Machine Learning-Enhanced Estimation of Cellular Protein Levels from Bright-Field Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Immunostaining
2.3. Imaging for AI Model
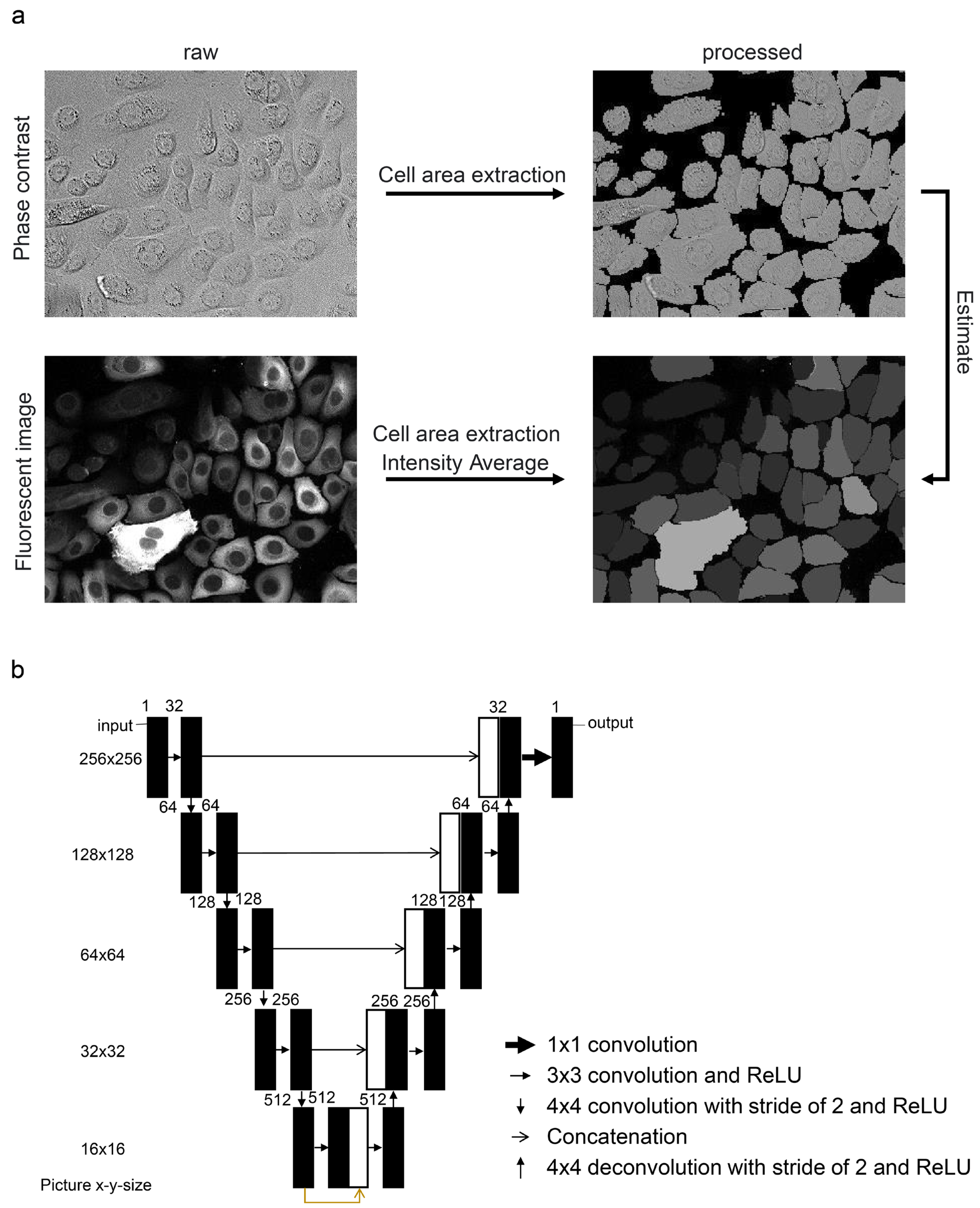
2.4. Image Processing and Machine Learning
2.5. Time-Lapse Imaging
2.6. Statistical Analysis
3. Results
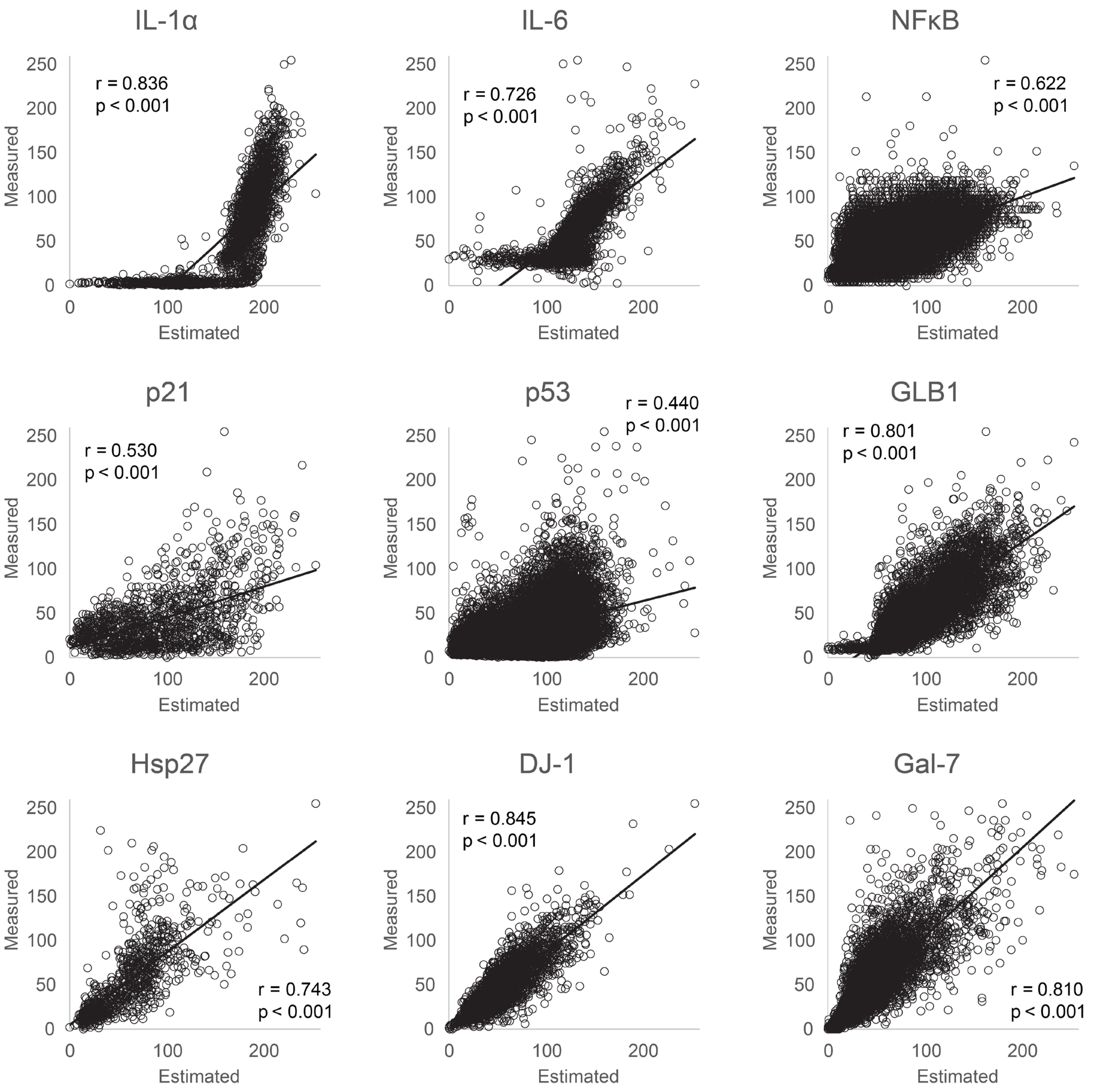
3.1. Evaluation of Protein Estimation Accuracy Using Machine Learning
3.2. Applicability of the Protein Expression Estimation Model to Live NHEKs before Fixation
3.3. Integration of Machine Learning Models into Live Imaging
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gorgoulis, V.G.; Pefani, D.-E.; Pateras, I.S.; Trougakos, I.P. Integrating the DNA Damage and Protein Stress Responses during Cancer Development and Treatment. J. Pathol. 2018, 246, 12–40. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Paterlini-Brechot, P.; Benali, N.L. Circulating Tumor Cells (CTC) Detection: Clinical Impact and Future Directions. Cancer Lett. 2007, 253, 180–204. [Google Scholar] [CrossRef] [PubMed]
- Proserpio, V.; Lönnberg, T. Single-Cell Technologies Are Revolutionizing the Approach to Rare Cells. Immunol. Cell Biol. 2016, 94, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G. Propagation of Tau Aggregates. Mol. Brain 2017, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-C.; Sun, X.-F.; Dyce, P.W.; Shen, W.; Chen, H. The Role of Germ Cell Loss During Primordial Follicle Assembly: A Review of Current Advances. Int. J. Biol. Sci. 2017, 13, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, M.; Hamm, J.P.; Peterka, D.S.; Yuste, R. Acute Focal Seizures Start as Local Synchronizations of Neuronal Ensembles. J. Neurosci. 2019, 39, 8562–8575. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, T.; Kakizuka, T.; Horikawa, K.; Seiriki, K.; Kasai, A.; Hashimoto, H.; Fujita, K.; Watanabe, T.M.; Nagai, T. Exploring Rare Cellular Activity in More Than One Million Cells by a Transscale Scope. Sci. Rep. 2021, 11, 16539. [Google Scholar] [CrossRef] [PubMed]
- Wiley, C.D.; Campisi, J. From Ancient Pathways to Aging Cells–Connecting Metabolism and Cellular Senescence. Cell Metab. 2016, 23, 1013–1021. [Google Scholar] [CrossRef]
- Chiche, A.; Le Roux, I.; von Joest, M.; Sakai, H.; Aguín, S.B.; Cazin, C.; Salam, R.; Fiette, L.; Alegria, O.; Flamant, P.; et al. Injury-Induced Senescence Enables In Vivo Reprogramming in Skeletal Muscle. Cell Stem Cell 2017, 20, 407–414.e4. [Google Scholar] [CrossRef]
- He, S.; Sharpless, N.E. Senescence in Health and Disease. Cell 2017, 169, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Faget, D.V.; Ren, Q.; Stewart, S.A. Unmasking Senescence: Context-Dependent Effects of SASP in Cancer. Nat. Rev. Cancer 2019, 19, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Fane, M.; Weeraratna, A.T. How the Ageing Microenvironment Influences Tumour Progression. Nat. Rev. Cancer 2020, 20, 89–106. [Google Scholar] [CrossRef] [PubMed]
- Loo, T.M.; Miyata, K.; Tanaka, Y.; Takahashi, A. Cellular Senescence and Senescence-Associated Secretory Phenotype via the cGAS-STING Signaling Pathway in Cancer. Cancer Sci. 2020, 111, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Salomon, R.; Kaczorowski, D.; Valdes-Mora, F.; Nordon, R.E.; Neild, A.; Farbehi, N.; Bartonicek, N.; Gallego-Ortega, D. Droplet-Based Single Cell RNAseq Tools: A Practical Guide. Lab Chip 2019, 19, 1706–1727. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks; Springer International Publishing: Midtown Manhattan, NY, USA, 2014; pp. 818–833. [Google Scholar]
- Christiansen, E.M.; Yang, S.J.; Ando, D.M.; Javaherian, A.; Skibinski, G.; Lipnick, S.; Mount, E.; O’Neil, A.; Shah, K.; Lee, A.K.; et al. In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images. Cell 2018, 173, 792–803.e19. [Google Scholar] [CrossRef]
- Tohgasaki, T.; Aihara, S.; Ikeda, M.; Takahashi, M.; Eto, M.; Kudo, R.; Taira, H.; Kido, A.; Kondo, S.; Ishiwatari, S. Investigation of Stratum Corneum Cell Morphology and Content Using Novel Machine-Learning Image Analysis. Skin Res. Technol. 2024, 30, e13565. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015, Proceedings, Part III 18; Springer: Berlin/Heidelberg, Germany, 2015; Volume 2015, pp. 234–241. [Google Scholar]
- Kusumoto, D.; Seki, T.; Sawada, H.; Kunitomi, A.; Katsuki, T.; Kimura, M.; Ito, S.; Komuro, J.; Hashimoto, H.; Fukuda, K.; et al. Anti-senescent Drug Screening by Deep Learning-Based Morphology Senescence Scoring. Nat. Commun. 2021, 12, 257. [Google Scholar] [CrossRef]
- Imai, Y.; Iida, M.; Kanie, K.; Katsuno, M.; Kato, R. Label-Free Morphological Sub-Population Cytometry for Sensitive Phenotypic Screening of Heterogenous Neural Disease Model Cells. Sci. Rep. 2022, 12, 9296. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar] [CrossRef]
- Wang, A.S.; Dreesen, O. Biomarkers of Cellular Senescence and Skin Aging. Front. Genet. 2018, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Quaresma, J.A.S. Organization of the Skin Immune System and Compartmentalized Immune Responses in Infectious Diseases. Clin. Microbiol. Rev. 2019, 32, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Kadono-Maekubo, N.; Suzuki, Y.; Furuichi, Y.; Shiraga, K.; Sasaki, H.; Ishida, A.; Takahashi, S.; Okada, T.; Toyooka, K.; et al. A Unique Mode of Keratinocyte Death Requires Intracellular Acidification. Proc. Natl. Acad. Sci. USA 2021, 118, e2020722118. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tohgasaki, T.; Touyama, A.; Kousai, S.; Imai, K. Machine Learning-Enhanced Estimation of Cellular Protein Levels from Bright-Field Images. Bioengineering 2024, 11, 774. https://doi.org/10.3390/bioengineering11080774
Tohgasaki T, Touyama A, Kousai S, Imai K. Machine Learning-Enhanced Estimation of Cellular Protein Levels from Bright-Field Images. Bioengineering. 2024; 11(8):774. https://doi.org/10.3390/bioengineering11080774
Chicago/Turabian StyleTohgasaki, Takeshi, Arisa Touyama, Shohei Kousai, and Kaita Imai. 2024. "Machine Learning-Enhanced Estimation of Cellular Protein Levels from Bright-Field Images" Bioengineering 11, no. 8: 774. https://doi.org/10.3390/bioengineering11080774
APA StyleTohgasaki, T., Touyama, A., Kousai, S., & Imai, K. (2024). Machine Learning-Enhanced Estimation of Cellular Protein Levels from Bright-Field Images. Bioengineering, 11(8), 774. https://doi.org/10.3390/bioengineering11080774





