Machine Learning-Driven Prediction of Brain Age for Alzheimer’s Risk: APOE4 Genotype and Gender Effects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data
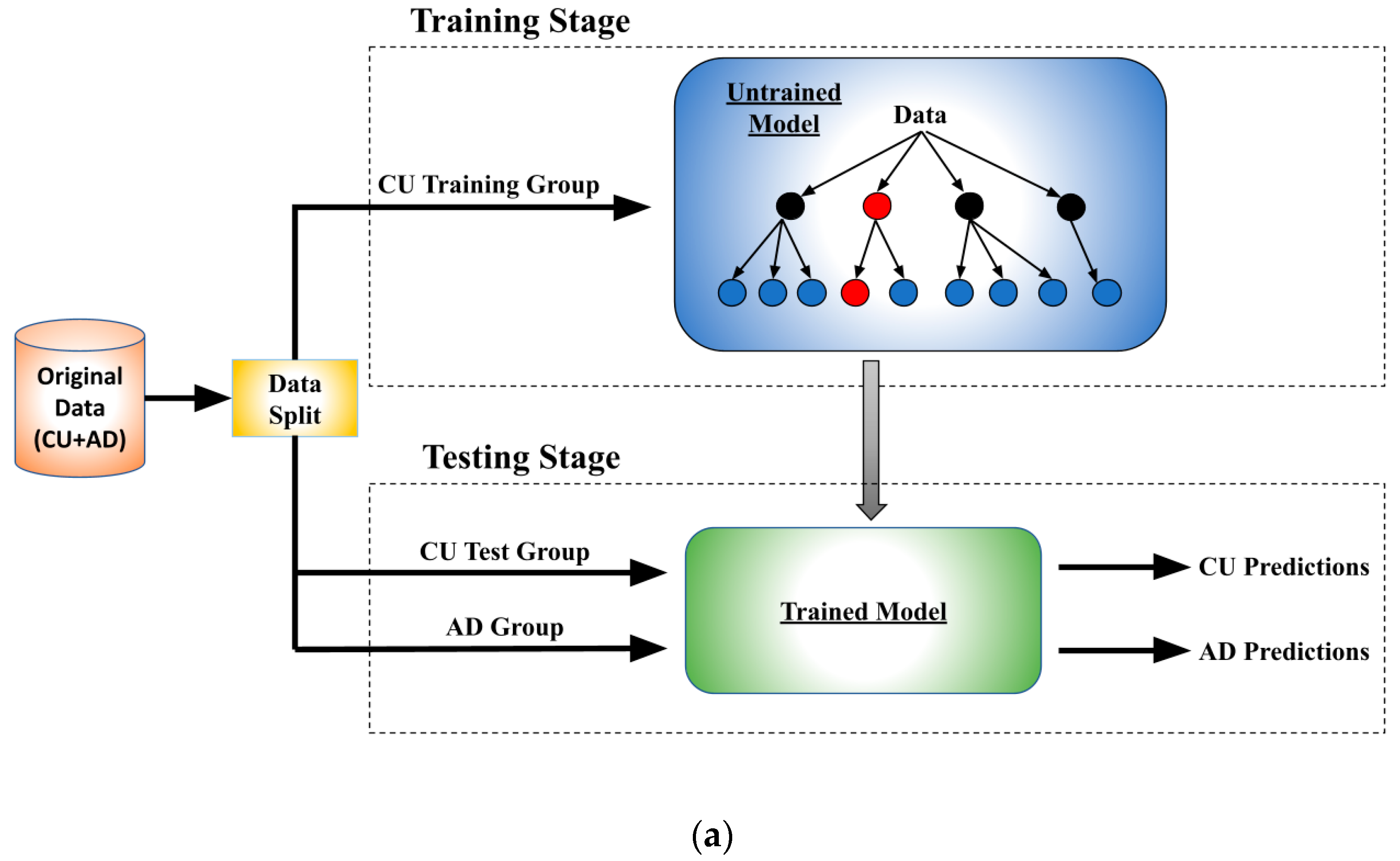
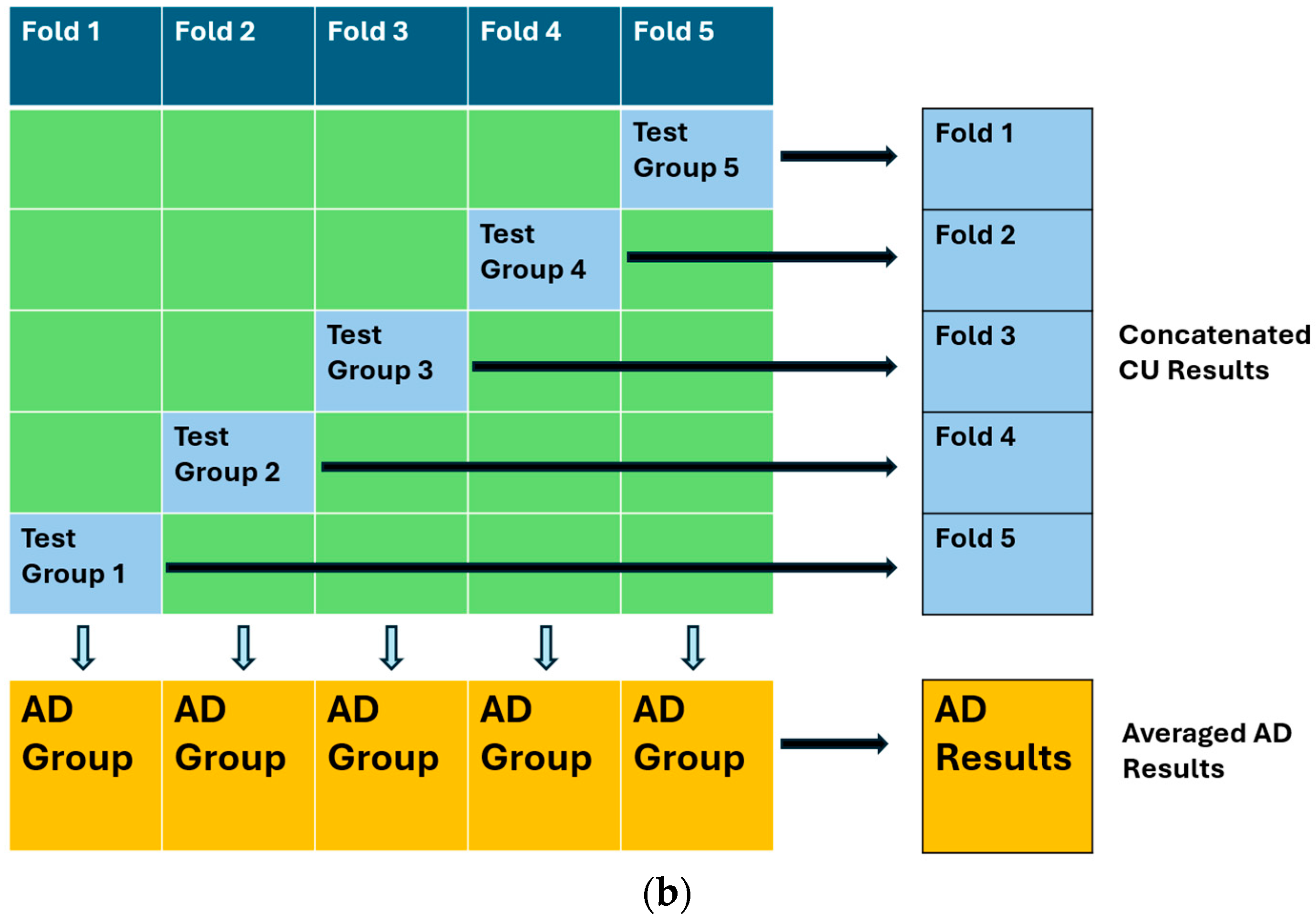
2.2. Architecture and Training Procedure
3. Results
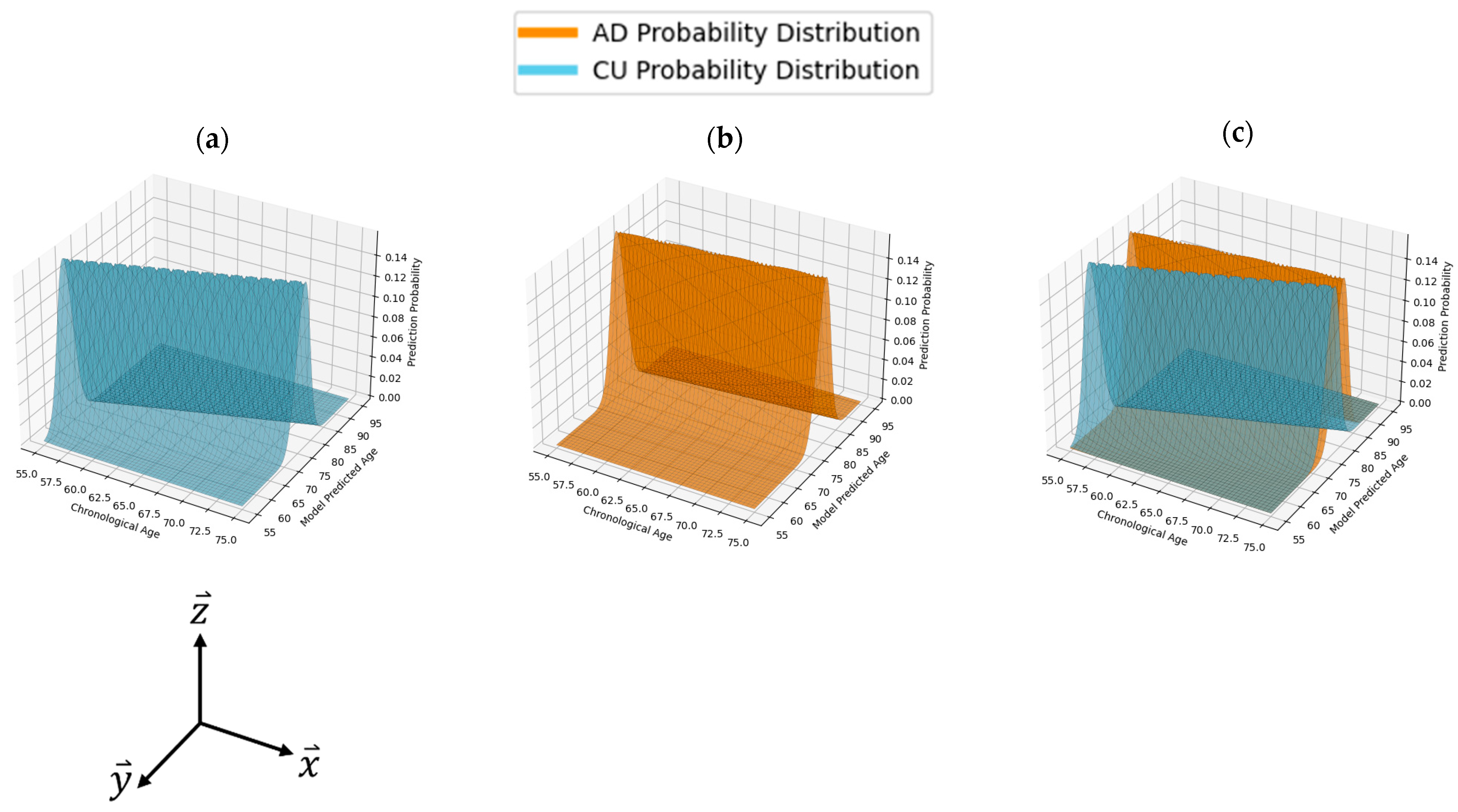
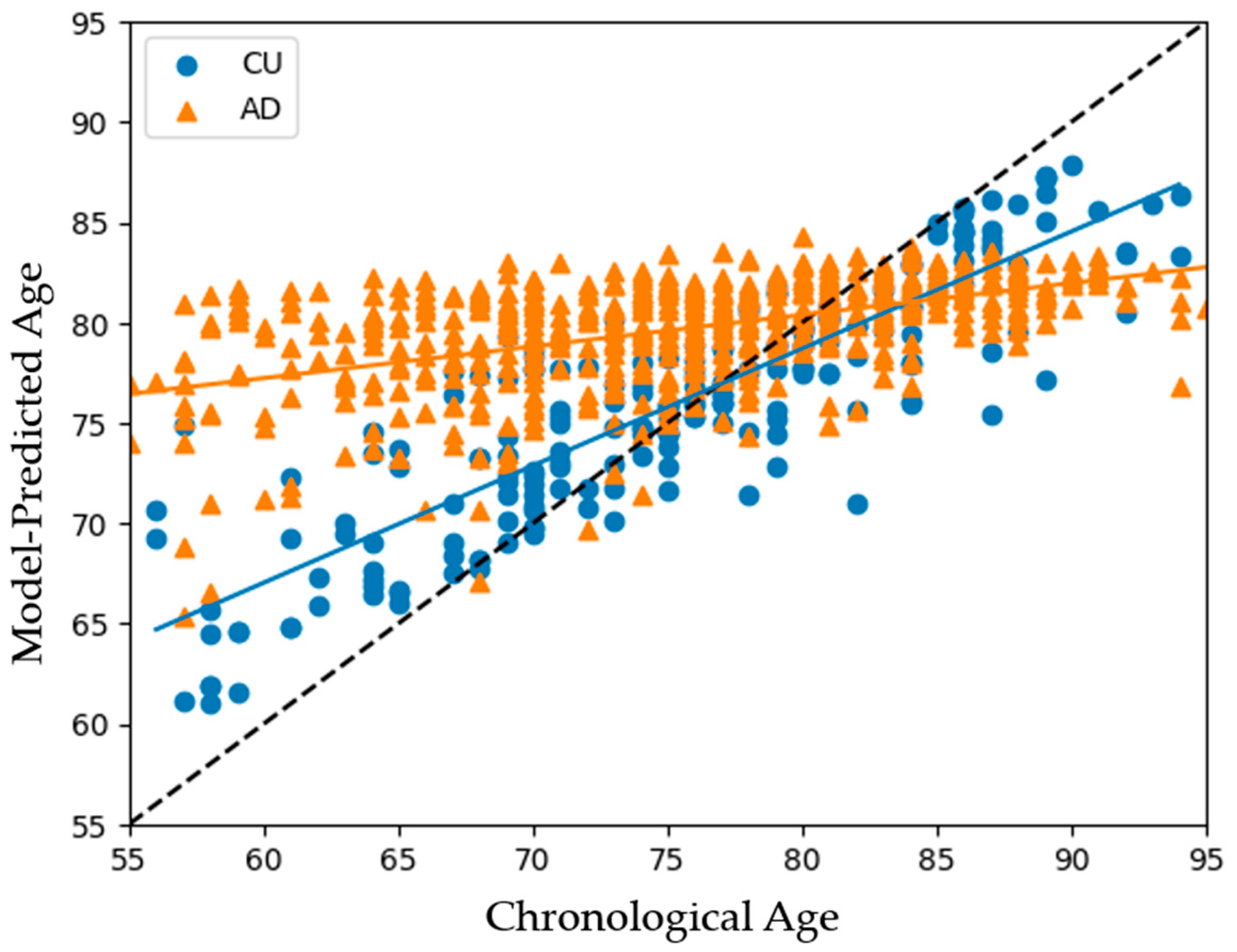
3.1. Model Performances on All Subjects
3.2. Comparisons among the Three ML Models
3.3. APOE4 Comparison
3.3.1. APOE4-Stratified Model Compositions
3.3.2. APOE4-Stratified Model Outcomes
3.4. Gender Comparison
3.4.1. Gender-Stratified Model Compositions
3.4.2. Gender-Stratified Model Outcomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gustavsson, A.; Norton, N.; Fast, T.; Frölich, L.; Georges, J.; Holzapfel, D.; Kirabali, T.; Krolak-Salmon, P.; Rossini, P.M.; Ferretti, M.T.; et al. Global Estimates on the Number of Persons across the Alzheimer’s Disease Continuum. Alzheimer’s Dement. 2023, 19, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A.; Counts, N.; Bröker, J.; Malik, S.; Chen, S.; Han, R.; Klusty, J.; Seligman, B.; Tortorice, D.; Vigo, D.; et al. Cost of Care for Alzheimer’s Disease and Related Dementias in the United States: 2016 to 2060. Npj Aging 2024, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Svennerholm, L.; Boström, K.; Jungbjer, B. Changes in Weight and Compositions of Major Membrane Components of Human Brain during the Span of Adult Human Life of Swedes. Acta Neuropathol. 1997, 94, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R.; Shiung, M.M.; Gunter, J.L.; O’Brien, P.C.; Weigand, S.D.; Knopman, D.S.; Boeve, B.F.; Ivnik, R.J.; Smith, G.E.; Cha, R.H.; et al. Comparison of Different MRI Brain Atrophy Rate Measures with Clinical Disease Progression in AD. Neurology 2004, 62, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; He, Y.; Rosa-Neto, P.; Gong, G.; Evans, A.C. Age-Related Alterations in the Modular Organization of Structural Cortical Network by Using Cortical Thickness from MRI. NeuroImage 2011, 56, 235–245. [Google Scholar] [CrossRef]
- Salat, D.H.; Lee, S.Y.; Van Der Kouwe, A.J.; Greve, D.N.; Fischl, B.; Rosas, H.D. Age-Associated Alterations in Cortical Gray and White Matter Signal Intensity and Gray to White Matter Contrast. NeuroImage 2009, 48, 21–28. [Google Scholar] [CrossRef]
- McGinnis, S.M.; Brickhouse, M.; Pascual, B.; Dickerson, B.C. Age-Related Changes in the Thickness of Cortical Zones in Humans. Brain Topogr. 2011, 24, 279–291. [Google Scholar] [CrossRef]
- Planche, V.; Manjon, J.V.; Mansencal, B.; Lanuza, E.; Tourdias, T.; Catheline, G.; Coupé, P. Structural Progression of Alzheimer’s Disease over Decades: The MRI Staging Scheme. Brain Commun. 2022, 4, fcac109. [Google Scholar] [CrossRef]
- Yanckello, L.M.; Hoffman, J.D.; Chang, Y.-H.; Lin, P.; Nehra, G.; Chlipala, G.; McCulloch, S.D.; Hammond, T.C.; Yackzan, A.T.; Lane, A.N.; et al. Apolipoprotein E Genotype-Dependent Nutrigenetic Effects to Prebiotic Inulin for Modulating Systemic Metabolism and Neuroprotection in Mice via Gut-Brain Axis. Nutr. Neurosci. 2022, 25, 1669–1679. [Google Scholar] [CrossRef]
- Ho, K.C.; Roessmann, U.; Straumfjord, J.V.; Monroe, G. Analysis of Brain Weight. I. Adult Brain Weight in Relation to Sex, Race, and Age. Arch. Pathol. Lab. Med. 1980, 104, 635–639. [Google Scholar]
- Takao, H.; Hayashi, N.; Ohtomo, K. A Longitudinal Study of Brain Volume Changes in Normal Aging. Eur. J. Radiol. 2012, 81, 2801–2804. [Google Scholar] [CrossRef] [PubMed]
- Peters, R. Ageing and the Brain. Postgrad. Med. J. 2006, 82, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.-Y.; Wang, Y.-J. Early Intervention in Alzheimer’s Disease: How Early Is Early Enough? Neurosci. Bull. 2020, 36, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.E.; Przybelski, S.A.; Lesnick, T.G.; Liesinger, A.M.; Spychalla, A.; Zhang, B.; Gunter, J.L.; Parisi, J.E.; Boeve, B.F.; Knopman, D.S.; et al. Early Alzheimer’s Disease Neuropathology Detected by Proton MR Spectroscopy. J. Neurosci. 2014, 34, 16247–16255. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, H.; Goldman, A.L.; Sambataro, F.; Verchinski, B.A.; Meyer-Lindenberg, A.; Weinberger, D.R.; Mattay, V.S. Normal Age-Related Brain Morphometric Changes: Nonuniformity across Cortical Thickness, Surface Area and Gray Matter Volume? Neurobiol. Aging 2012, 33, 617.e1–617.e9. [Google Scholar] [CrossRef]
- Tang, A.S.; Rankin, K.P.; Cerono, G.; Miramontes, S.; Mills, H.; Roger, J.; Zeng, B.; Nelson, C.; Soman, K.; Woldemariam, S.; et al. Leveraging Electronic Health Records and Knowledge Networks for Alzheimer’s Disease Prediction and Sex-Specific Biological Insights. Nat. Aging 2024, 4, 379–395. [Google Scholar] [CrossRef]
- Popuri, K.; Ma, D.; Wang, L.; Beg, M.F. Using Machine Learning to Quantify Structural MRI Neurodegeneration Patterns of Alzheimer’s Disease into Dementia Score: Independent Validation on 8,834 Images from ADNI, AIBL, OASIS, and MIRIAD Databases. Hum. Brain Mapp. 2020, 41, 4127–4147. [Google Scholar] [CrossRef]
- Qiu, S.; Miller, M.I.; Joshi, P.S.; Lee, J.C.; Xue, C.; Ni, Y.; Wang, Y.; De Anda-Duran, I.; Hwang, P.H.; Cramer, J.A.; et al. Multimodal Deep Learning for Alzheimer’s Disease Dementia Assessment. Nat. Commun. 2022, 13, 3404. [Google Scholar] [CrossRef]
- Park, J.H.; Cho, H.E.; Kim, J.H.; Wall, M.M.; Stern, Y.; Lim, H.; Yoo, S.; Kim, H.S.; Cha, J. Machine Learning Prediction of Incidence of Alzheimer’s Disease Using Large-Scale Administrative Health Data. Npj Digit. Med. 2020, 3, 46. [Google Scholar] [CrossRef]
- Li, Q.; Yang, X.; Xu, J.; Guo, Y.; He, X.; Hu, H.; Lyu, T.; Marra, D.; Miller, A.; Smith, G.; et al. Early Prediction of Alzheimer’s Disease and Related Dementias Using Real-world Electronic Health Records. Alzheimer’s Dement. 2023, 19, 3506–3518. [Google Scholar] [CrossRef]
- Diogo, V.S.; Ferreira, H.A.; Prata, D.; for the Alzheimer’s Disease Neuroimaging Initiative. Early Diagnosis of Alzheimer’s Disease Using Machine Learning: A Multi-Diagnostic, Generalizable Approach. Alzheimer’s Res. Ther. 2022, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, F.; Xu, Z.; Adekkanattu, P.; Brandt, P.; Jiang, G.; Kiefer, R.C.; Luo, Y.; Mao, C.; Pacheco, J.A.; et al. Data-Driven Discovery of Probable Alzheimer’s Disease and Related Dementia Subphenotypes Using Electronic Health Records. Learn. Health Syst. 2020, 4, e10246. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Rafique, M.U.; Liang, G.; Blanton, H.; Zhang, Y.; Wang, C.; Jacobs, N.; Lin, A.-L. Efficient Training on Alzheimer’s Disease Diagnosis with Learnable Weighted Pooling for 3D PET Brain Image Classification. Electronics 2023, 12, 467. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Liang, G.; Zhang, Y.; Khanal, S.; Lin, A.-L.; Jacobs, N. Advit: Vision Transformer On Multi-Modality Pet Images For Alzheimer Disease Diagnosis. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022; pp. 1–4. [Google Scholar]
- Verma, G.; Jacob, Y.; Jha, M.; Morris, L.S.; Delman, B.N.; Marcuse, L.; Fields, M.; Balchandani, P. Quantification of Brain Age Using High-Resolution 7 Tesla MR Imaging and Implications for Patients with Epilepsy. Epilepsy Behav. Rep. 2022, 18, 100530. [Google Scholar] [CrossRef]
- NACC Alzheimer’s Data. Available online: https://naccdata.org/ (accessed on 5 September 2024).
- Hammond, T.C.; Xing, X.; Yanckello, L.M.; Stromberg, A.; Chang, Y.-H.; Nelson, P.T.; Lin, A.-L. Human Gray and White Matter Metabolomics to Differentiate APOE and Stage Dependent Changes in Alzheimer’s Disease. J. Cell. Immunol. 2021, 3, 397–412. [Google Scholar] [CrossRef]
- Jack, C.R.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Feldman, H.H.; Frisoni, G.B.; Hampel, H.; Jagust, W.J.; Johnson, K.A.; Knopman, D.S.; et al. A/T/N: An Unbiased Descriptive Classification Scheme for Alzheimer Disease Biomarkers. Neurology 2016, 87, 539–547. [Google Scholar] [CrossRef]
- Jack, C.R.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a Biological Definition of Alzheimer’s Disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Hammond, T.C.; Lin, A.-L. Glucose Metabolism Is a Better Marker for Predicting Clinical Alzheimer’s Disease than Amyloid or Tau. J. Cell. Immunol. 2022, 4, 15–18. [Google Scholar]
- Hammond, T.C.; Xing, X.; Wang, C.; Ma, D.; Nho, K.; Crane, P.K.; Elahi, F.; Ziegler, D.A.; Liang, G.; Cheng, Q.; et al. β-Amyloid and Tau Drive Early Alzheimer’s Disease Decline While Glucose Hypometabolism Drives Late Decline. Commun. Biol. 2020, 3, 352. [Google Scholar] [CrossRef]
- Sanganahalli, B.G.; Mihailovic, J.M.; Vekaria, H.J.; Coman, D.; Yackzan, A.T.; Flemister, A.; Aware, C.; Wenger, K.; Hubbard, W.B.; Sullivan, P.G.; et al. mTOR Inhibition Enhances Synaptic and Mitochondrial Function in Alzheimer’s Disease in an APOE Genotype-Dependent Manner. J. Cereb. Blood Flow Metab. 2024. [Google Scholar] [CrossRef]
- Lin, A.-L.; Parikh, I.; Yanckello, L.M.; White, R.S.; Hartz, A.M.S.; Taylor, C.E.; McCulloch, S.D.; Thalman, S.W.; Xia, M.; McCarty, K.; et al. APOE Genotype-Dependent Pharmacogenetic Responses to Rapamycin for Preventing Alzheimer’s Disease. Neurobiol. Dis. 2020, 139, 104834. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yanckello, L.M.; Ma, D.; Hoffman, J.D.; Parikh, I.; Thalman, S.; Bauer, B.; Hartz, A.M.S.; Hyder, F.; Lin, A.-L. Neuroimaging Biomarkers of mTOR Inhibition on Vascular and Metabolic Functions in Aging Brain and Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 225. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Wang, A.C.; Parikh, I.; Green, S.J.; Hoffman, J.D.; Chlipala, G.; Murphy, M.P.; Sokola, B.S.; Bauer, B.; Hartz, A.M.S.; et al. Ketogenic Diet Enhances Neurovascular Function with Altered Gut Microbiome in Young Healthy Mice. Sci. Rep. 2018, 8, 6670. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.-L.; Jahrling, J.B.; Zhang, W.; DeRosa, N.; Bakshi, V.; Romero, P.; Galvan, V.; Richardson, A. Rapamycin Rescues Vascular, Metabolic and Learning Deficits in Apolipoprotein E4 Transgenic Mice with Pre-Symptomatic Alzheimer’s Disease. J. Cereb. Blood Flow Metab. 2017, 37, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.-L.; Parikh, I.; Hoffman, J.D.; Ma, D. Neuroimaging Biomarkers of Caloric Restriction on Brain Metabolic and Vascular Functions. Curr. Nutr. Rep. 2017, 6, 41–48. [Google Scholar] [CrossRef]
- Chang, Y.-H.; Yanckello, L.M.; Chlipala, G.E.; Green, S.J.; Aware, C.; Runge, A.; Xing, X.; Chen, A.; Wenger, K.; Flemister, A.; et al. Prebiotic Inulin Enhances Gut Microbial Metabolism and Anti-Inflammation in Apolipoprotein E4 Mice with Sex-Specific Implications. Sci. Rep. 2023, 13, 15116. [Google Scholar] [CrossRef]
- Parikh, I.; Guo, J.; Chuang, K.-H.; Zhong, Y.; Rempe, R.G.; Hoffman, J.D.; Armstrong, R.; Bauer, B.; Hartz, A.M.S.; Lin, A.-L. Caloric Restriction Preserves Memory and Reduces Anxiety of Aging Mice with Early Enhancement of Neurovascular Functions. Aging 2016, 8, 2814–2826. [Google Scholar] [CrossRef]
- Lin, A.-L.; Zhang, W.; Gao, X.; Watts, L. Caloric Restriction Increases Ketone Bodies Metabolism and Preserves Blood Flow in Aging Brain. Neurobiol. Aging 2015, 36, 2296–2303. [Google Scholar] [CrossRef]

| Subject Characteristic | CU | AD | p-Value |
|---|---|---|---|
| Number | 1100 | 602 | |
| APOE4 (% Carrier) | 32% | 58% | <0.001 |
| Age | 76.1 ± 8.3 | 76.1 ± 8.5 | 0.93 |
| Gender (% Female) | 64% | 47% | <0.001 |
| Education | 15.5 ± 3.6 | 14.7 ± 3.8 | <0.001 |
| Feature Rank | Feature Description | CU (mean ± STD) | AD (mean ± STD) | p-Value |
|---|---|---|---|---|
| 1 | Right entorhinal mean cortical thickness (mm) | 3.76 ± 0.58 | 2.80 ± 0.86 | <0.001 |
| 2 | Left entorhinal mean cortical thickness (mm) | 3.56 ± 0.62 | 2.73 ± 0.80 | <0.001 |
| 3 | Segmented total hippocampi volume (cc) | 6.28 ± 0.39 | 5.37 ± 1.00 | <0.001 |
| 4 | Segmented left hippocampus volume (cc) | 3.11 ± 0.30 | 2.63 ± 0.52 | <0.001 |
| 5 | Left isthmus cingulate mean cortical thickness (mm) | 2.30 ± 0.30 | 1.97 ± 0.35 | <0.001 |
| 6 | Segmented right hippocampus volume (cc) | 3.19 ± 0.39 | 2.73 ± 0.53 | <0.001 |
| 7 | Right superior temporal mean cortical thickness (mm) | 2.23 ± 0.30 | 1.90 ± 0.30 | <0.001 |
| 8 | Right isthmus cingulate mean cortical thickness (mm) | 2.33 ± 0.31 | 2.00 ± 0.38 | <0.001 |
| 9 | Right fusiform mean cortical thickness (mm) | 2.56 ± 0.48 | 2.13 ± 0.42 | <0.001 |
| 10 | Left superior temporal mean cortical thickness (mm) | 2.12 ± 0.25 | 1.85 ± 0.33 | <0.001 |
| Model Type | BAD | CU (STD) | AD (STD) | ID |
|---|---|---|---|---|
| Linear Regression | 9.70 | 4.9 | 6.1 | 0.618 |
| XGBoost | 9.72 | 4.0 | 3.6 | 0.768 |
| Random Forest | 8.2 | 3.2 | 3.0 | 0.782 |
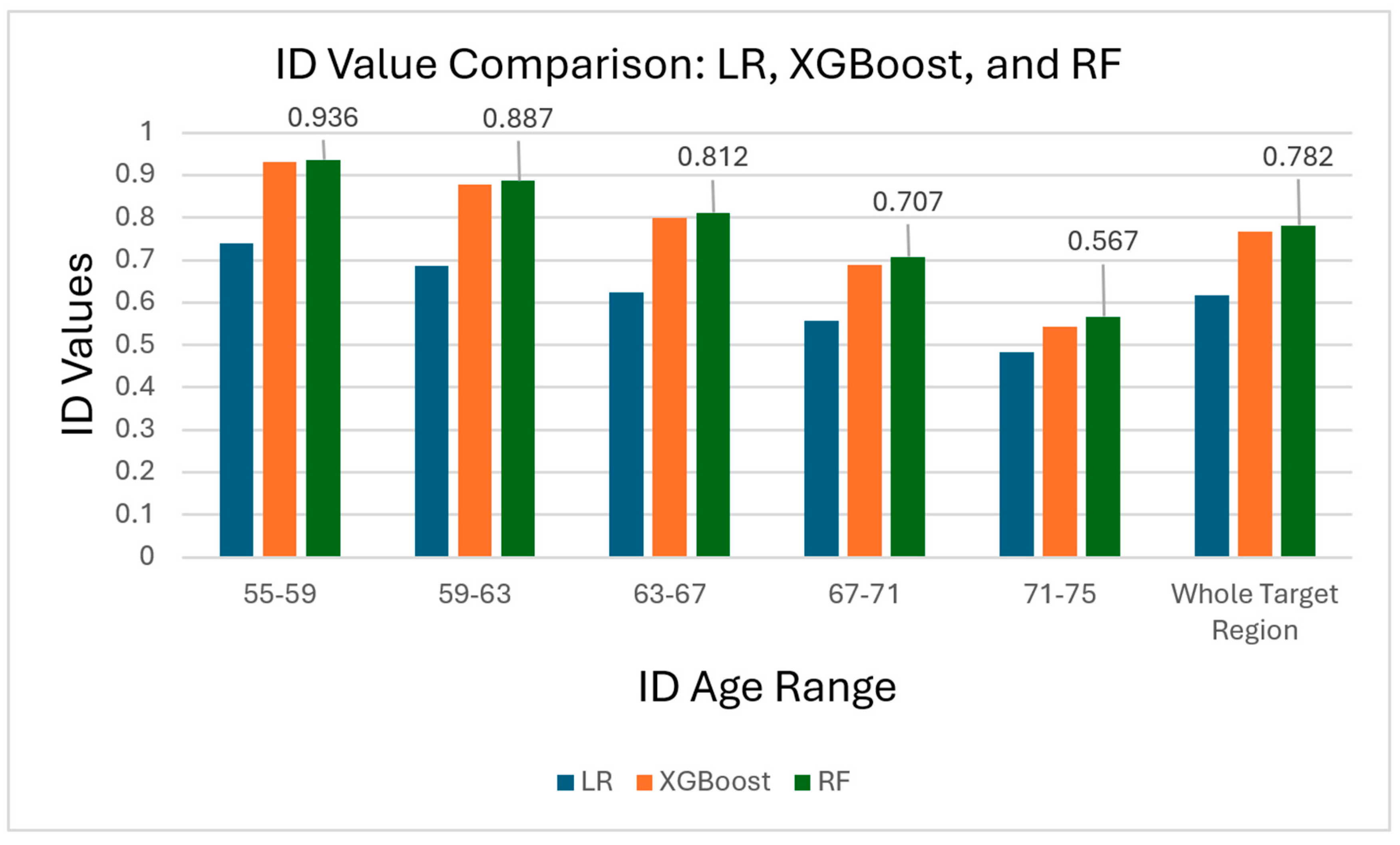
| Years of Age | 55–59 | 59–63 | 63–67 | 67–71 | 71–75 |
|---|---|---|---|---|---|
| Linear Regression ID | 0.739 | 0.686 | 0.625 | 0.558 | 0.484 |
| XGBoost ID | 0.930 | 0.878 | 0.799 | 0.689 | 0.544 |
| Random Forest ID | 0.936 | 0.887 | 0.812 | 0.707 | 0.567 |
| Training Group | Training Method | Training Size | Training Group Makeup |
|---|---|---|---|
| A | E4-Specific | 351 | 351 E4-carriers, 0 E4-NCs |
| B | E4-Specific | 749 | 0 E4-carriers, 749 E4-NCs |
| C | Mixed | 1100 | 351 E4-carriers, 749 E4-NCs |
| D | Mixed-Condensed | 352 | 176 E4-carriers, 176 E4-NCs |
| E | Mixed-Condensed | 749 | 351 E4-carriers, 398 E4-NCs |
| Training Group | Test Group | BAD | CU (STD) | AD (STD) | ID |
|---|---|---|---|---|---|
| A | E4-carriers | 7.1 | 2.8 | 2.3 | 0.787 |
| B | E4-NCs | 7.4 | 3.0 | 3.3 | 0.738 |
| C | E4-carriers | 8.3 | 3.2 | 2.7 | 0.807 |
| C | E4-NCs | 7.9 | 3.1 | 3.5 | 0.733 |
| D | E4-carriers | 7.1 | 2.9 | 2.3 | 0.787 |
| D | E4-NCs | 5.1 | 3.2 | 3.0 | 0.581 |
| E | E4-carriers | 7.8 | 3.1 | 2.5 | 0.804 |
| E | E4-NCs | 7.05 | 3.5 | 3.2 | 0.687 |
| Training Group | Training Method | Training Size | Training Group Makeup |
|---|---|---|---|
| A | Gender-Specific | 705 | 705 Females, 0 Males |
| B | Gender-Specific | 395 | 0 Females, 395 Males |
| C | Mixed | 1100 | 705 Females, 395 Males |
| D | Condensed-Mixed | 706 | 353 Females, 353 Males |
| E | Condensed-Mixed | 396 | 198 Females, 198 Males |
| Training Group | Test Group | BAD | CU (STD) | AD (STD) | ID |
|---|---|---|---|---|---|
| A | Females | 8.0 | 3.2 | 2.9 | 0.789 |
| B | Males | 7.7 | 2.9 | 3.0 | 0.779 |
| C | Females | 8.5 | 3.3 | 2.7 | 0.816 |
| C | Males | 8.1 | 2.9 | 3.2 | 0.783 |
| D | Females | 7.4 | 3.4 | 2.6 | 0.766 |
| D | Males | 8.0 | 3.2 | 3.3 | 0.743 |
| E | Females | 6.3 | 3.0 | 2.5 | 0.746 |
| E | Males | 7.1 | 2.8 | 3.1 | 0.756 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woods, C.; Xing, X.; Khanal, S.; Lin, A.-L. Machine Learning-Driven Prediction of Brain Age for Alzheimer’s Risk: APOE4 Genotype and Gender Effects. Bioengineering 2024, 11, 943. https://doi.org/10.3390/bioengineering11090943
Woods C, Xing X, Khanal S, Lin A-L. Machine Learning-Driven Prediction of Brain Age for Alzheimer’s Risk: APOE4 Genotype and Gender Effects. Bioengineering. 2024; 11(9):943. https://doi.org/10.3390/bioengineering11090943
Chicago/Turabian StyleWoods, Carter, Xin Xing, Subash Khanal, and Ai-Ling Lin. 2024. "Machine Learning-Driven Prediction of Brain Age for Alzheimer’s Risk: APOE4 Genotype and Gender Effects" Bioengineering 11, no. 9: 943. https://doi.org/10.3390/bioengineering11090943
APA StyleWoods, C., Xing, X., Khanal, S., & Lin, A.-L. (2024). Machine Learning-Driven Prediction of Brain Age for Alzheimer’s Risk: APOE4 Genotype and Gender Effects. Bioengineering, 11(9), 943. https://doi.org/10.3390/bioengineering11090943






