Micropatterning Decellularized ECM as a Bioactive Surface to Guide Cell Alignment, Proliferation, and Migration
Abstract
:1. Introduction
2. Experimental Procedures
2.1. Micropatterning Polydimethyl Siloxane Substrates
2.2. Substrate Preparation and Cell Culture
2.3. Preparation of Decellularized ECM and Culture
2.4. Immunofluorescence and Microscopy
2.5. Quantification and Analysis
2.6. Statistics
3. Results
3.1. Patterning Micron-Level Physical Constraints Guides Fibroblast Adhesion, Alignment, and Proliferation
3.2. Micropatterns Facilitate Aligned ECM Assembly
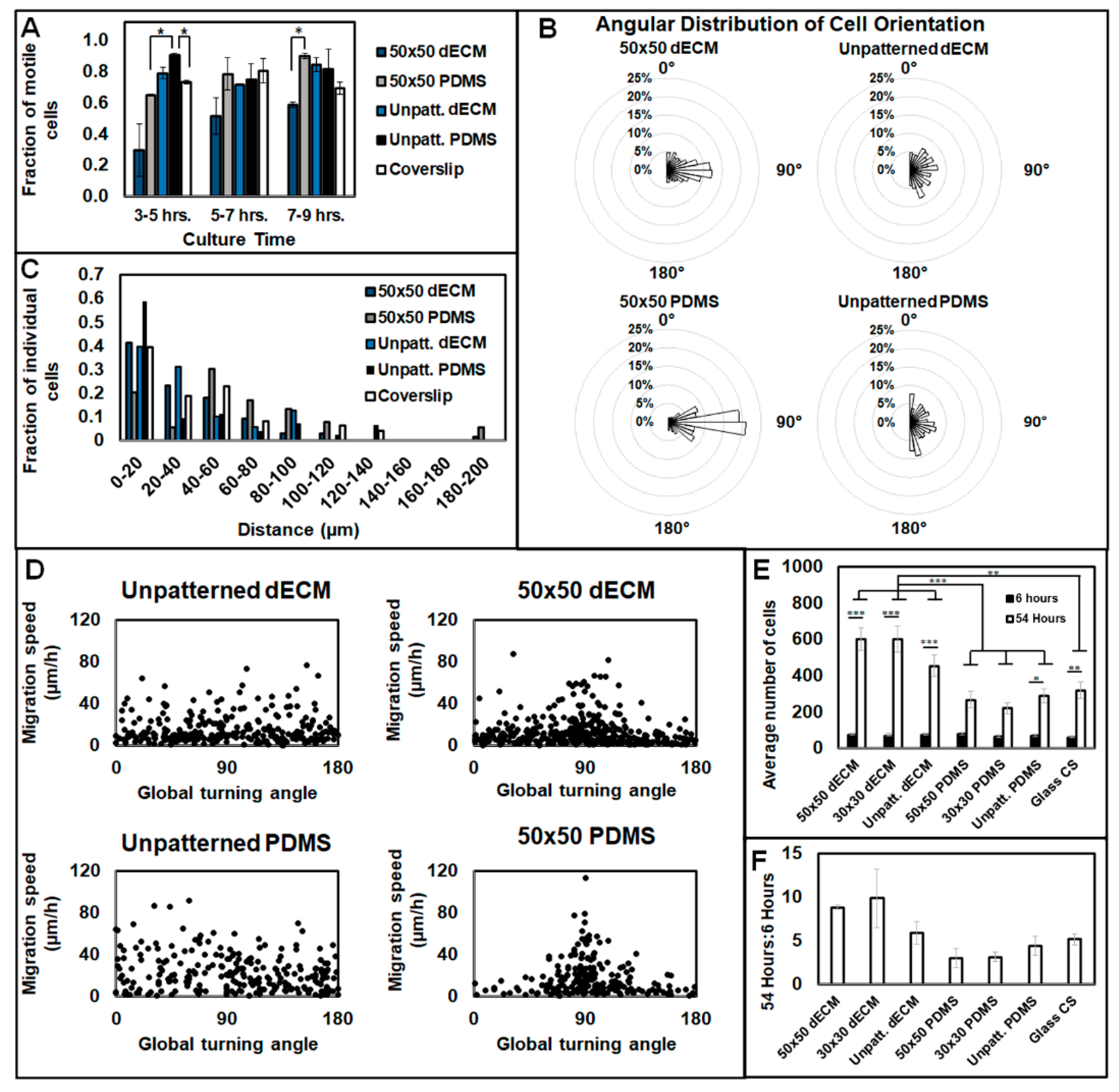
3.3. Aligned ECM Promotes Guided Cell Adhesion and Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dankers, P.Y.W.; Harmsen, M.C.; Brouwer, L.A.; Van Luyn, M.J.A.; Meijer, E.W. A modular and supramolecular approach to bioactive scaffolds for tissue engineering. Nat. Mater. 2005, 4, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Liu, D.; Qi, X.; Yue, Z.; Cao, H.; Zhang, K.; Lei, X.; Wang, Y.; Kong, D.; Gao, J.; et al. Self-assembled GFFYK peptide hydrogel enhances the therapeutic efficacy of mesenchymal stem cells in a mouse hindlimb ischemia model. Acta Biomater. 2019, 85, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Koffler, J.; Kaufman-Francis, K.; Shandalov, Y.; Egozi, D.; Pavlov, D.A.; Landesberg, A.; Levenberg, S. Improved vascular organization enhances functional integration of engineered skeletal muscle grafts. Proc. Natl. Acad. Sci. USA 2011, 108, 14789–14794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Cheng, R.; Sun, Z.; Su, W.; Pan, G.; Zhao, S.; Zhao, J.; Cui, W. Flexible bipolar nanofibrous membranes for improving gradient microstructure in tendon-to-bone healing. Acta Biomater. 2017, 61, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.J.; Hyder, N.; Quadir, M.A.; Courchesne, N.-M.D.; Seeherman, H.J.; Nevins, M.; Spector, M.; Hammond, P.T. Adaptive growth factor delivery from a polyelectrolyte coating promotes synergistic bone tissue repair and reconstruction. Proc. Natl. Acad. Sci. USA 2014, 111, 12847–12852. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Long, N.J.; Li, L.; Lu, Y.; Li, M.; Cao, J.; Zhang, Y.; Zhang, Q.; Xu, S.; Yang, Z.; et al. Multi-functional bismuth-doped bioglasses: Combining bioactivity and photothermal response for bone tumor treatment and tissue repair. Light. Sci. Appl. 2018, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Peng, J.; Dong, X.; Xu, Y.; Li, H.; Chang, J. Combined chemical and structural signals of biomaterials synergistically activate cell-cell communications for improving tissue regeneration. Acta Biomater. 2017, 55, 249–261. [Google Scholar] [CrossRef]
- Zhao, W.; Michalik, D.; Ferguson, S.; Hofstetter, W.; Lemaître, J.; Von Rechenberg, B.; Bowen, P. Rapid evaluation of bioactive Ti-based surfaces using an in vitro titration method. Nat. Commun. 2019, 10, 2062. [Google Scholar] [CrossRef] [Green Version]
- Jeffries, E.M.; Wang, Y. Biomimetic micropatterned multi-channel nerve guides by templated electrospinning. Biotechnol. Bioeng. 2012, 109, 1571–1582. [Google Scholar] [CrossRef] [Green Version]
- Cornwell, K.G.; Landsman, A.; James, K.S. Extracellular Matrix Biomaterials for Soft Tissue Repair. Clin. Podiatr. Med. Surg. 2009, 26, 507–523. [Google Scholar] [CrossRef]
- Harris, G.M.; Madigan, N.N.; Lancaster, K.Z.; Enquist, L.W.; Windebank, A.J.; Schwartz, J.; Schwarzbauer, J.E. Nerve Guidance by a Decellularized Fibroblast Extracellular Matrix. Matrix Boil. 2016, 60, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Kwok, J.C.F.; Dick, G.; Wang, D.; Fawcett, J.W. Extracellular matrix and perineuronal nets in CNS repair. Dev. Neurobiol. 2011, 71, 1073–1089. [Google Scholar] [CrossRef] [PubMed]
- Harris, G.M.; Raitman, I.; Schwarzbauer, J.E. Cell-derived decellularized extracellular matrices. Methods Cell Biol. 2018, 143, 97–114. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, P.; Carraher, C.; Schwarzbauer, J.E. Assembly of Fibronectin Extracellular Matrix. Annu. Rev. Cell Dev. Boil. 2010, 26, 397–419. [Google Scholar] [CrossRef] [Green Version]
- Marastoni, S.; Ligresti, G.; Lorenzon, E.; Colombatti, A.; Mongiat, M. Extracellular Matrix: A Matter of Life and Death. Connect. Tissue Res. 2008, 49, 203–206. [Google Scholar] [CrossRef]
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- Mao, Y.; Schwarzbauer, J.E. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Boil. 2005, 24, 389–399. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Zhang, Z.; Lou, K.; Zhang, Q.; Wang, S.; Ni, J.; Liu, W.; Fan, S.; Lin, X. Current advances in the development of natural meniscus scaffolds: Innovative approaches to decellularization and recellularization. Cell Tissue Res. 2017, 370, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Cukierman, E.; Blanco, P.; Palucka, A.K.; Gill, M.; Pascual, V.; Banchereau, J.F. Taking Cell-Matrix Adhesions to the Third Dimension. Science 2001, 294, 1708–1712. [Google Scholar] [CrossRef]
- Hellewell, A.L.; Rosini, S.; Adams, J.C. A Rapid, Scalable Method for the Isolation, Functional Study, and Analysis of Cell-derived Extracellular Matrix. J. Vis. Exp. 2017, 119, 55051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.; Schwarzbauer, J.E. Accessibility to the fibronectin synergy site in a 3D matrix regulates engagement of alpha5beta1 versus alphavbeta3 integrin receptors. Cell Commun. Adhes. 2006, 13, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.; Vogt, C.D.; Leong, K.W.; Zhao, F. Highly Aligned Nanofibrous Scaffold Derived from Decellularized Human Fibroblasts. Adv. Funct. Mater. 2014, 24, 3027–3035. [Google Scholar] [CrossRef] [Green Version]
- Zhong, S.P.; Zhang, Y.; Lim, C.T. Tissue scaffolds for skin wound healing and dermal reconstruction. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 510–525. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, B.; Ao, M.; White, L.M.; Means, A.L.; Brewer, B.M.; Yang, L.; Washington, M.K.; Shi, C.; Franco, O.E.; Weaver, A.M.; et al. Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J. Cell Boil. 2017, 216, 3799–3816. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Bae, W.-G.; Kim, Y.J.; Seonwoo, H.; Choung, H.-W.; Jang, K.-J.; Park, S.; Kim, B.H.; Kim, H.-N.; Choi, K.S.; et al. Directional Matrix Nanotopography with Varied Sizes for Engineering Wound Healing. Adv. Heal. Mater. 2017, 6, 1700297. [Google Scholar] [CrossRef]
- Oakley, C.; Jaeger, N.A.; Brunette, D.M. Sensitivity of Fibroblasts and Their Cytoskeletons to Substratum To pographies: Topographic Guidance and Topographic Compensation by Micromachined Grooves of Different Dimensions. Exp. Cell Res. 1997, 234, 413–424. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Scheideler, L.; Reichl, R.; Geis-Gerstorfer, J. Effects of topography and composition of titanium surface oxides on osteoblast responses. Biomaterials 2004, 25, 4087–4103. [Google Scholar] [CrossRef]
- Harris, G.M.; Piroli, M.E.; Jabbarzadeh, E. Deconstructing the Effects of Matrix Elasticity and Geometry in Mesenchymal Stem Cell Lineage Commitment. Adv. Funct. Mater. 2013, 24, 2396–2403. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Orkwis, J.A.; Harris, G.M. Preparation of Tunable Extracellular Matrix Microenvironments to Evaluate Schwann Cell Phenotype Specification. J. Vis. Exp. 2020, 160, 61496. [Google Scholar] [CrossRef]
- Fu, J.; Wang, Y.-K.; Yang, M.T.; Desai, R.; Yu, X.; Liu, Z.; Chen, C.S. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 2010, 7, 733–736. [Google Scholar] [CrossRef] [PubMed]
- Luu, T.; Gott, S.C.; Woo, B.W.K.; Rao, M.P.; Liu, W.F. Micro- and Nanopatterned Topographical Cues for Regulating Macrophage Cell Shape and Phenotype. ACS Appl. Mater. Interfaces 2015, 7, 28665–28672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, J.R.F.; Wu, N.; Porter, D.; Buford, M.; Wolfarth, M.; Holian, A. Particle length-dependent titanium dioxide nanomaterials toxicity and bioactivity. Part. Fibre Toxicol. 2009, 6, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Orkwis, J.A.; Devine, B.M.; Harris, G.M. Extracellular matrix cues modulate Schwann cell morphology, proliferation, and protein expression. J. Tissue Eng. Regen. Med. 2019, 14, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Demidova-Rice, T.N.; Hamblin, M.R.; Herman, I.M. Acute and Impaired Wound Healing: Pathophysiology and Current Methods for Drug Delivery, Part 2: Role of Growth Factors in Normal and Pathological Wound Healing Therapeutic Potential and Methods of Delivery. Adv. Ski. Wound Care 2012, 25, 349–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yim, E.K.F.; Darling, E.M.; Kulangara, K.; Guilak, F.; Leong, K.W. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials 2010, 31, 1299–1306. [Google Scholar] [CrossRef] [Green Version]
- Yim, E.K.F.; Reano, R.M.; Pang, S.W.; Yee, A.F.; Chen, C.S.; Leong, K.W. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials 2005, 26, 5405–5413. [Google Scholar] [CrossRef] [Green Version]
- Ohara, P.T.; Buck, R.C. Contact guidance in vitro: A light, transmission, and scanning electron microscopic study. Exp. Cell Res. 1979, 121, 235–249. [Google Scholar] [CrossRef]
- Braber, E.D.; De Ruijter, J.; Smits, H.; Ginsel, L.; Von Recum, A.; Jansen, J. Quantitative analysis of cell proliferation and orientation on substrata with uniform parallel surface micro-grooves. Biomaterials 1996, 17, 1093–1099. [Google Scholar] [CrossRef] [Green Version]
- Brunette, D. Spreading and orientation of epithelial cells on grooved substrata. Exp. Cell Res. 1986, 167, 203–217. [Google Scholar] [CrossRef]
- Dunn, G.A.; Brown, A.F. Alignment of fibroblasts on grooved surfaces described by a simple geometric transformation. J. Cell Sci. 1986, 83, 313–340. [Google Scholar] [PubMed]
- Hamilton, D.W.; Chehroudi, B.; Brunette, D.M. Comparative response of epithelial cells and osteoblasts to microfabricated tapered pit topographies in vitro and in vivo. Biomaterials 2007, 28, 2281–2293. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicka-Patynowski, I.; Endo, T.; Yamamoto, H.; Esaki, M. The ins and outs of fibronectin matrix assembly. J. Cell Sci. 2003, 116, 3269–3276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, A.; Nuschke, A.; Yates, C.C. Skin tissue repair: Matrix microenvironmental influences. Matrix Boil. 2015, 49, 25–36. [Google Scholar] [CrossRef] [Green Version]
- Vaz, R.; Martins, G.G.; Thorsteinsdóttir, S.; Rodrigues, G. Fibronectin promotes migration, alignment and fusion in an in vitro myoblast cell model. Cell Tissue Res. 2012, 348, 569–578. [Google Scholar] [CrossRef]
- Bershadsky, A.D.; Balaban, N.Q.; Geiger, B. Adhesion-dependent cell mechanosensitivity. Annu. Rev. Cell Dev. Biol. 2003, 19, 677–695. [Google Scholar] [CrossRef] [Green Version]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Boil. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [Green Version]
- Lehnert, D.; Wehrle-Haller, B.; David, C.; Weiland, U.; Ballestrem, C.; Imhof, B.A.; Bastmeyer, M. Cell behaviour on micropatterned substrata: Limits of extracellular matrix geometry for spreading and adhesion. J. Cell Sci. 2004, 117, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Li, W.; Dong, X.; Yuan, X.; Midgley, A.C.; Chang, H.; Wang, Y.; Wang, H.; Wang, K.; Ma, P.X.; et al. In vivo engineered extracellular matrix scaffolds with instructive niches for oriented tissue regeneration. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cady, E.; Orkwis, J.A.; Weaver, R.; Conlin, L.; Madigan, N.N.; Harris, G.M. Micropatterning Decellularized ECM as a Bioactive Surface to Guide Cell Alignment, Proliferation, and Migration. Bioengineering 2020, 7, 102. https://doi.org/10.3390/bioengineering7030102
Cady E, Orkwis JA, Weaver R, Conlin L, Madigan NN, Harris GM. Micropatterning Decellularized ECM as a Bioactive Surface to Guide Cell Alignment, Proliferation, and Migration. Bioengineering. 2020; 7(3):102. https://doi.org/10.3390/bioengineering7030102
Chicago/Turabian StyleCady, Emily, Jacob A. Orkwis, Rachel Weaver, Lia Conlin, Nicolas N. Madigan, and Greg M. Harris. 2020. "Micropatterning Decellularized ECM as a Bioactive Surface to Guide Cell Alignment, Proliferation, and Migration" Bioengineering 7, no. 3: 102. https://doi.org/10.3390/bioengineering7030102
APA StyleCady, E., Orkwis, J. A., Weaver, R., Conlin, L., Madigan, N. N., & Harris, G. M. (2020). Micropatterning Decellularized ECM as a Bioactive Surface to Guide Cell Alignment, Proliferation, and Migration. Bioengineering, 7(3), 102. https://doi.org/10.3390/bioengineering7030102





