Forced Degradation Testing as Complementary Tool for Biosimilarity Assessment
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Consumables
2.2. Generation of Stressed mAb Samples
2.3. Digestion and Reduction of mAb Samples
2.4. Liquid Chromatography and Mass Spectrometry
2.5. Data Processing
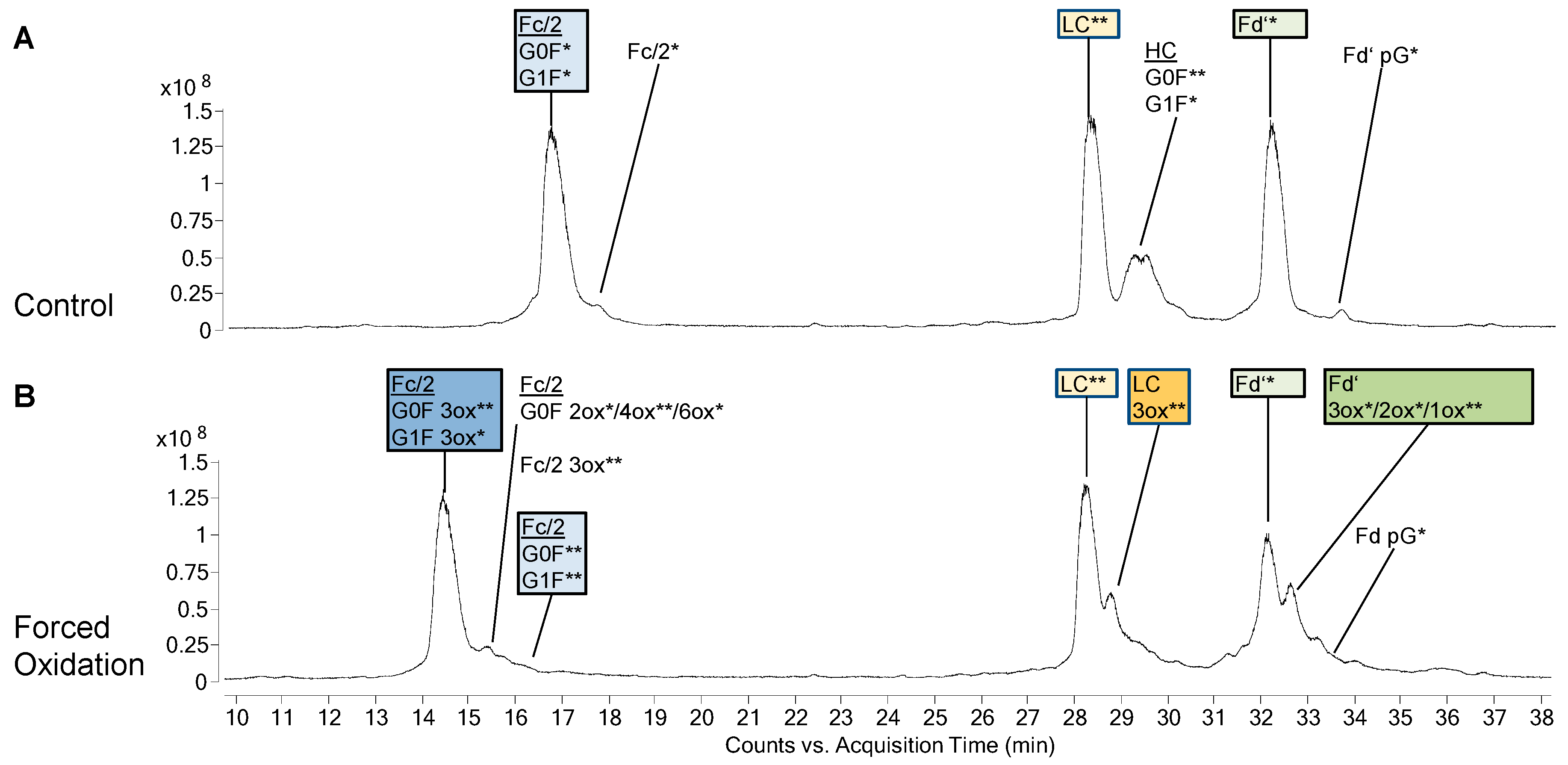
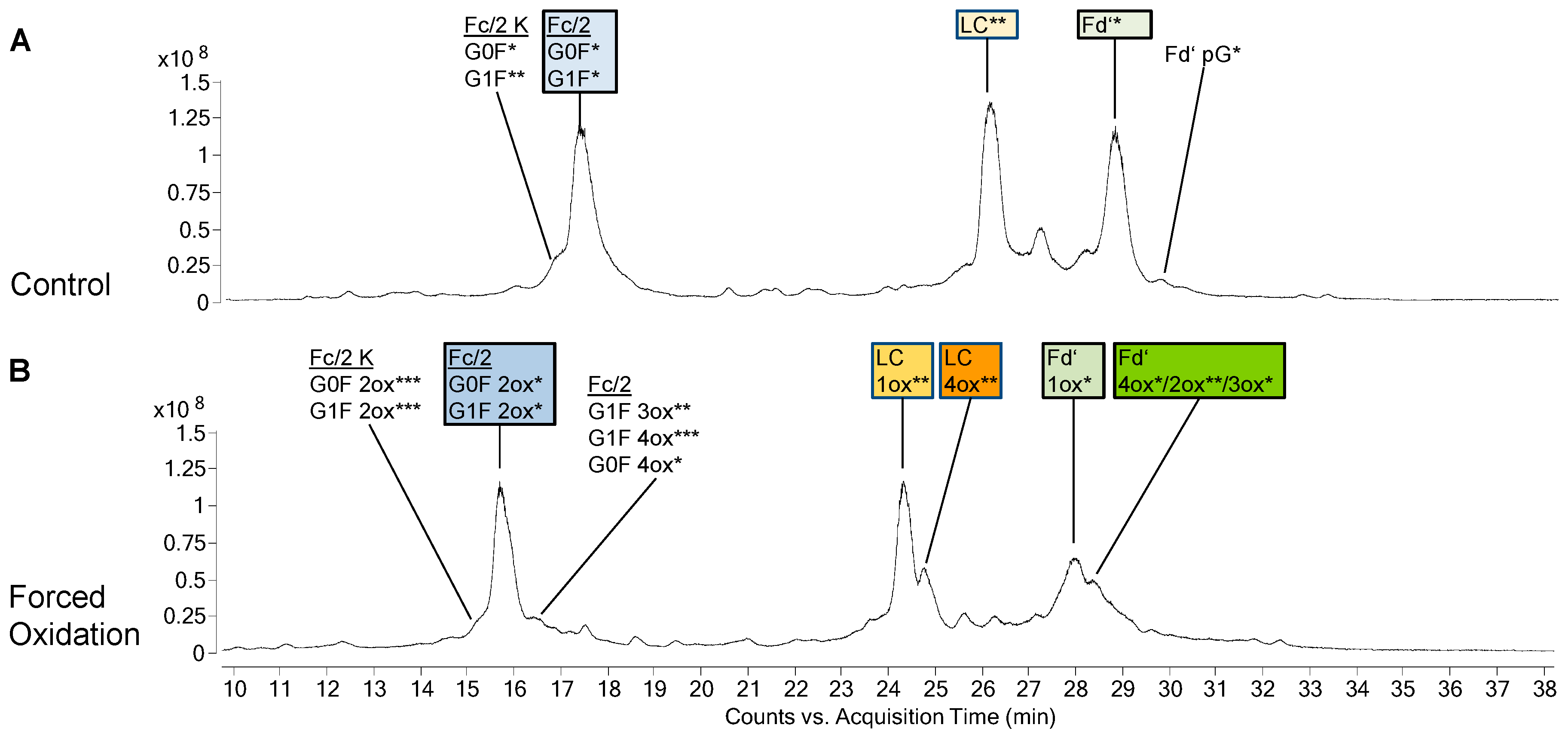
3. Results
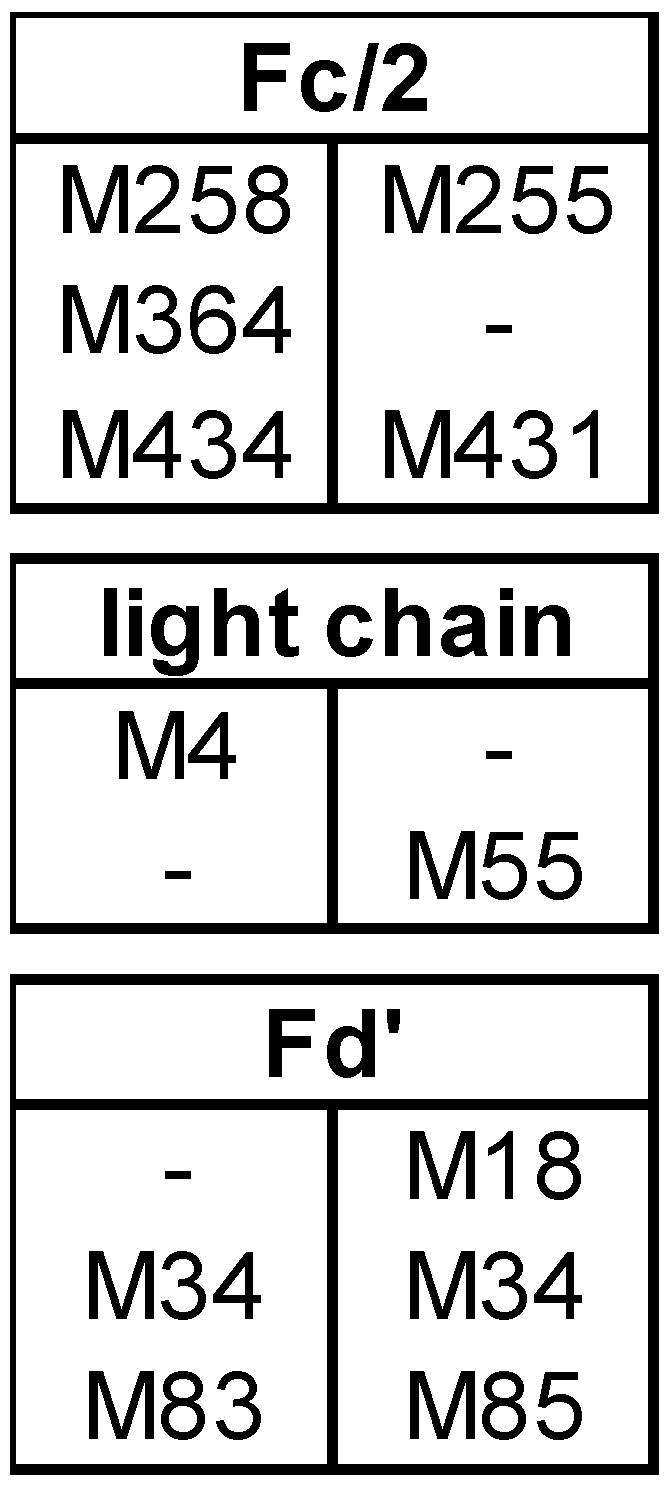
3.1. Subunit Mass and PTMs of Unstressed Bevacizumab and Infliximab
3.2. Forced Oxidation
3.3. Time Course of Forced Oxidation for Comparison of BS and RP
4. Discussion
4.1. PTMs in Unstressed Samples
4.2. Subunit Analysis after Forced Oxidation
4.3. CQA Considerations
4.4. Different Susceptibility of Bevacizumab BS and RP during Forced Oxidation
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Mora, F. Biosimilar: What it is not. Br. J. Clin. Pharmacol. 2015, 80, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Rader, R.A. Nomenclature of new biosimilars will be highly controversial. BioProcess Int. 2011, 9, 28–32. [Google Scholar]
- Miranda-Hernandez, M.P.; Lopez-Morales, C.A.; Pina-Lara, N.; Perdomo-Abundez, F.C.; Perez, N.O.; Revilla-Beltri, J.; Molina-Perez, A.; Estrada-Marin, L.; Flores-Ortiz, L.F.; Ruiz-Arguelles, A.; et al. Pharmacokinetic comparability of a biosimilar trastuzumab anticipated from its physicochemical and biological characterization. BioMed Res. Int. 2015, 2015, 874916. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, D.C.; Misery, L.; Naeyaert, S.; Taylor, P.C. Biological therapies in immune-mediated inflammatory diseases: Can biosimilars reduce access inequities? Front Pharm. 2019, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Grilo, A.L.; Mantalaris, A. The increasingly human and profitable monoclonal antibody market. Trends Biotechnol. 2019, 37, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Parr, M.K.; Montacir, O.; Montacir, H. Physicochemical characterization of biopharmaceuticals. J. Pharm. Biomed. Anal. 2016, 130, 366–389. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Debaene, F.; Diemer, H.; Wagner-Rousset, E.; Colas, O.; Dorsselaer, A.V.; Cianférani, S. Cutting-edge mass spectrometry characterization of originator, biosimilar and biobetter antibodies. J. Mass Spectrom. 2015, 50, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Wagner-Rousset, E.; Ayoub, D.; Van Dorsselaer, A.; Sanglier-Cianférani, S. Characterization of therapeutic antibodies and related products. Anal. Chem. 2013, 85, 715–736. [Google Scholar] [CrossRef]
- Dashivets, T.; Stracke, J.; Dengl, S.; Knaupp, A.; Pollmann, J.; Buchner, J.; Schlothauer, T. Oxidation in the complementarity-determining regions differentially influences the properties of therapeutic antibodies. mAbs 2016, 8, 1525–1535. [Google Scholar] [CrossRef]
- Liu, D.; Ren, D.; Huang, H.; Dankberg, J.; Rosenfeld, R.; Cocco, M.J.; Li, L.; Brems, D.N.; Remmele, R.L. Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry 2008, 47, 5088–5100. [Google Scholar] [CrossRef]
- Pan, H.; Chen, K.; Chu, L.; Kinderman, F.; Apostol, I.; Huang, G. Methionine oxidation in human IgG1 Fc decreases binding affinities to protein A and FcRn. Protein Sci. 2009, 18, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Gaza-Bulseco, G.; Faldu, S.; Hurkmans, K.; Chumsae, C.; Liu, H. Effect of methionine oxidation of a recombinant monoclonal antibody on the binding affinity to protein A and protein G. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008, 870, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.; Yan, Q.; So, C.K.; Soden, T.; Lewis, M.J.; Hu, P. Understanding the impact of methionine oxidation on the biological functions of IgG1 antibodies using hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 2016, 88, 9495–9502. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.A.; Zhang, B.; Cheng, W.; Wang, Y.J. Methionine, tryptophan, and histidine oxidation in a model protein, PTH: Mechanisms and stabilization. J. Pharm. Sci. 2009, 98, 4485–4500. [Google Scholar] [CrossRef] [PubMed]
- Nowak, C.K.; Cheung, J.M.; Dellatore, S.; Katiyar, A.; Bhat, R.; Sun, J.; Liu, H. Forced degradation of recombinant monoclonal antibodies: A practical guide. mAbs 2017, 9, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Jeong, J.; Burgess, B.; Jazayri, M.; Tang, Y.; Taylor Zhang, Y. Development of a rapid RP-UHPLC-MS method for analysis of modifications in therapeutic monoclonal antibodies. J. Chromatogr. B 2016, 1032, 172–181. [Google Scholar] [CrossRef]
- International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. ICH Q1A Guideline, Stability Testing of New Drug Substances and Drug Products. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/ich-q-1-r2-stability-testing-new-drug-substances-products-step-5_en.pdf (accessed on 21 July 2019).
- Folzer, E.; Diepold, K.; Bomans, K.; Finkler, C.; Schmidt, R.; Bulau, P.; Huwyler, J.; Mahler, H.C.; Koulov, A.V. Selective oxidation of methionine and tryptophan residues in a therapeutic IgG1 molecule. J. Pharm. Sci. 2015, 104, 2824–2831. [Google Scholar] [CrossRef]
- Shah, D.D.; Zhang, J.; Hsieh, M.C.; Sundaram, S.; Maity, H.; Mallela, K.M.G. Effect of peroxide- versus alkoxyl-induced chemical oxidation on the structure, stability, aggregation, and function of a therapeutic monoclonal antibody. J. Pharm. Sci. 2018, 107, 2789–2803. [Google Scholar] [CrossRef]
- Hawe, A.; Wiggenhorn, M.; van de Weert, M.; Garbe, J.H.O.; Mahler, H.-C.; Jiskoot, W. Forced degradation of therapeutic proteins. J. Pharm. Sci. 2012, 101, 895–913. [Google Scholar] [CrossRef]
- Zhang, Z.; Pan, H.; Chen, X. Mass spectrometry for structural characterization of therapeutic antibodies. Mass Spectrom. Rev. 2009, 28, 147–176. [Google Scholar] [CrossRef]
- Mouchahoir, T.; Schiel, J.E. Development of an LC-MS/MS peptide mapping protocol for the NISTMab. Anal. Bioanal. Chem. 2018, 410, 2111–2126. [Google Scholar] [CrossRef]
- An, Y.; Zhang, Y.; Mueller, H.M.; Shameem, M.; Chen, X. A new tool for monoclonal antibody analysis application of Ides proteolysis in IgG domain-specific characterization. mAbs 2014, 6, 879–893. [Google Scholar] [CrossRef]
- Fornelli, L.; Ayoub, D.; Aizikov, K.; Beck, A.; Tsybin, Y.O. Middle-down analysis of monoclonal antibodies with electron transfer dissociation orbitrap fourier transform mass spectrometry. Anal. Chem. 2014, 86, 3005–3012. [Google Scholar] [CrossRef]
- Pavon, J.A.; Li, X.; Chico, S.; Kishnani, U.; Soundararajan, S.; Cheung, J.; Li, H.; Richardson, D.; Shameem, M.; Yang, X. Analysis of monoclonal antibody oxidation by simple mixed mode chromatography. J. Chromatogr. A 2016, 1431, 154–165. [Google Scholar] [CrossRef]
- Sokolowska, I.; Mo, J.; Dong, J.; Lewis, M.J.; Hu, P. Subunit mass analysis for monitoring antibody oxidation. mAbs 2017, 9, 498–505. [Google Scholar] [CrossRef]
- Regl, C.; Wohlschlager, T.; Holzmann, J.; Huber, C.G. A generic HPLC method for absolute quantification of oxidation in monoclonal antibodies and Fc-fusion proteins using UV and MS detection. Anal. Chem. 2017, 89, 8391–8398. [Google Scholar] [CrossRef]
- Pisupati, K.; Benet, A.; Tian, Y.; Okbazghi, S.; Kang, J.; Ford, M.; Saveliev, S.; Sen, K.I.; Carlson, E.; Tolbert, T.J.; et al. Biosimilarity under stress: A forced degradation study of remicade® and remsima™. mAbs 2017, 0862, 1–13. [Google Scholar] [CrossRef]
- Kim, J.; Chung, J.; Park, S.; Jung, S.; Kang, D. Evaluation of the physicochemical and biological stability of reconstituted and diluted SB2 (infliximab). Eur. J. Hosp. Pharm. Sci. Pract. 2018, 25, 157–164. [Google Scholar] [CrossRef]
- Hernandez-Jimenez, J.; Martinez-Ortega, A.; Salmeron-Garcia, A.; Cabeza, J.; Prados, J.C.; Ortiz, R.; Navas, N. Study of aggregation in therapeutic monoclonal antibodies subjected to stress and long-term stability tests by analyzing size exclusion liquid chromatographic profiles. Int. J. Biol. Macromol. 2018, 118, 511–524. [Google Scholar] [CrossRef]
- Martinez-Ortega, A.; Herrera, A.; Salmeron-Garcia, A.; Cabeza, J.; Cuadros-Rodriguez, L.; Navas, N. Validated reverse phase HPLC diode array method for the quantification of intact bevacizumab, infliximab and trastuzumab for long-term stability study. Int. J. Biol. Macromol. 2018, 116, 993–1003. [Google Scholar] [CrossRef]
- Suarez, I.; Salmeron-Garcia, A.; Cabeza, J.; Capitan-Vallvey, L.F.; Navas, N. Development and use of specific ELISA methods for quantifying the biological activity of bevacizumab, cetuximab and trastuzumab in stability studies. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1032, 155–164. [Google Scholar] [CrossRef]
- Hermosilla, J.; Sanchez-Martin, R.; Perez-Robles, R.; Salmeron-Garcia, A.; Casares, S.; Cabeza, J.; Cuadros-Rodriguez, L.; Navas, N. Comparative stability studies of different infliximab and biosimilar CT-P13 clinical solutions by combined use of physicochemical analytical techniques and enzyme-linked immunosorbent assay (ELISA). BioDrugs 2019, 33, 193–205. [Google Scholar] [CrossRef]
- Pisupati, K.; Tian, Y.; Okbazghi, S.; Benet, A.; Ackermann, R.; Ford, M.; Saveliev, S.; Hosfield, C.M.; Urh, M.; Carlson, E.; et al. A multidimensional analytical comparison of remicade and the biosimilar remsima. Anal. Chem. 2017, 89, 4838–4846. [Google Scholar] [CrossRef]
- Edelman, G.M.; Cunningham, B.A.; Gall, W.E.; Gottlieb, P.D.; Rutishauser, U.; Waxdal, M.J. The covalent structure of an entire gammaG immunoglobulin molecule. Proc. Natl. Acad. Sci. USA 1969, 63, 78–85. [Google Scholar] [CrossRef]
- Chumsae, C.; Gaza-Bulseco, G.; Sun, J.; Liu, H. Comparison of methionine oxidation in thermal stability and chemically stressed samples of a fully human monoclonal antibody. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 850, 285–294. [Google Scholar] [CrossRef]
- Luo, Q.; Joubert, M.K.; Stevenson, R.; Ketchem, R.R.; Narhi, L.O.; Wypych, J. Chemical modifications in therapeutic protein aggregates generated under different stress conditions. J. Biol. Chem. 2011, 286, 25134–25144. [Google Scholar] [CrossRef]
- Lim, H.; Lee, S.H.; Lee, H.T.; Lee, J.U.; Son, J.Y.; Shin, W.; Heo, Y.S. Structural biology of the TNFalpha antagonists used in the treatment of rheumatoid arthritis. Int. J. Mol. Sci. 2018, 19, 768. [Google Scholar] [CrossRef]
- Bertolotti-Ciarlet, A.; Wang, W.; Lownes, R.; Pristatsky, P.; Fang, Y.; McKelvey, T.; Li, Y.; Li, Y.; Drummond, J.; Prueksaritanont, T.; et al. Impact of methionine oxidation on the binding of human IgG1 to Fc Rn and Fc gamma receptors. Mol. Immunol. 2009, 46, 1878–1882. [Google Scholar] [CrossRef]
- Seo, N.; Polozova, A.; Zhang, M.; Yates, Z.; Cao, S.; Li, H.; Kuhns, S.; Maher, G.; McBride, H.J.; Liu, J. Analytical and functional similarity of amgen biosimilar ABP 215 to bevacizumab. mAbs 2018, 10, 678–691. [Google Scholar] [CrossRef]
- Hageman, T.; Wei, H.; Kuehne, P.; Fu, J.; Ludwig, R.; Tao, L.; Leone, A.; Zocher, M.; Das, T.K. Impact of tryptophan oxidation in complementarity-determining regions of two monoclonal antibodies on structure-function characterized by hydrogen-deuterium exchange mass spectrometry and surface plasmon resonance. Pharm. Res. 2018, 36, 24. [Google Scholar] [CrossRef]
- Kerwin, B.A. Polysorbates 20 and 80 used in the formulation of protein biotherapeutics: Structure and degradation pathways. J. Pharm. Sci. 2008, 97, 2924–2935. [Google Scholar] [CrossRef]

| mAb | Product | Duration of Forced Oxidation or Control Treatment | |||
|---|---|---|---|---|---|
| 0 h | 24 h | 48 h | 72 h | ||
| Bevacizumab | RP | con - | con oxi | con oxi | con oxi |
| BS | con - | con oxi | con oxi | con oxi | |
| Infliximab | RP | con - | con oxi | con oxi | con oxi |
| BS | con - | con oxi | con oxi | con oxi | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyck, Y.F.K.; Rehm, D.; Joseph, J.F.; Winkler, K.; Sandig, V.; Jabs, W.; Parr, M.K. Forced Degradation Testing as Complementary Tool for Biosimilarity Assessment. Bioengineering 2019, 6, 62. https://doi.org/10.3390/bioengineering6030062
Dyck YFK, Rehm D, Joseph JF, Winkler K, Sandig V, Jabs W, Parr MK. Forced Degradation Testing as Complementary Tool for Biosimilarity Assessment. Bioengineering. 2019; 6(3):62. https://doi.org/10.3390/bioengineering6030062
Chicago/Turabian StyleDyck, Yan Felix Karl, Daniel Rehm, Jan Felix Joseph, Karsten Winkler, Volker Sandig, Wolfgang Jabs, and Maria Kristina Parr. 2019. "Forced Degradation Testing as Complementary Tool for Biosimilarity Assessment" Bioengineering 6, no. 3: 62. https://doi.org/10.3390/bioengineering6030062
APA StyleDyck, Y. F. K., Rehm, D., Joseph, J. F., Winkler, K., Sandig, V., Jabs, W., & Parr, M. K. (2019). Forced Degradation Testing as Complementary Tool for Biosimilarity Assessment. Bioengineering, 6(3), 62. https://doi.org/10.3390/bioengineering6030062






