Predictive Monitoring of Shake Flask Cultures with Online Estimated Growth Models
Abstract
:1. Introduction
- 1.
- Unlike static models, which cannot be applied without preexisting data, the proposed PF workflow extends predictive monitoring to shake flask cultures, which assess novel strains and substrate compositions.
- 2.
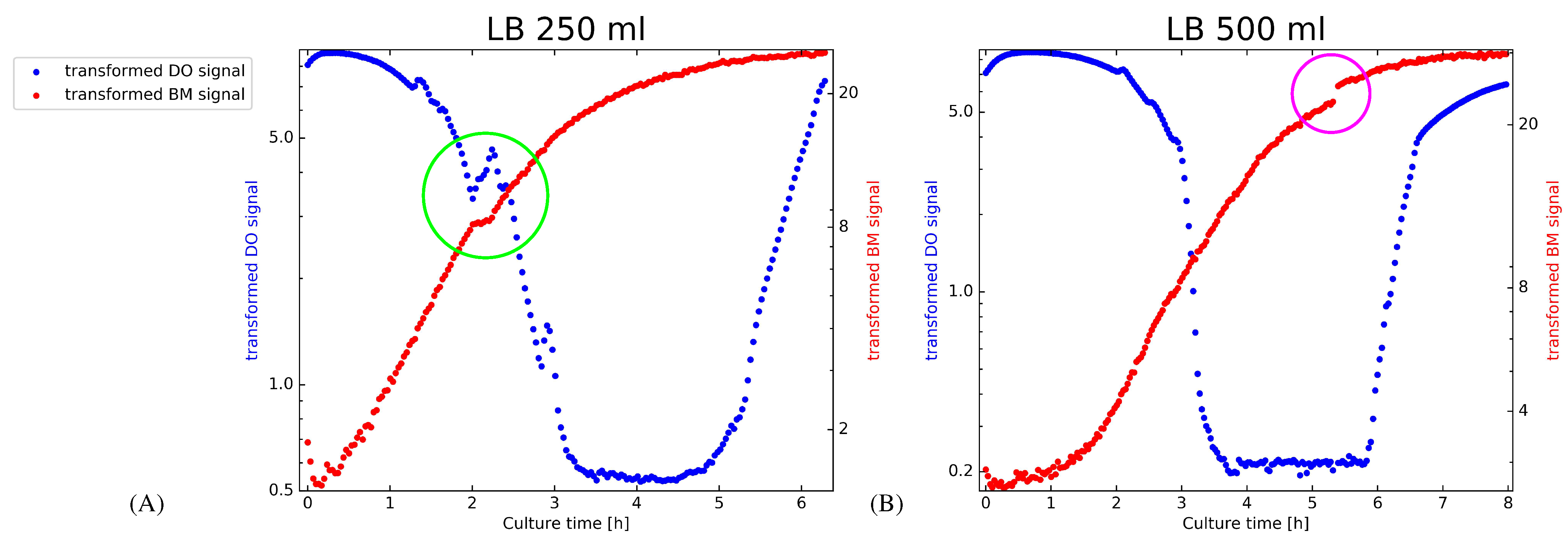
- The ability of the PF to adapt swiftly to altered growth characteristics without user intervention allows predictive monitoring to cope with non-stationary situations, which are illustrated in Figure 1.
2. Materials and Methods
2.1. Shake Flask Experiments
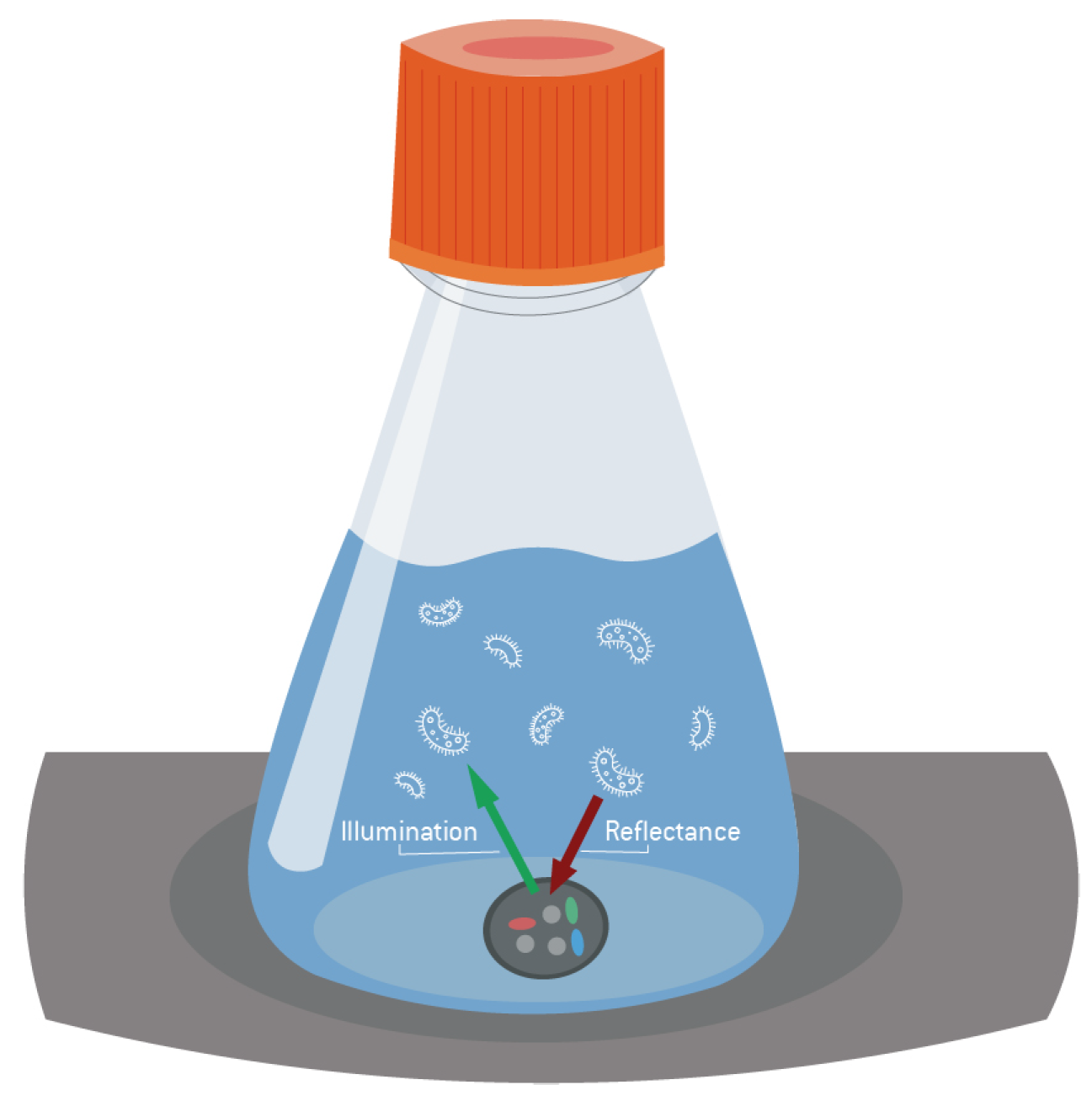
2.2. Sensor Technology
2.3. Data
2.4. Software
2.5. Models for Growth and Decline
2.6. Particle Filter Workflow
2.7. Initialization of the Particle Filter and Static Model Fitting
- The time of sample collection and the respective transformed BM or DO signal of a complete culture are randomly reshuffled and used as training data.
- After initializing the samples with randomly drawn Gompertz or Logistic model parameters, MCMC itself is just an application of the PF updates in Figure 3, however, on reshuffled growth profiles.
- By reshuffling the growth profiles, the sample window of the PF will always capture the dynamics of the entire growth profile. The generated samples are thus adapted to capture the dynamics of the entire profile and infer indeed a static model.
2.8. Predictive Accuracy
2.9. Statistical Significance
2.10. Event Time Assessment
2.11. Hyperparameter Tuning
2.11.1. Unbiased Evaluation
2.11.2. Validation Experiments
- 1.
- To demonstrate the importance of adapting the state noise levels in the proposed PF implementation, we assess the filter internals while adapting to the transformed BM signal of the LB-500 mL-2 experiment (Figure 1, subplot (B)). The depicted results are obtained after initializing the PF on the LB-250ml-2 experiment before we switch to the LB-500 mL-2 experiment. The implications of estimating appropriate state noise levels are best captured in a synchronized view of how the particle filter operates. We illustrate to this end how the state noise levels of the filter, the transformed BM signal and a windowed evolve. The values are in this case estimated from one step ahead predictions with a sliding window of 15 samples.
- 2.
- Evaluating PF inferred growth models for predictive monitoring during strain and substrate optimization has to mimic situations where little is known about how the culture evolves. To obtain unbiased assessments for such use cases, we use both LB 250 mL cultures for PF initialization and switch the PF for evaluation purposes to the E. coli cultures that were grown in 500 mL of LB and TB medium (Table 2). Strain and substrate optimization implies that data of a culture that was obtained under respective conditions for fitting SMs is not yet available. Such situations hence prevent us in practice from using statically fit models for predictive monitoring. For evaluation purposes, it is however still informative to see how well PF based inference competes with SMs.
2.12. Derivation, Code and Data Availability
3. Results
3.1. Tracking Performance
3.2. Predictions and Accuracy
3.3. Event Notification
4. Discussion and Conclusions
- Different scales of growth model parameters are explicitly considered and allow for a parameter specific adaptation between convergence and tracking.
- Some phases of microbial growth follow static patterns and allow precise parameter inference. At other times, randomly occurring factors cause the growth dynamics to change considerably. Considering a window for estimating state noise levels adapts inference automatically to different transient dynamics. Predictive monitoring thus reacts efficiently to changing regimes.
- By adapting the state noise levels automatically, tedious validation experiments for calibrating the state noise levels are avoided and the proposed PF can be applied immediately.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BM | Biomass |
| DAG | Directed acyclic graph |
| DO | Dissolved oxygen |
| EKF | Extended Kalman filter |
| FDA | Food and drug administration |
| FDR | False discovery rate |
| GPL | GNU public license |
| KF | Kalman filter |
| LB | Lysogeny broth |
| ML | Machine learning |
| Mean square error | |
| MCMC | Markov Chain Monte Carlo |
| O2 | Oxygen |
| OD | Optical density at |
| PAT | Process analytical technology |
| PF | Particle filter |
| QbD | Quality by design |
| SF | Static fit |
| SFR | shake flask reader |
| Sum of squared differences | |
| TB | Terrific broth |
References
- Büchs, J. Introduction to advantages and problems of shaken cultures. Biochem. Eng. J. 2001, 7, 91–98. [Google Scholar] [CrossRef]
- Winkler, K.; Socher, M.L. Shake Flask Technology. In Encyclopedia of Industrial Biotechnology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; pp. 1–16. [Google Scholar] [CrossRef]
- Vasala, A.; Panula, J.; Bollok, M.; Illmann, L.; Hälsig, C.; Neubauer, P. A new wireless system for decentralised measurement of physiological parameters from shake flasks. Microb. Cell Fact. 2006, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Anderlei, T.; Büchs, J. Device for sterile online measurement of the oxygen transfer rate in shaking flasks. Biochem. Eng. J. 2001, 7, 157–162. [Google Scholar] [CrossRef]
- Bruder, S.; Reifenrath, M.; Thomik, T.; Boles, E.; Herzog, K. Parallelised online biomass monitoring in shake flasks enables efficient strain and carbon source dependent growth characterisation of Saccharomyces cerevisiae. Microb. Cell Fact. 2016, 15, 127. [Google Scholar] [CrossRef] [Green Version]
- Schiefelbein, S.; Fröhlich, A.; John, G.T.; Beutler, F.; Wittmann, C.; Becker, J. Oxygen supply in disposable shake-flasks: Prediction of oxygen transfer rate, oxygen saturation and maximum cell concentration during aerobic growth. Biotechnol. Lett. 2013, 35, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Findeis, M.; John, G.T. Accurate Insight into Oxygen Content of Shake Cultures. Available online: https://www.presens.de/knowledge/publications/application-note/accurate-insight-into-oxygen-content-of-shake-cultures-582 (accessed on 9 April 2021).
- Scheidle, M.; Klinger, J.; Büchs, J. Combination of On-line pH and Oxygen Transfer Rate Measurement in Shake Flasks by Fiber Optical Technique and Respiration Activity MOnitoring System (RAMOS). Sensors 2007, 7, 3472–3480. [Google Scholar] [CrossRef] [Green Version]
- Food and Drug Administration. Guidance for Industry PAT—A Framework for Innovative Pharmaceutical Manufacturing and Quality Assurance; Food and Drug Administration: Washington, DC, USA, 2004. Available online: https://www.fda.gov/media/71012/download (accessed on 9 April 2021).
- Food and Drug Administration. Q8(R2)-Pharmaceutical-Development; ICH: Rockville, MD, USA, 2009. [Google Scholar]
- Trautmann, H. PAT in Bioprocessing. BioWorld EUROPE. 2005. Available online: http://www.abiotec.ch/downloads/e_PAT_in_Bioprocessing.pdf (accessed on 9 April 2021).
- Rathore, A.S.; Winkle, H. Quality by design for biopharmaceuticals. Nat. Biotechnol. 2009, 27, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.S.; Mhatre, R. (Eds.) Quality by Design for Biopharmaceuticals: Principles and Case Studies; Wiley: Hoboken, NJ, USA, 2009. [Google Scholar]
- Golabgir, A.; Herwig, C. Combining Mechanistic Modeling and Raman Spectroscopy for Real-Time Monitoring of Fed-Batch Penicillin Production. Chem. Ing. Tech. 2016, 88, 764–776. [Google Scholar] [CrossRef]
- Kager, J.; Herwig, C.; Stelzer, I.V. State estimation for a penicillin fed-batch process combining particle filtering methods with online and time delayed offline measurements. Chem. Eng. Sci. 2018, 177, 234–244. [Google Scholar] [CrossRef]
- Montague, G.A.; Martin, E.B.; O’Malley, C.J. Forecasting for fermentation operational decision making. Biotechnol. Prog. 2008, 24, 1033–1041. [Google Scholar] [CrossRef]
- Karim, M.; Hodge, D.; Simon, L. Data-Based Modeling and Analysis of Bioprocesses: Some Real Experiences. Biotechnol. Prog. 2003, 19, 1591–1605. [Google Scholar] [CrossRef]
- Trelea, I.C.; Titica, M.; Landaud, S.; Latrille, E.; Corrieu, G.; Cheruy, A. Predictive modelling of brewing fermentation: From knowledge-based to black-box models. Math. Comput. Simul. 2001, 56, 405–424. [Google Scholar] [CrossRef]
- Losen, M.; Frölich, B.; Pohl, M.; Büchs, J. Effect of oxygen limitation and medium composition on Escherichia coli fermentation in shake-flask cultures. Biotechnol. Prog. 2004, 20, 1062–1068. [Google Scholar] [CrossRef]
- Russell, J.B.; Diez-Gonzalez, F. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 1998, 39, 205–234. [Google Scholar]
- Peterson, C.N.; Mandel, M.J.; Silhavy, T.J. Escherichia coli starvation diets: Essential nutrients weigh in distinctly. J. Bacteriol. 2005, 187, 7549–7553. [Google Scholar] [CrossRef] [Green Version]
- Ripley, B.D. Pattern Recognition and Neural Networks; Cambridge University Press: Cambridge, UK, 1996. [Google Scholar]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Monod, J. The Growth of Bacterial Cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef] [Green Version]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; van’t Riet, K. Modeling of the Bacterial Growth Curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef] [Green Version]
- Kalman, R.E. A new approach to linear filtering and prediction problems. J. Basic Eng. 1960, 82, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Doucet, A. (Ed.) Sequential Monte Carlo Methods in Practice; OCLC: 867161886; Statistics for Engineering and Information Science; Springer: New York, NY, USA, 2010. [Google Scholar]
- Gordon, N.; Salmond, D.; Smith, A. Novel approach to nonlinear/non-Gaussian Bayesian state estimation. IEE Proc. F-Radar Signal Process. 1993, 140, 107. [Google Scholar] [CrossRef] [Green Version]
- Maschke, R.W.; Seidel, S.; Bley, T.; Eibl, R.; Eibl, D. Determination of culture design spaces in shaken disposable cultivation systems for CHO suspension cell cultures. Biochem. Eng. J. 2022, 177, 108224. [Google Scholar] [CrossRef]
- Lennox, E. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1955, 1, 190–206. [Google Scholar] [CrossRef]
- Terrific Broth (TB) Medium. 2015. Available online: http://cshprotocols.cshlp.org/content/2015/9/pdb.rec085894.short (accessed on 9 April 2021). [CrossRef]
- Ude, C.; Schmidt-Hager, J.; Findeis, M.; John, G.; Scheper, T.; Beutel, S. Application of an Online-Biomass Sensor in an Optical Multisensory Platform Prototype for Growth Monitoring of Biotechnical Relevant Microorganism and Cell Lines in Single-Use Shake Flasks. Sensors 2014, 14, 17390–17405. [Google Scholar] [CrossRef] [Green Version]
- Ebert, F.V.; Reitz, C.; Cruz-Bournazou, M.N.; Neubauer, P. Characterization of a noninvasive on-line turbidity sensor in shake flasks for biomass measurements. Biochem. Eng. J. 2018, 132, 20–28. [Google Scholar] [CrossRef]
- Patmanidis, S.; Charalampidis, A.C.; Kordonis, I.; Mitsis, G.D.; Papavassilopoulos, G.P. Comparing Methods for Parameter Estimation of the Gompertz Tumor Growth Model. IFAC-PapersOnLine 2017, 50, 12203–12209. [Google Scholar] [CrossRef]
- Gompertz, B. On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies. Philos. Trans. R. Soc. 1825, 115, 513–583. [Google Scholar] [CrossRef]
- Skinner, G.E.; Larkin, J.W.; Rhodehamel, E.J. Mathematical modeling of microbial growth: A review. J. Food Saf. 1994, 14, 175–217. [Google Scholar] [CrossRef]
- Gibson, A.M.; Bratchell, N.; Roberts, T.A. The effect of sodium chloride and temperature on the rate and extent of growth of Clostridium botulinum type A in pasteurized pork slurry. J. Appl. Bacteriol. 1987, 62, 479–490. [Google Scholar] [CrossRef]
- Masreliez, C.J.; Martin, R.D. Robust Bayesian Estimation for the Linear Model and Robustifying the Kalman Filter. IEEE Trans. Autom. Control 1977, 22, 361–371. [Google Scholar] [CrossRef]
- Smith, G.L.; Schmidt, S.F.; McGee, L.A. Application of Statistical Filter Theory to the Optimal Estimation of Position and Velocity on Board a Circumlunar Vehicle; Technical Report R-135; NASA Ames Research Center: Moffet Field, CA, USA, 1962. [Google Scholar]
- McElhoe, B.A. An Assessment of the Navigation and Course Corrections for a Manned Flyby of Mars or Venus. IEEE Trans. Aerosp. Electron. Syst. 1966, 2, 613–623. [Google Scholar] [CrossRef]
- Julier, S.J.; Uhlmann, J.K. A New Extension of the Kalman Filter to Nonlinear Systems. In Proceedings of Signal Processing, Sensor Fusion, and Target Recognition VI; Kadar, I., Ed.; SPIE: Bellingham, WA, USA, 1997. [Google Scholar] [CrossRef]
- Julier, S.J.; Uhlmann, J.K. Unscented filtering and nonlinear estimation. Proc. IEEE 2004, 92, 401–422. [Google Scholar] [CrossRef] [Green Version]
- Sykacek, P.; Roberts, S. Adaptive Classification by Variational Kalman Filtering. In Advances in Neural Information Processing Systems; Becker, S., Thrun, S., Obermayer, K., Eds.; MIT Press: Cambridge, MA, USA, 2003; Volume 15, pp. 753–760. [Google Scholar]
- Sykacek, P.; Roberts, S.J.; Stokes, M. Adaptive BCI based on variational Bayesian Kalman filtering: An empirical evaluation. IEEE Trans. Biomed. Eng. 2004, 51, 719–727. [Google Scholar] [CrossRef]
- Ripley, B.D. Stochastic Simulation; Wiley: New York, NY, USA, 1987. [Google Scholar]
- Kitagawa, G. A Monte Carlo Filtering and Smoothing Method for Non–Gaussian Nonlinear State Space Models. In Proceedings of the 2nd US-Japan Joint Seminar on Statistical Time Series Analysis, Honolulu, HI, USA, 25–29 January 1993; Volume 2, pp. 110–131. [Google Scholar]
- Doucet, A.; Barat, E.; Duvaut, P. Monte Carlo approach to recursive Bayesian state estimation. In Proceedings of the IEEE Signal Processing/Athos Workshop on Higher Order Statistics, Girona, Spain, 12–14 June 1995; pp. 12–14. [Google Scholar]
- Del Moral, P. Non Linear Filtering: Interacting Particle Solution. Markov Process. Relat. Fields 1996, 2, 555–580. [Google Scholar]
- Del Moral, P.; Doucet, A.; Jasra, A. On adaptive resampling strategies for sequential Monte Carlo methods. Bernoulli 2012, 18, 252–278. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, T. Controlling the False Discovery Rate: A practical and powerful approach for multiple testing. J. R. Stat. Soc. Ser. B 1995, 85, 289–300. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural Networks for Pattern Recognition; Clarendon Press: Oxford, UK, 1995. [Google Scholar]
- Neal, R.M. Bayesian Learning for Neural Networks; Springer: New York, NY, USA, 1996. [Google Scholar]






| Name | Media | Flask Size [mL] | Filling Volume [mL] | Shaking Rate [rpm] | OD600 | Gly [g L] |
|---|---|---|---|---|---|---|
| LB-250 mL-1 | LB | 250 | 40 | 180 | 0.109 | - |
| LB-250 mL-2 | LB | 250 | 40 | 180 | 0.109 | - |
| LB-500 mL-1 | LB | 500 | 80 | 200 | 0.128 | - |
| LB-500 mL-2 | LB | 500 | 80 | 200 | 0.130 | - |
| TB-500 mL-3 | TB | 500 | 50 | 200 | 0.074 | 5.43 |
| TB-500 mL-4 | TB | 500 | 50 | 200 | 0.076 | 5.45 |
| TB-500 mL-5 | TB | 500 | 80 | 200 | 0.122 | 5.21 |
| TB-500 mL-6 | TB | 500 | 80 | 200 | 0.116 | 5.26 |
| SM Training | PF Initialization | SM & PF Testing |
|---|---|---|
| LB-500 mL-1 | LB-250 mL-1 & 2 | LB-500 mL-2 |
| LB-500 mL-2 | LB-250 mL-1 & 2 | LB-500 mL-1 |
| TB-500 mL-3 | LB-250 mL-1 & 2 | TB-500 mL-4 |
| TB-500 mL-4 | LB-250 mL-1 & 2 | TB-500 mL-5 |
| TB-500 mL-5 | LB-250 mL-1 & 2 | TB-500 mL-6 |
| Time [%] | MSE | MSE | % sig | td [h] | td [h] | Loss [%] | Loss [%] |
|---|---|---|---|---|---|---|---|
| Gompertz growth model on LB-500 mL | |||||||
| 30 | |||||||
| 40 | |||||||
| 50 | |||||||
| 60 | |||||||
| 70 | |||||||
| 80 | |||||||
| Gompertz growth model on TB-500 mL | |||||||
| 30 | |||||||
| 40 | |||||||
| 50 | |||||||
| 60 | |||||||
| 70 | |||||||
| 80 | |||||||
| Logistic growth model on LB-500 mL | |||||||
| 30 | |||||||
| 40 | |||||||
| 50 | |||||||
| 60 | |||||||
| 70 | |||||||
| 80 | |||||||
| Logistic growth model on TB-500 mL | |||||||
| 30 | |||||||
| 40 | |||||||
| 50 | |||||||
| 60 | |||||||
| 70 | |||||||
| 80 | |||||||
| Time [%] | MSE | MSE | % sig | td [h] | td [h] | Loss [%] | Loss [%] |
|---|---|---|---|---|---|---|---|
| Gompertz growth model on LB-500 mL | |||||||
| 30 | |||||||
| 40 | |||||||
| 50 | |||||||
| 60 | |||||||
| Gompertz growth model on TB-500 mL | |||||||
| 30 | |||||||
| 40 | |||||||
| 50 | |||||||
| 60 | |||||||
| Logistic growth model on LB-500 mL | |||||||
| 30 | |||||||
| 40 | |||||||
| 50 | |||||||
| 60 | |||||||
| Logistic growth model on TB-500 mL | |||||||
| 30 | |||||||
| 40 | |||||||
| 50 | |||||||
| 60 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pretzner, B.; Maschke, R.W.; Haiderer, C.; John, G.T.; Herwig, C.; Sykacek, P. Predictive Monitoring of Shake Flask Cultures with Online Estimated Growth Models. Bioengineering 2021, 8, 177. https://doi.org/10.3390/bioengineering8110177
Pretzner B, Maschke RW, Haiderer C, John GT, Herwig C, Sykacek P. Predictive Monitoring of Shake Flask Cultures with Online Estimated Growth Models. Bioengineering. 2021; 8(11):177. https://doi.org/10.3390/bioengineering8110177
Chicago/Turabian StylePretzner, Barbara, Rüdiger W. Maschke, Claudia Haiderer, Gernot T. John, Christoph Herwig, and Peter Sykacek. 2021. "Predictive Monitoring of Shake Flask Cultures with Online Estimated Growth Models" Bioengineering 8, no. 11: 177. https://doi.org/10.3390/bioengineering8110177
APA StylePretzner, B., Maschke, R. W., Haiderer, C., John, G. T., Herwig, C., & Sykacek, P. (2021). Predictive Monitoring of Shake Flask Cultures with Online Estimated Growth Models. Bioengineering, 8(11), 177. https://doi.org/10.3390/bioengineering8110177









