templating Fibrous Hydrogels Promote Ovarian Tissue Remodeling and Oocyte Growth. Bioact. Mater. 2024, 32, 292–303. [Google Scholar] [CrossRef]
- Lam, K.S.; Lebl, M.; Krchňák, V. The “One-Bead-One-Compound” Combinatorial Library Method. Chem. Rev. 1997, 97, 411–448. [Google Scholar] [CrossRef]
- Hao, D.; Xiao, W.; Liu, R.; Kumar, P.; Li, Y.; Zhou, P.; Guo, F.; Farmer, D.L.; Lam, K.S.; Wang, F.; et al. Discovery and Characterization of a Potent and Specific Peptide Ligand Targeting Endothelial Progenitor Cells and Endothelial Cells for Tissue Regeneration. ACS Chem. Biol. 2017, 12, 1075–1086. [Google Scholar] [CrossRef]
- Hao, D.; Liu, R.; Fernandez, T.G.; Pivetti, C.; Jackson, J.E.; Kulubya, E.S.; Jiang, H.-J.; Ju, H.-Y.; Liu, W.-L.; Panitch, A.; et al. A Bioactive Material with Dual Integrin-Targeting Ligands Regulates Specific Endogenous Cell Adhesion and Promotes Vascularized Bone Regeneration in Adult and Fetal Bone Defects. Bioact. Mater. 2023, 20, 179–193. [Google Scholar] [CrossRef]
- Sarig, U.; Sarig, H.; Gora, A.; Krishnamoorthi, M.K.; Au-Yeung, G.C.T.; de-Berardinis, E.; Chaw, S.Y.; Mhaisalkar, P.; Bogireddi, H.; Ramakrishna, S.; et al. Biological and Mechanical Interplay at the Macro- and Microscales Modulates the Cell-Niche Fate. Sci. Rep. 2018, 8, 3937. [Google Scholar] [CrossRef] [PubMed]
- Assunção, M.; Dehghan-Baniani, D.; Yiu, C.H.K.; Später, T.; Beyer, S.; Blocki, A. Cell-Derived Extracellular Matrix for Tissue Engineering and Regenerative Medicine. Front. Bioeng. Biotechnol. 2020, 8, 602009. [Google Scholar] [CrossRef] [PubMed]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An Overview of Tissue and Whole Organ Decellularization Processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef]
- Abaci, A.; Guvendiren, M. Designing Decellularized Extracellular Matrix-Based Bioinks for 3D Bioprinting. Adv. Healthc. Mater. 2020, 9, 2000734. [Google Scholar] [CrossRef]
- Hussey, G.S.; Nascari, D.G.; Saldin, L.T.; Kolich, B.; Lee, Y.C.; Crum, R.J.; El-Mossier, S.O.; D’Angelo, W.; Dziki, J.L.; Badylak, S.F. Ultrasonic Cavitation to Prepare ECM Hydrogels. Acta Biomater. 2020, 108, 77–86. [Google Scholar] [CrossRef] [PubMed]
- VeDepo, M.C.; Detamore, M.S.; Hopkins, R.A.; Converse, G.L. Recellularization of Decellularized Heart Valves: Progress toward the Tissue-Engineered Heart Valve. J. Tissue Eng. 2017, 8, 2041731417726327. [Google Scholar] [CrossRef] [PubMed]
- Tenreiro, M.F.; Almeida, H.V.; Calmeiro, T.; Fortunato, E.; Ferreira, L.; Alves, P.M.; Serra, M. Interindividual Heterogeneity Affects the Outcome of Human Cardiac Tissue Decellularization. Sci. Rep. 2021, 11, 20834. [Google Scholar] [CrossRef] [PubMed]
- Simsa, R.; Rothenbücher, T.; Gürbüz, H.; Ghosheh, N.; Emneus, J.; Jenndahl, L.; Kaplan, D.L.; Bergh, N.; Serrano, A.M.; Fogelstrand, P. Brain Organoid Formation on Decellularized Porcine Brain ECM Hydrogels. PLoS ONE 2021, 16, e0245685. [Google Scholar] [CrossRef] [PubMed]
- Chani, B.; Puri, V.; Sobti, R.C.; Jha, V.; Puri, S. Decellularized Scaffold of Cryopreserved Rat Kidney Retains Its Recellularization Potential. PLoS ONE 2017, 12, e0173040. [Google Scholar] [CrossRef] [PubMed]
- Uday Chandrika, K.; Tripathi, R.; Kameshwari, Y.; Rangaraj, N.; Mahesh Kumar, J.; Singh, S. Refunctionalization of Decellularized Organ Scaffold of Pancreas by Recellularization: Whole Organ Regeneration into Functional Pancreas. Tissue Eng. Regen. Med. 2020, 18, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Sarmin, A.M.; Connelly, J.T. Fabrication of Human Skin Equivalents Using Decellularized Extracellular Matrix. Curr. Protoc. 2022, 2, e393. [Google Scholar] [CrossRef] [PubMed]
- Marques-Magalhães; Cruz, T.; Costa, M.; Estêvão, D.; Rios, E.; Canão, P.A.; Velho, S.; Carneiro, F.; Oliveira, M.J.; Cardoso, A.P. Decellularized Colorectal Cancer Matrices as Bioactive Scaffolds for Studying Tumor-Stroma Interactions. Cancers 2022, 14, 359. [Google Scholar] [CrossRef] [PubMed]
- van Tienderen, G.S.; van Beek, M.E.A.; Schurink, I.J.; Rosmark, O.; Roest, H.P.; Tieleman, J.; Demmers, J.; Muntz, I.; Conboy, J.; Westergren-Thorsson, G.; et al. Modelling Metastatic Colonization of Cholangiocarcinoma Organoids in Decellularized Lung and Lymph Nodes. Front. Oncol. 2022, 12, 1101901. [Google Scholar] [CrossRef] [PubMed]
- Dahms, S.E.; Piechota, H.J.; Dahiya, R.; Lue, T.F.; Tanagho, E.A. Composition and Biomechanical Properties of the Bladder Acellular Matrix Graft: Comparative Analysis in Rat, Pig and Human. Br. J. Urol. 1998, 82, 411–419. [Google Scholar] [CrossRef]
- Golebiowska, A.A.; Intravaia, J.T.; Sathe, V.M.; Kumbar, S.G.; Nukavarapu, S.P. Decellularized Extracellular Matrix Biomaterials for Regenerative Therapies: Advances, Challenges and Clinical Prospects. Bioact. Mater. 2024, 32, 98–123. [Google Scholar] [CrossRef]
- McCabe, M.C.; Hill, R.C.; Calderone, K.; Cui, Y.; Yan, Y.; Quan, T.; Fisher, G.J.; Hansen, K.C. Alterations in Extracellular Matrix Composition during Aging and Photoaging of the Skin. Matrix Biol. Plus 2020, 8, 100041. [Google Scholar] [CrossRef] [PubMed]
- Neishabouri, A.; Soltani Khaboushan, A.; Daghigh, F.; Kajbafzadeh, A.-M.; Majidi Zolbin, M. Decellularization in Tissue Engineering and Regenerative Medicine: Evaluation, Modification, and Application Methods. Front. Bioeng. Biotechnol. 2022, 10, 805299. [Google Scholar] [CrossRef] [PubMed]
- Sutlive, J.; Xiu, H.; Chen, Y.; Gou, K.; Xiong, F.; Guo, M.; Chen, Z. Generation, Transmission, and Regulation of Mechanical Forces in Embryonic Morphogenesis. Small 2022, 18, e2103466. [Google Scholar] [CrossRef] [PubMed]
- Ogoke, O.; Guiggey, D.; Mon, T.; Shamul, C.; Ross, S.; Rao, S.; Parashurama, N. Spatiotemporal Imaging and Analysis of Mouse and Human Liver Bud Morphogenesis. Dev. Dyn. 2022, 251, 662–686. [Google Scholar] [CrossRef] [PubMed]
- Shellard, A.; Mayor, R. Collective Durotaxis along a Self-Generated Stiffness Gradient in Vivo. Nature 2021, 600, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Abdel Fattah, A.R.; Daza, B.; Rustandi, G.; Berrocal-Rubio, M.Á.; Gorissen, B.; Poovathingal, S.; Davie, K.; Barrasa-Fano, J.; Cóndor, M.; Cao, X.; et al. Actuation Enhances Patterning in Human Neural Tube Organoids. Nat. Commun. 2021, 12, 3192. [Google Scholar] [CrossRef] [PubMed]
- Qazi, T.H.; Blatchley, M.R.; Davidson, M.D.; Yavitt, F.M.; Cooke, M.E.; Anseth, K.S.; Burdick, J.A. Programming Hydrogels to Probe Spatiotemporal Cell Biology. Cell Stem Cell 2022, 29, 678–691. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, D.D.; Brown, T.E.; Kyburz, K.A.; Kiyotake, E.; Anseth, K.S. Design and Characterization of a Synthetically Accessible, Photodegradable Hydrogel for User-Directed Formation of Neural Networks. Biomacromolecules 2014, 15, 2808–2816. [Google Scholar] [CrossRef] [PubMed]
- Yavitt, F.M.; Brown, T.E.; Hushka, E.A.; Brown, M.E.; Gjorevski, N.; Dempsey, P.J.; Lutolf, M.P.; Anseth, K.S. The Effect of Thiol Structure on Allyl Sulfide Photodegradable Hydrogels and Their Application as a Degradable Scaffold for Organoid Passaging. Adv. Mater. 2020, 32, e1905366. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.E.; Anseth, K.S. Spatiotemporal Hydrogel Biomaterials for Regenerative Medicine. Chem. Soc. Rev. 2017, 46, 6532–6552. [Google Scholar] [CrossRef]
- Lee, S.-H.; Moon, J.J.; West, J.L. Three-Dimensional Micropatterning of Bioactive Hydrogels via Two-Photon Laser Scanning Photolithography for Guided 3D Cell Migration. Biomaterials 2008, 29, 2962–2968. [Google Scholar] [CrossRef] [PubMed]
- DeForest, C.A.; Polizzotti, B.D.; Anseth, K.S. Sequential Click Reactions for Synthesizing and Patterning Three-Dimensional Cell Microenvironments. Nat. Mater. 2009, 8, 659–664. [Google Scholar] [CrossRef]
- Ao, Z.; Cai, H.; Wu, Z.; Ott, J.; Wang, H.; Mackie, K.; Guo, F. Controllable Fusion of Human Brain Organoids Using Acoustofluidics. Lab. Chip 2021, 21, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Abdel Fattah, A.R.; Kolaitis, N.; Van Daele, K.; Daza, B.; Rustandi, A.G.; Ranga, A. Targeted Mechanical Stimulation via Magnetic Nanoparticles Guides in Vitro Tissue Development. Nat. Commun. 2023, 14, 5281. [Google Scholar] [CrossRef] [PubMed]
- Blatchley, M.R.; Anseth, K.S. Middle-out Methods for Spatiotemporal Tissue Engineering of Organoids. Nat. Rev. Bioeng. 2023, 1, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Saunders, T.E. A Matter of Time: Formation and Interpretation of the Bicoid Morphogen Gradient. Curr. Top. Dev. Biol. 2020, 137, 79–117. [Google Scholar] [CrossRef] [PubMed]
- Lucas, T.; Tran, H.; Perez Romero, C.A.; Guillou, A.; Fradin, C.; Coppey, M.; Walczak, A.M.; Dostatni, N. 3 Minutes to Precisely Measure Morphogen Concentration. PLoS Genet. 2018, 14, e1007676. [Google Scholar] [CrossRef] [PubMed]
- DeForest, C.A.; Anseth, K.S. Photoreversible Patterning of Biomolecules within Click-Based Hydrogels. Angew. Chem. Int. Ed Engl. 2012, 51, 1816–1819. [Google Scholar] [CrossRef]
- Attayek, P.J.; Ahmad, A.A.; Wang, Y.; Williamson, I.; Sims, C.E.; Magness, S.T.; Allbritton, N.L. In Vitro Polarization of Colonoids to Create an Intestinal Stem Cell Compartment. PLoS ONE 2016, 11, e0153795. [Google Scholar] [CrossRef]
- Nader, G.P.; de, F.; Williart, A.; Piel, M. Nuclear Deformations, from Signaling to Perturbation and Damage. Curr. Opin. Cell Biol. 2021, 72, 137–145. [Google Scholar] [CrossRef]
- Guo, Q.; Wang, X.; Tibbitt, M.W.; Anseth, K.S.; Montell, D.J.; Elisseeff, J.H. Light Activated Cell Migration in Synthetic Extracellular Matrices. Biomaterials 2012, 33, 8040–8046. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ara, G.; Taberner, N.; Takayama, M.; Sandaltzopoulou, E.; Villava, C.E.; Bosch-Padrós, M.; Takata, N.; Trepat, X.; Eiraku, M.; Ebisuya, M. Optogenetic Control of Apical Constriction Induces Synthetic Morphogenesis in Mammalian Tissues. Nat. Commun. 2022, 13, 5400. [Google Scholar] [CrossRef] [PubMed]
- Chrisnandy, A.; Blondel, D.; Rezakhani, S.; Broguiere, N.; Lutolf, M.P. Synthetic Dynamic Hydrogels Promote Degradation-Independent in Vitro Organogenesis. Nat. Mater. 2022, 21, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Puppi, J.; Strom, S.C.; Hughes, R.D.; Bansal, S.; Castell, J.V.; Dagher, I.; Ellis, E.C.S.; Nowak, G.; Ericzon, B.-G.; Fox, I.J.; et al. Improving the Techniques for Human Hepatocyte Transplantation: Report from a Consensus Meeting in London. Cell Transplant. 2012, 21, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lanzoni, G.; Hani, H.; Overi, D.; Cardinale, V.; Simpson, S.; Pitman, W.; Allen, A.; Yi, X.; Wang, X.; et al. Patch Grafting, Strategies for Transplantation of Organoids into Solid Organs Such as Liver. Biomaterials 2021, 277, 121067. [Google Scholar] [CrossRef]
- Chatterjee, D.; Ing, R.J.; Gien, J. Update on Congenital Diaphragmatic Hernia. Anesth. Analg. 2020, 131, 808. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.M.J.; Lewandowski, A.J. The Transitional Heart: From Early Embryonic and Fetal Development to Neonatal Life. Fetal Diagn. Ther. 2020, 47, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Baschat, A.A.; Galan, H.L.; Lee, W.; DeVore, G.R.; Mari, G.; Hobbins, J.; Vintzileos, A.; Platt, L.D.; Manning, F.A. The Role of the Fetal Biophysical Profile in the Management of Fetal Growth Restriction. Am. J. Obstet. Gynecol. 2022, 226, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Thapa, B.R.; Le Suer, J.A.; Tilston-Lünel, A.; Herriges, M.J.; Berical, A.; Beermann, M.L.; Wang, F.; Bawa, P.S.; Kohn, A.; et al. Airway Stem Cell Reconstitution by the Transplantation of Primary or Pluripotent Stem Cell-Derived Basal Cells. Cell Stem Cell 2023, 30, 1199–1216.e7. [Google Scholar] [CrossRef]
- Search for: Other Terms: Organoid|List Results|ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/search?term=organoid&viewType=Table (accessed on 5 February 2024).
- Guan, X.; Huang, S. Advances in the Application of 3D Tumor Models in Precision Oncology and Drug Screening. Front. Bioeng. Biotechnol. 2022, 10, 1021966. [Google Scholar] [CrossRef]
- Kagan, B.J.; Kitchen, A.C.; Tran, N.T.; Habibollahi, F.; Khajehnejad, M.; Parker, B.J.; Bhat, A.; Rollo, B.; Razi, A.; Friston, K.J. In Vitro Neurons Learn and Exhibit Sentience When Embodied in a Simulated Game-World. Neuron 2022, 110, 3952–3969.e8. [Google Scholar] [CrossRef] [PubMed]
- Li, T.L.; Liu, Y.; Forro, C.; Yang, X.; Beker, L.; Bao, Z.; Cui, B.; Pașca, S.P. Stretchable Mesh Microelectronics for the Biointegration and Stimulation of Human Neural Organoids. Biomaterials 2022, 290, 121825. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, X.; Zhang, X.; Wang, Y.; Pei, W. Implantable Intracortical Microelectrodes: Reviewing the Present with a Focus on the Future. Microsyst. Nanoeng. 2023, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, W.; Zhou, W. Computational Methods for Single-Cell DNA Methylome Analysis. Genom. Proteom. Bioinform. 2023, 21, 48–66. [Google Scholar] [CrossRef] [PubMed]
- Hennig, K.; Wang, I.; Moreau, P.; Valon, L.; DeBeco, S.; Coppey, M.; Miroshnikova, Y.A.; Albiges-Rizo, C.; Favard, C.; Voituriez, R.; et al. Stick-Slip Dynamics of Cell Adhesion Triggers Spontaneous Symmetry Breaking and Directional Migration of Mesenchymal Cells on One-Dimensional Lines. Sci. Adv. 2020, 6, eaau5670. [Google Scholar] [CrossRef] [PubMed]
- Rapp, T.L.; DeForest, C.A. Tricolor Visible Wavelength-Selective Photodegradable Hydrogel Biomaterials. Nat. Commun. 2023, 14, 5250. [Google Scholar] [CrossRef] [PubMed]
- Blūms, V.; Piotrowski, M.; Hussain, M.I.; Norton, B.G.; Connell, S.C.; Gensemer, S.; Lobino, M.; Streed, E.W. A Single-Atom 3D Sub-Attonewton Force Sensor. Sci. Adv. 2018, 4, eaao4453. [Google Scholar] [CrossRef] [PubMed]
- Salari, V.; Moradi, N.; Sajadi, M.; Fazileh, F.; Shahbazi, F. Quantum Decoherence Time Scales for Ionic Superposition States in Ion Channels. Phys. Rev. E 2015, 91, 032704. [Google Scholar] [CrossRef]
- Barjas Qaswal, A. Quantum Tunneling of Ions through the Closed Voltage-Gated Channels of the Biological Membrane: A Mathematical Model and Implications. Quantum Rep. 2019, 1, 219–225. [Google Scholar] [CrossRef]
- Qaswal, A. A Theoretical Study to Explain the Referred Pain Phenomenon and Its Characteristics via Quantum Tunneling of Potassium Ions Through the Channels of Neuronal Membrane. NeuroQuantology 2019, 17, 43–52. [Google Scholar] [CrossRef]
- Kim, Y.; Bertagna, F.; D’Souza, E.M.; Heyes, D.J.; Johannissen, L.O.; Nery, E.T.; Pantelias, A.; Sanchez-Pedreño Jimenez, A.; Slocombe, L.; Spencer, M.G.; et al. Quantum Biology: An Update and Perspective. Quantum Rep. 2021, 3, 80–126. [Google Scholar] [CrossRef]
- Hameroff, S.; Penrose, R. Consciousness in the Universe: A Review of the “Orch OR” Theory. Phys. Life Rev. 2014, 11, 39–78. [Google Scholar] [CrossRef] [PubMed]
- Contreras, M.; Artigas-Jerónimo, S.; Pastor Comín, J.J.; de la Fuente, J. A Quantum Vaccinomics Approach Based on Protein-Protein Interactions. In Methods in Molecular Biology; MIMB: Salt Lake City, UT, USA, 2022; Volume 2411, pp. 287–305. [Google Scholar] [CrossRef]
- Boeger, H. Kinetic Proofreading. Annu. Rev. Biochem. 2022, 91, 423–447. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, A.; Stumpf, M.P. Non-equilibrium statistical physics, transitory epigenetic landscapes, and cell fate decision dynamics. Math Biosci Eng. 2020, 17, 7916–7930. [Google Scholar] [CrossRef]
- Orlandini, E.; Micheletti, C. Topological and Physical Links in Soft Matter Systems. J. Phys. Condens. Matter 2021, 34, 013002. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wyle, Y.; Lu, N.; Hepfer, J.; Sayal, R.; Martinez, T.; Wang, A. The Role of Biophysical Factors in Organ Development: Insights from Current Organoid Models. Bioengineering 2024, 11, 619. https://doi.org/10.3390/bioengineering11060619
Wyle Y, Lu N, Hepfer J, Sayal R, Martinez T, Wang A. The Role of Biophysical Factors in Organ Development: Insights from Current Organoid Models. Bioengineering. 2024; 11(6):619. https://doi.org/10.3390/bioengineering11060619
Chicago/Turabian StyleWyle, Yofiel, Nathan Lu, Jason Hepfer, Rahul Sayal, Taylor Martinez, and Aijun Wang. 2024. "The Role of Biophysical Factors in Organ Development: Insights from Current Organoid Models" Bioengineering 11, no. 6: 619. https://doi.org/10.3390/bioengineering11060619
APA StyleWyle, Y., Lu, N., Hepfer, J., Sayal, R., Martinez, T., & Wang, A. (2024). The Role of Biophysical Factors in Organ Development: Insights from Current Organoid Models. Bioengineering, 11(6), 619. https://doi.org/10.3390/bioengineering11060619








