Evolution of Retinal Neuron Fractality When Interfacing with Carbon Nanotube Electrodes
Abstract
1. Introduction
2. Background
3. Materials and Methods
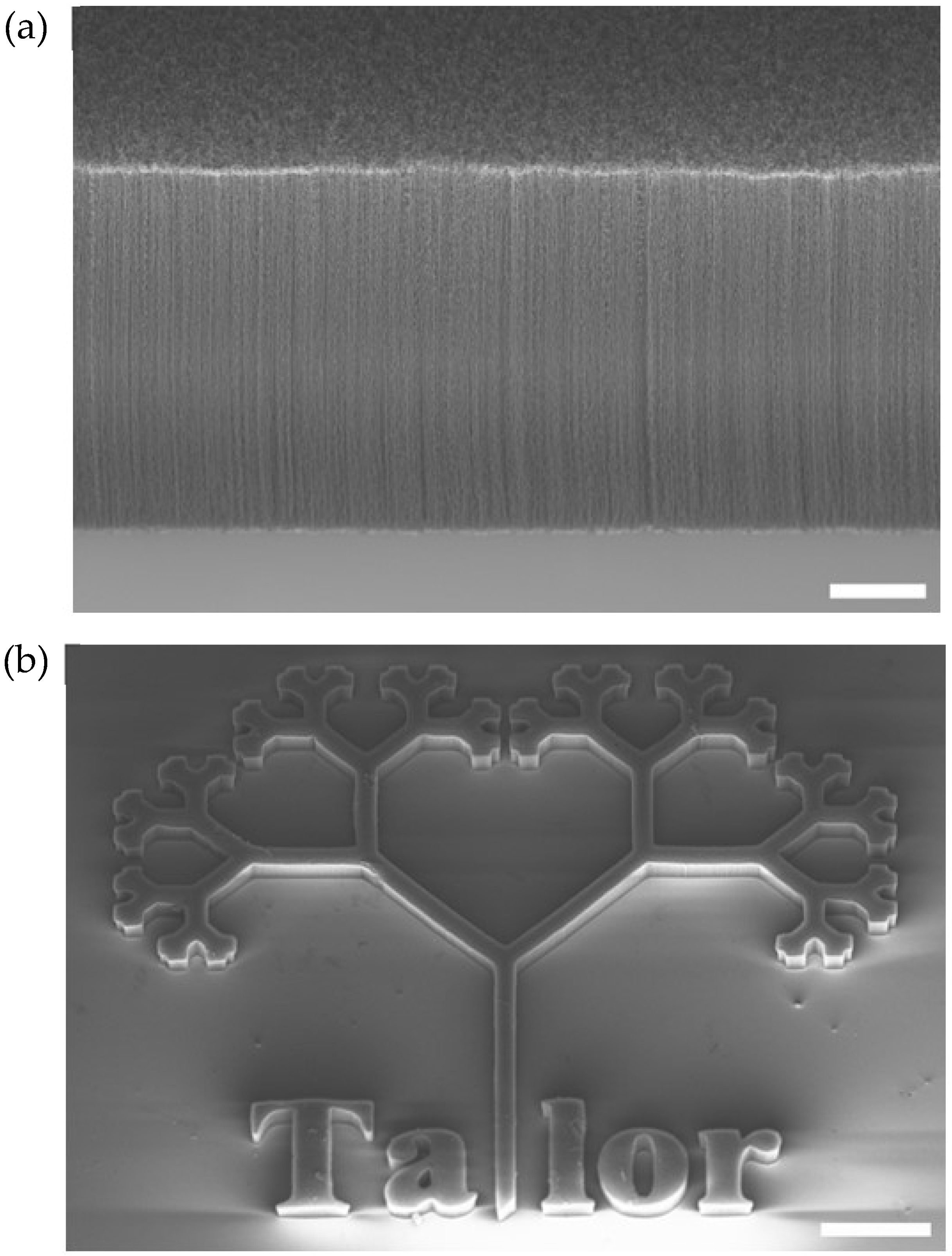
3.1. Electrode Fabrication and Characterization
3.2. Dissociated Retinal Cell Cultures
3.3. Immunocytochemistry
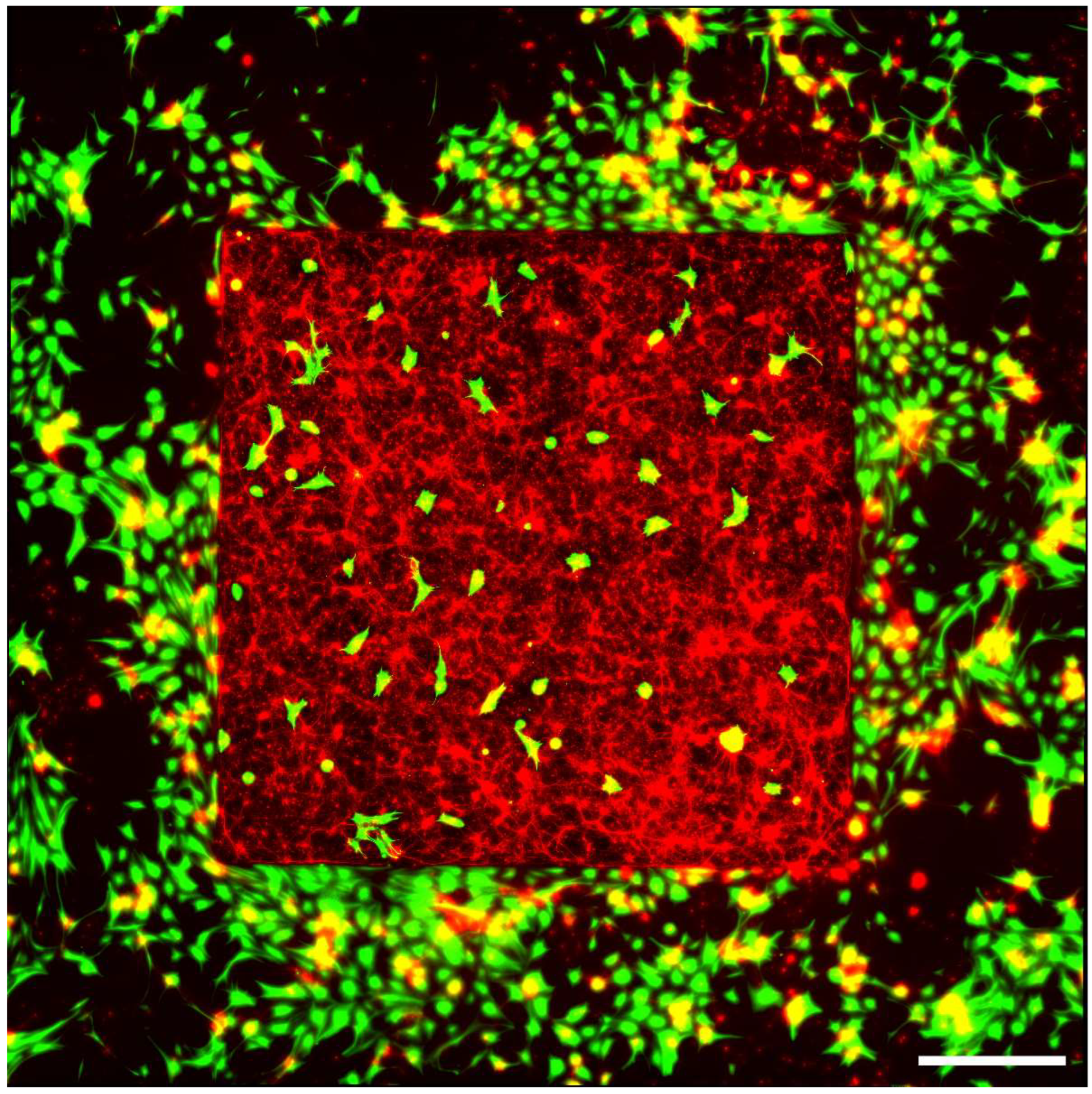
3.4. Fluorescence Microscopy
3.5. Post-Culture SEM Imaging
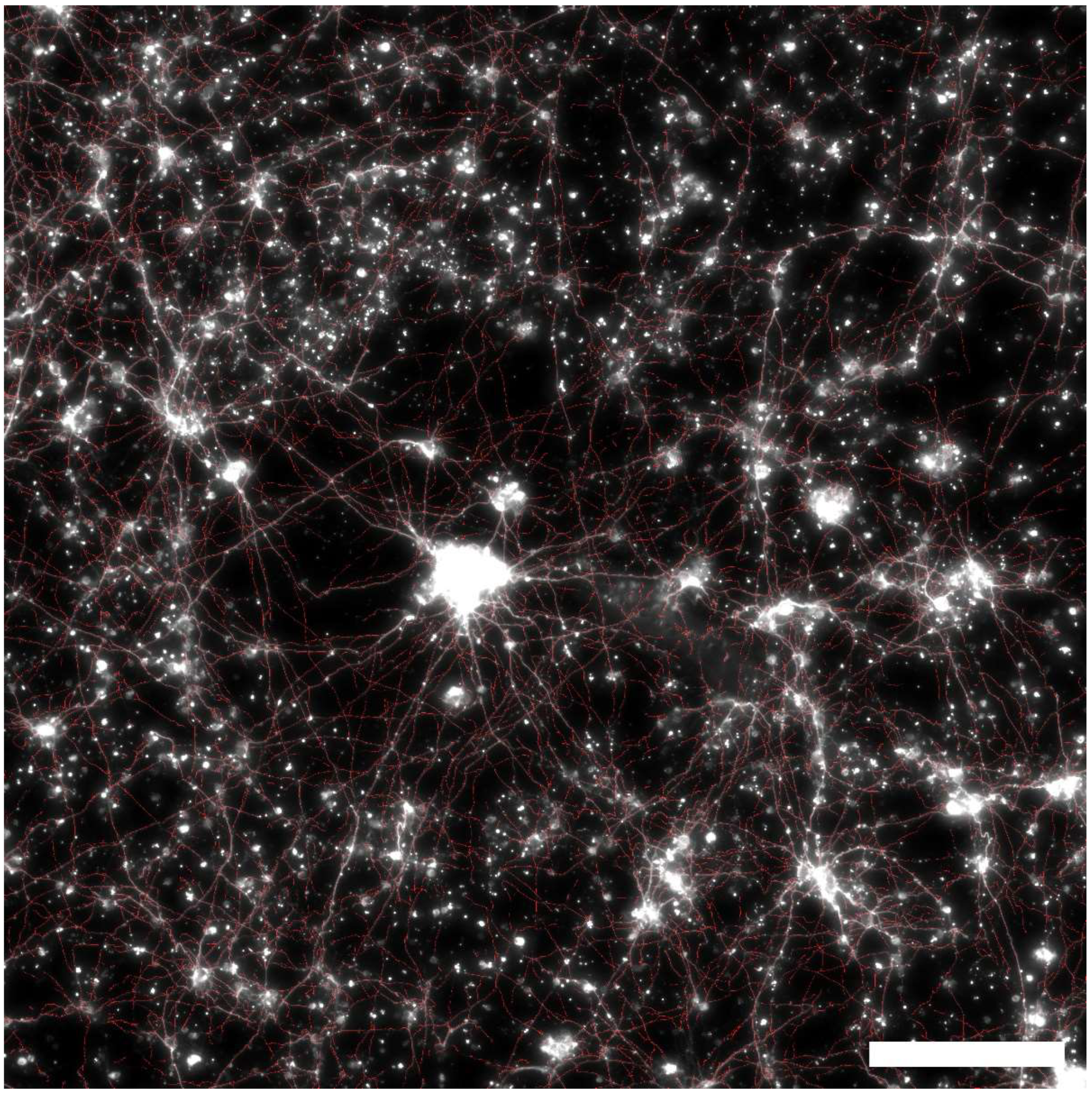
3.6. Alignment Analysis of Cultured Neurons
3.7. Quantitative Measurement of Neuron Process Length NCNT
3.8. Quantitative Measurement of Fractal Dimension D and Neuron Arbor Coverage S
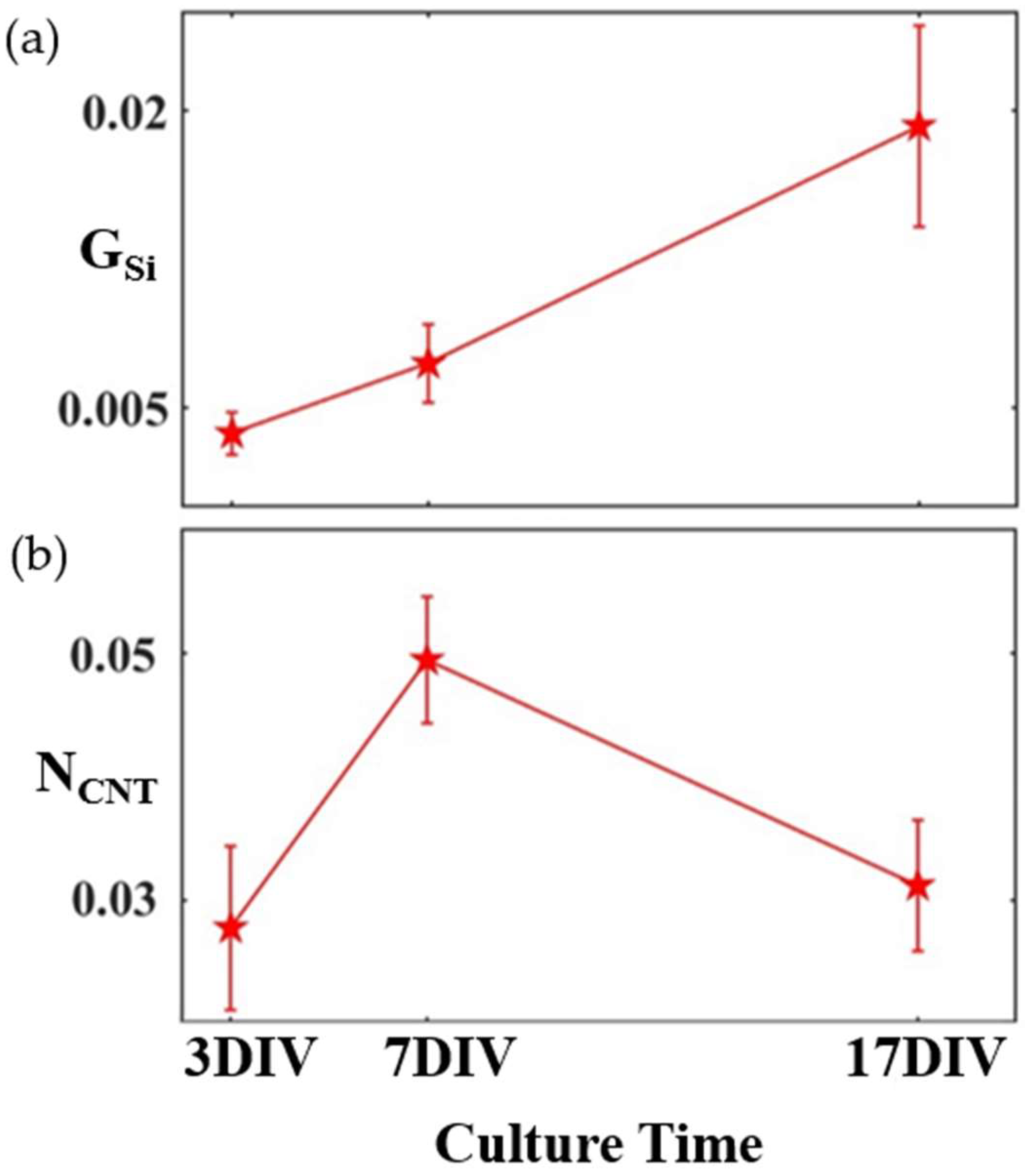
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, D.J.; Lozano, C.S.; Dallapiazza, R.F.; Lozano, A.M. Current and future directions of deep brain stimulation for neurological and psychiatric disorders. J. Neurosurg. 2019, 131, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Kim, H.; Song, Y.M.; Park, J.-U. Implantation of electronic visual prosthesis for blindness restoration. Opt. Mater. Express 2019, 9, 3878. [Google Scholar] [CrossRef]
- Chenais, N.A.L.; Airaghi Leccardi, M.J.I.; Ghezzi, D. Photovoltaic retinal prosthesis restores high-resolution responses to single-pixel stimulation in blind retinas. Commun. Mater. 2021, 2, 28. [Google Scholar] [CrossRef]
- Prévot, P.-H.; Gehere, K.; Arcizet, F.; Akolkar, H.; Khoei, M.A.; Blaize, K.; Oubari, O.; Daye, P.; Lanoë, M.; Valet, M.; et al. Behavioural responses to a photovoltaic subretinal prosthesis implanted in non-human primates. Nat. Biomed. Eng. 2019, 4, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Meffin, H.; Garrett, D.J.; Ibbotson, M.R. Stimulation Strategies for Improving the Resolution of Retinal Prostheses. Front. Neurosci. 2020, 14, 262. [Google Scholar] [CrossRef]
- Lorach, H.; Goetz, G.; Smith, R.; Lei, X.; Mandel, Y.; Kamins, T.; Mathieson, K.; Huie, P.; Harris, J.; Sher, A.; et al. Photovoltaic restoration of sight with high visual acuity. Nat. Med. 2015, 21, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Palanker, D.; Le Mer, Y.; Mohand-Said, S.; Muqit, M.; Sahel, J.A. Photovoltaic Restoration of Central Vision in Atrophic Age-Related Macular Degeneration. Ophthalmology 2020, 127, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Palanker, D.; Le Mer, Y.; Mohand-Said, S.; Sahel, J.A. Simultaneous perception of prosthetic and natural vision in AMD patients. Nat. Commun. 2022, 13, 513. [Google Scholar] [CrossRef] [PubMed]
- Edwards, T.L.; Cottriall, C.L.; Xue, K.; Simunovic, M.P.; Ramsden, J.D.; Zrenner, E.; Maclaren, R.E. Assessment of the Electronic Retinal Implant Alpha AMS in Restoring Vision to Blind Patients with End-Stage Retinitis Pigmentosa. Ophthalmology 2018, 125, 432–443. [Google Scholar] [CrossRef]
- Yang, R.; Zhao, P.; Wang, L.; Feng, C.; Peng, C.; Wang, Z.; Zhang, Y.; Shen, M.; Shi, K.; Weng, S.; et al. Assessment of visual function in blind mice and monkeys with subretinally implanted nanowire arrays as artificial photoreceptors. Nat. Biomed. Eng. 2023, 1–22. [Google Scholar] [CrossRef]
- Wang, B.-Y.; Chen, Z.C.; Bhuckory, M.; Huang, T.; Shin, A.; Zuckerman, V.; Ho, E.; Rosenfeld, E.; Galambos, L.; Kamins, T.; et al. Electronic photoreceptors enable prosthetic visual acuity matching the natural resolution in rats. Nat. Commun. 2022, 13, 6627. [Google Scholar] [CrossRef]
- Von Bartheld, C.S.; Bahney, J.; Herculano-Houzel, S. The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. J. Comp. Neurol. 2016, 524, 3865–3895. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Ben, A. Astrocyte heterogeneity: An underappreciated topic in neurobiology. Curr. Opin. Neurobiol. 2010, 20, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, A.; Pannicke, T.; Grosche, J.; Francke, M.; Wiedemann, P.; Skatchkov, S.N.; Osborne, N.N.; Reichenbach, A. Müller cells in the healthy and diseased retina. Prog. Retin. Eye Res. 2006, 25, 397–424. [Google Scholar] [CrossRef]
- Verkhratsky, A.; Butt, A. Morphology of Glial Cells. In Glial Neurobiology; John Wiley & Sons: Hoboken, NJ, USA, 2007; pp. 21–28. [Google Scholar]
- Moslehi, S.; Rowland, C.; Smith, J.H.; Watterson, W.J.; Griffiths, W.; Montgomery, R.D.; Philliber, S.; Marlow, C.A.; Perez, M.T.; Taylor, R.P. Fractal Electronics for Stimulating and Sensing Neural Networks: Enhanced Electrical, Optical, and Cell Interaction Properties; Springer International Publishing: Berlin/Heidelberg, Germany, 2024; pp. 849–875. [Google Scholar]
- Smith, J.H.; Rowland, C.; Harland, B.; Moslehi, S.; Montgomery, R.D.; Schobert, K.; Watterson, W.J.; Dalrymple-Alford, J.; Taylor, R.P. How neurons exploit fractal geometry to optimize their network connectivity. Sci. Rep. 2021, 11, 2332. [Google Scholar] [CrossRef]
- Rowland, C.; Smith, J.H.; Moslehi, S.; Harland, B.; Dalrymple-Alford, J.; Taylor, R.P. Neuron arbor geometry is sensitive to the limited-range fractal properties of their dendrites. Front. Netw. Physiol. 2023, 3, 1072815. [Google Scholar] [CrossRef] [PubMed]
- Rowland, C.; Harland, B.; Smith, J.H.; Moslehi, S.; Dalrymple-Alford, J.; Taylor, R.P. Investigating Fractal Analysis as a Diagnostic Tool That Probes the Connectivity of Hippocampal Neurons. Front. Physiol. 2022, 13, 932598. [Google Scholar] [CrossRef]
- Rowland, C.; Moslehi, S.; Smith, J.H.; Harland, B.; Dalrymple-Alford, J.; Taylor, R.P. Fractal Resonance: Can Fractal Geometry Be Used to Optimize the Connectivity of Neurons to Artificial Implants? Springer International Publishing: Berlin/Heidelberg, Germany, 2024; pp. 877–906. [Google Scholar]
- Cajal, S.R.Y. Texture of the Nervous System of Man and the Vertebrates; Springer: Vienna, Austria, 1999. [Google Scholar]
- Watterson, W.J.; Moslehi, S.; Rowland, C.; Zappitelli, K.M.; Smith, J.H.; Miller, D.; Chouinard, J.E.; Golledge, S.L.; Taylor, R.P.; Perez, M.-T.; et al. The Roles of an Aluminum Underlayer in the Biocompatibility and Mechanical Integrity of Vertically Aligned Carbon Nanotubes for Interfacing with Retinal Neurons. Micromachines 2020, 11, 546. [Google Scholar] [CrossRef] [PubMed]
- Mata, D.; Amaral, M.; Fernandes, A.J.S.; Colaço, B.; Gama, A.; Paiva, M.C.; Gomes, P.S.; Silva, R.F.; Fernandes, M.H. Diels–Alder functionalized carbon nanotubes for bone tissue engineering: In vitro/in vivo biocompatibility and biodegradability. Nanoscale 2015, 7, 9238–9251. [Google Scholar] [CrossRef]
- Pampaloni, N.P.; Scaini, D.; Perissinotto, F.; Bosi, S.; Prato, M.; Ballerini, L. Sculpting neurotransmission during synaptic development by 2D nanostructured interfaces. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 2521–2532. [Google Scholar] [CrossRef]
- Abu-Saude, M.; Morshed, B. Characterization of a Novel Polypyrrole (PPy) Conductive Polymer Coated Patterned Vertical CNT (pvCNT) Dry ECG Electrode. Chemosensors 2018, 6, 27. [Google Scholar] [CrossRef]
- Simitzi, C.R.A.; Stratakis, E. Controlling the morphology and outgrowth of nerve and neuroglial cells: The effect of surface topography. Acta Biomater. 2017, 51, 21–52. [Google Scholar] [CrossRef] [PubMed]
- Smeal, R.M.; Rabbitt, R.; Biran, R.; Tresco, P.A. Substrate Curvature Influences the Direction of Nerve Outgrowth. Ann. Biomed. Eng. 2005, 33, 376–382. [Google Scholar] [CrossRef]
- Smeal, R.M.T.; Patrick, A. The influence of substrate curvature on neurite outgrowth is cell type dependent. Exp. Neurol. 2008, 213, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Clark, P.; Connolly, P.; Curtis, A.S.G.; Dow, J.A.T.; Wilkinson, C.D.W. Topographical control of cell behaviour: II. multiple grooved substrata. Development 1990, 108, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Rajnicek, A.M.; Britland, S.; McCaig, C.D. Contact guidance of CNS neurites on grooved quartz: Influence of groove dimensions, neuronal age and cell type. J. Cell Sci. 1997, 110, 2905–2913. [Google Scholar] [CrossRef] [PubMed]
- Goldner, J.S.B.; Jan, M. Neurite bridging across micropatterned grooves. Biomaterials 2006, 27, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Johansson, F.; Carlberg, P.; Danielsen, N.; Montelius, L.; Kanje, M. Axonal outgrowth on nano-imprinted patterns. Biomaterials 2006, 27, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tang, Q.Y.; Jadhav, A.D.; Narang, A.; Qian, W.X.; Shi, P.; Pang, S.W. Large-scale Topographical Screen for Investigation of Physical Neural-Guidance Cues. Sci. Rep. 2015, 5, 8644. [Google Scholar] [CrossRef]
- Lee, J.W.; Lee, K.S.; Cho, N.; Ju, B.K.; Lee, K.B.; Lee, S.H. Topographical guidance of mouse neuronal cell on SiO2 microtracks. Sens. Actuators B Chem. 2007, 128, 252–257. [Google Scholar] [CrossRef]
- Saba, M.; William, J.W.; Conor, R.; Julian, H.S.; Maria-Thereza, P.; Richard, P.T. Physical Guidance of Cultured Retinal Neurons Using Zig-zag Surface Patterns. Am. J. Biomed. Sci. Res. 2020, 11, 219–221. [Google Scholar] [CrossRef]
- Gomez, N.; Lu, Y.; Chen, S.; Schmidt, C.E. Immobilized nerve growth factor and microtopography have distinct effects on polarization versus axon elongation in hippocampal cells in culture. Biomaterials 2007, 28, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Bareket-Keren, L.; Hanein, Y. Carbon nanotube-based multi electrode arrays for neuronal interfacing: Progress and prospects. Front. Neural Circuits 2013, 6, 35286. [Google Scholar] [CrossRef] [PubMed]
- Fabbro, A.; Bosi, S.; Ballerini, L.; Prato, M. Carbon Nanotubes: Artificial Nanomaterials to Engineer Single Neurons and Neuronal Networks. ACS Chem. Neurosci. 2012, 3, 611–618. [Google Scholar] [CrossRef]
- Krukiewicz, K.; Janas, D.; Vallejo-Giraldo, C.; Biggs, M.J.P. Self-supporting carbon nanotube films as flexible neural interfaces. Electrochim. Acta 2019, 295, 253–261. [Google Scholar] [CrossRef]
- Hu, H.; Ni, Y.; Montana, V.; Haddon, R.C.; Parpura, V. Chemically Functionalized Carbon Nanotubes as Substrates for Neuronal Growth. Nano Lett. 2004, 4, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Nick, C.; Yadav, S.; Joshi, R.; Thielemann, C.; Schneider, J.J. Growth and structural discrimination of cortical neurons on randomly oriented and vertically aligned dense carbon nanotube networks. Beilstein J. Nanotechnol. 2014, 5, 1575–1579. [Google Scholar] [CrossRef] [PubMed]
- Sorkin, R.; Greenbaum, A.; David-Pur, M.; Anava, S.; Ayali, A.; Ben-Jacob, E.; Hanein, Y. Process entanglement as a neuronal anchorage mechanism to rough surfaces. Nanotechnology 2009, 20, 015101. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, L.A.; Ju, Y.-E.; Marg, B.; Osterfield, M.; Janmey, P.A. Neurite branching on deformable substrates. NeuroReport 2002, 13, 2411–2415. [Google Scholar] [CrossRef]
- Georges, P.C.; Miller, W.J.; Meaney, D.F.; Sawyer, E.S.; Janmey, P.A. Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. Biophys. J. 2006, 90, 3012–3018. [Google Scholar] [CrossRef]
- Wang, K.; Fishman, H.A.; Dai, H.; Harris, J.S. Neural stimulation with a carbon nanotube microelectrode array. Nano Lett. 2006, 6, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Mazzatenta, A.; Giugliano, M.; Campidelli, S.; Gambazzi, L.; Businaro, L.; Markram, H.; Prato, M.; Ballerini, L. Interfacing Neurons with Carbon Nanotubes: Electrical Signal Transfer and Synaptic Stimulation in Cultured Brain Circuits. J. Neurosci. 2007, 27, 6931–6936. [Google Scholar] [CrossRef] [PubMed]
- Cellot, G.; Cilia, E.; Cipollone, S.; Rancic, V.; Sucapane, A.; Giordani, S.; Gambazzi, L.; Markram, H.; Grandolfo, M.; Scaini, D.; et al. Carbon nanotubes might improve neuronal performance by favouring electrical shortcuts. Nat. Nanotechnol. 2009, 4, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Lovat, V.; Pantarotto, D.; Lagostena, L.; Cacciari, B.; Grandolfo, M.; Righi, M.; Spalluto, G.; Prato, M.; Ballerini, L. Carbon nanotube substrates boost neuronal electrical signaling. Nano Lett. 2005, 5, 1107–1110. [Google Scholar] [CrossRef] [PubMed]
- Butterwick, A.; Huie, P.; Jones, B.W.; Marc, R.E.; Marmor, M.; Palanker, D. Effect of shape and coating of a subretinal prosthesis on its integration with the retina. Exp. Eye Res. 2009, 88, 22–29. [Google Scholar] [CrossRef]
- Piret, G.; Perez, M.-T.; Prinz, C.N. Support of Neuronal Growth over Glial Growth and Guidance of Optic Nerve Axons by Vertical Nanowire Arrays. ACS Appl. Mater. Interfaces 2015, 7, 18944–18948. [Google Scholar] [CrossRef]
- Chapman, C.A.; Chen, H.; Stamou, M.; Lein, P.J.; Seker, E. Mechanisms of Reduced Astrocyte Surface Coverage in Cortical Neuron-Glia Co-cultures on Nanoporous Gold Surfaces. Cell. Mol. Bioeng. 2016, 9, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Moslehi, S.; Rowland, C.; Smith, J.H.; Watterson, W.J.; Miller, D.; Niell, C.M.; Alemán, B.J.; Perez, M.-T.; Taylor, R.P. Controlled assembly of retinal cells on fractal and Euclidean electrodes. PLoS ONE 2022, 17, e0265685. [Google Scholar] [CrossRef] [PubMed]
- Moslehi, S.; Rowland, C.; Smith, J.H.; Griffiths, W.; Watterson, W.J.; Niell, C.M.; Alemán, B.J.; Perez, M.T.; Taylor, R.P. Comparison of fractal and grid electrodes for studying the effects of spatial confinement on dissociated retinal neuronal and glial behavior. Sci. Rep. 2022, 12, 17513. [Google Scholar] [CrossRef]
- Zalis, M.C.; Johansson, S.; Englund-Johansson, U. Immunocytochemical Profiling of Cultured Mouse Primary Retinal Cells. J. Histochem. Cytochem. 2017, 65, 223–239. [Google Scholar] [CrossRef]
- Sharma, R.K.; Netland, P.A. Early born lineage of retinal neurons express class III β-tubulin isotype. Brain Res. 2007, 1176, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Latora, V.; Marchiori, M. Economic small-world behavior in weighted networks. Eur. Phys. J. B-Condens. Matter Complex Syst. 2003, 32, 249–263. [Google Scholar] [CrossRef]
- Achard, S.; Bullmore, E. Efficiency and Cost of Economical Brain Functional Networks. PLoS Comput. Biol. 2007, 3, e17. [Google Scholar] [CrossRef] [PubMed]
- Woiterski, L.; Claudepierre, T.; Luxenhofer, R.; Jordan, R.; Käs, J.A. Stages of neuronal network formation. New J. Phys. 2013, 15, 025029. [Google Scholar] [CrossRef]
- Anava, S.; Greenbaum, A.; Jacob, E.B.; Hanein, Y.; Ayali, A. The Regulative Role of Neurite Mechanical Tension in Network Development. Biophys. J. 2009, 96, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- De Santos-Sierra, D.; Sendiña-Nadal, I.; Leyva, I.; Almendral, J.A.; Anava, S.; Ayali, A.; Papo, D.; Boccaletti, S. Emergence of Small-World Anatomical Networks in Self-Organizing Clustered Neuronal Cultures. PLoS ONE 2014, 9, e85828. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Appaix, F.; Bibari, O.; Marchand, G.; Benabid, A.-L.; Sauter-Starace, F.; Waard, M.D. Control of neuronal network organization by chemical surface functionalization of multi-walled carbon nanotube arrays. Nanotechnology 2011, 22, 195101. [Google Scholar] [CrossRef] [PubMed]
- Watterson, W.J.; Montgomery, R.D.; Taylor, R.P. Fractal Electrodes as a Generic Interface for Stimulating Neurons. Sci. Rep. 2017, 7, 6717. [Google Scholar] [CrossRef]
- Watterson, W.J.; Montgomery, R.D.; Taylor, R.P. Modeling the Improved Visual Acuity Using Photodiode Based Retinal Implants Featuring Fractal Electrodes. Front. Neurosci. 2018, 12, 277. [Google Scholar] [CrossRef]


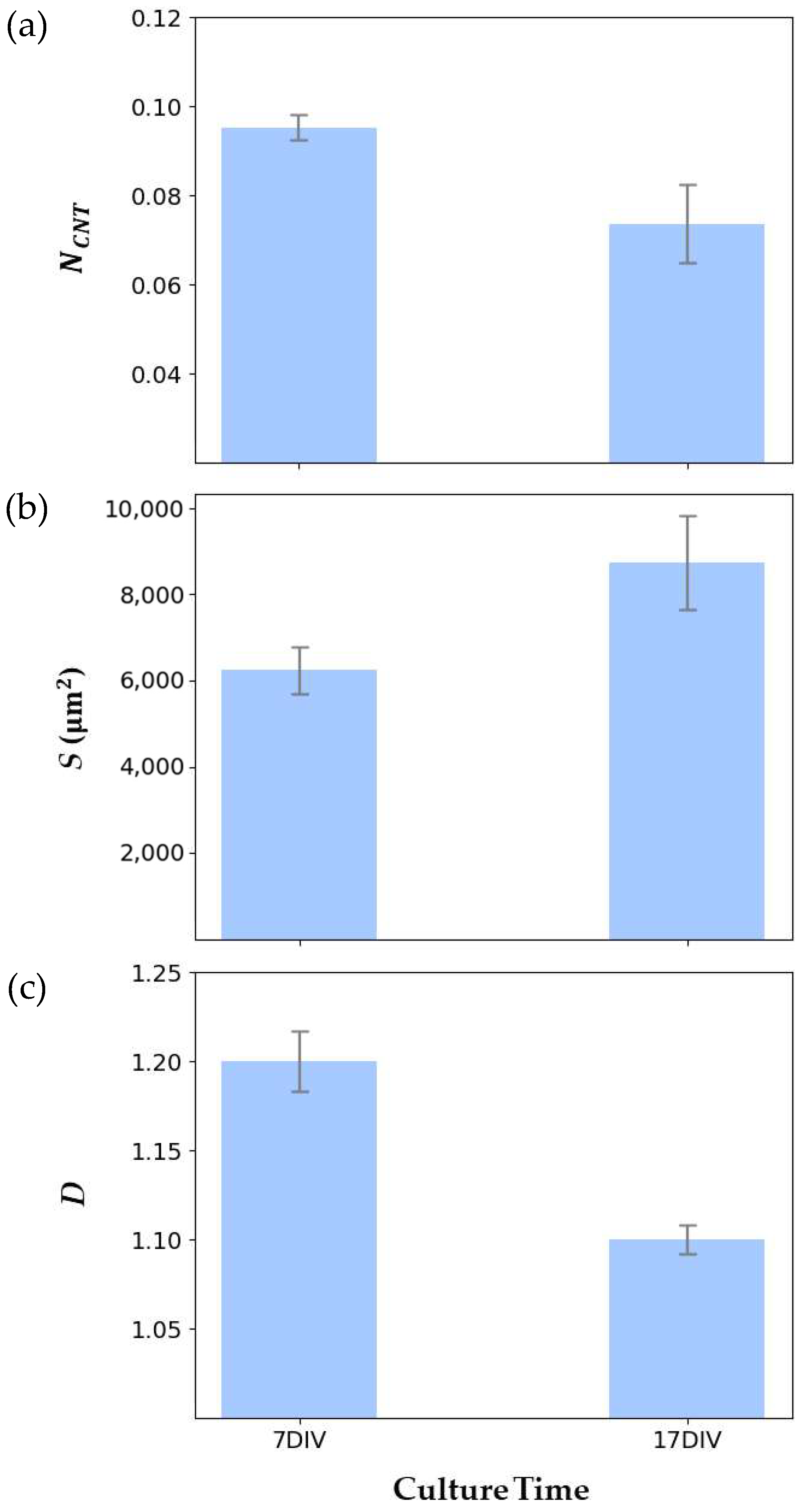
| Measure | 7 DIV | 17 DIV | t | p |
|---|---|---|---|---|
| NCNT | 0.095 ± 0.003 | 0.074 ± 0.009 | 2.072 | 0.174 |
| S | 6240 ± 552.5 μm2 | 8750 ± 1088 μm2 | 2.031 | 0.072 |
| D | 1.20 ± 0.017 | 1.10 ± 0.008 | 4.586 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dillon, A.P.; Moslehi, S.; Brouse, B.; Keremane, S.; Philliber, S.; Griffiths, W.; Rowland, C.; Smith, J.H.; Taylor, R.P. Evolution of Retinal Neuron Fractality When Interfacing with Carbon Nanotube Electrodes. Bioengineering 2024, 11, 823. https://doi.org/10.3390/bioengineering11080823
Dillon AP, Moslehi S, Brouse B, Keremane S, Philliber S, Griffiths W, Rowland C, Smith JH, Taylor RP. Evolution of Retinal Neuron Fractality When Interfacing with Carbon Nanotube Electrodes. Bioengineering. 2024; 11(8):823. https://doi.org/10.3390/bioengineering11080823
Chicago/Turabian StyleDillon, Aiden P., Saba Moslehi, Bret Brouse, Saumya Keremane, Sam Philliber, Willem Griffiths, Conor Rowland, Julian H. Smith, and Richard P. Taylor. 2024. "Evolution of Retinal Neuron Fractality When Interfacing with Carbon Nanotube Electrodes" Bioengineering 11, no. 8: 823. https://doi.org/10.3390/bioengineering11080823
APA StyleDillon, A. P., Moslehi, S., Brouse, B., Keremane, S., Philliber, S., Griffiths, W., Rowland, C., Smith, J. H., & Taylor, R. P. (2024). Evolution of Retinal Neuron Fractality When Interfacing with Carbon Nanotube Electrodes. Bioengineering, 11(8), 823. https://doi.org/10.3390/bioengineering11080823






