Pichia pastoris (Komagataella phaffii) as a Cost-Effective Tool for Vaccine Production for Low- and Middle-Income Countries (LMICs)
Abstract
:1. Introduction
2. Versatility of Yeasts as an Expression System Platform
3. P. pastoris as a Powerful Protein Production Host System
3.1. Generalised Research Application Examples of the Overall Improved Features of P. pastoris
3.1.1. Strain Engineering
3.1.2. Process Engineering
3.2. P. pastoris as an Expression System for the Production of Human Subunit Vaccines
3.2.1. Examples of General Diseases
3.2.2. Examples of Tropical Diseases
3.2.3. Disease Outbreaks and Pandemics
4. Some Economic Considerations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amanna, I.J.; Slifka, M.K. Successful Vaccines. Curr. Top Microbiol. Immunol. 2020, 428, 1–30. [Google Scholar] [CrossRef]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New Vaccine Technologies to Combat Outbreak Situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappuoli, R.; Hanon, E. Sustainable Vaccine Development: A Vaccine Manufacturer’s Perspective. Curr. Opin. Immunol. 2018, 53, 111–118. [Google Scholar] [CrossRef]
- Kaslow, D.C.; Biernaux, S. RTS, S: Toward a First Landmark on the Malaria Vaccine Technology Roadmap. Vaccine 2015, 33, 7425–7432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Malaria Vaccine Rainbow Tables. Available online: http://www.who.int/vaccine_research/links/Rainbow/en/index.html (accessed on 19 September 2020).
- McGuire, E.P.; Fong, Y.; Toote, C.; Cunningham, C.K.; McFarland, E.J.; Borkowsky, W.; Barnett, S.; Itell, H.L.; Kumar, A.; Gray, G.; et al. HIV-exposed infants vaccinated with an MF59/recombinant gp120 vaccine have higher-magnitude anti-V1V2 IgG responses than adults immunized with the same vaccine. J. Virol. 2018, 92, e01070-17. [Google Scholar] [CrossRef] [Green Version]
- Marzi, A.; Reynolds, P.; Mercado-Hernandez, R.; Callison, J.; Feldmann, F.; Rosenke, R.; Thomas, T.; Scott, D.P.; Hanley, P.W.; Haddock, E.; et al. Single Low-Dose VSV-EBOV Vaccination Protects Cynomolgus Macaques from Lethal Ebola Challenge. EBioMedicine 2019, 49, 223–231. [Google Scholar] [CrossRef] [Green Version]
- Shen, A.K.; Cooke, M.T. Infectious Disease Vaccines. Nat. Rev. Drug Discov. 2019, 18, 169–170. [Google Scholar] [CrossRef]
- Le, T.T.; Andreadakis, Z.; Kumar, A.; Gómez Román, R.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID-19 Vaccine Development Landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, X.; Zhang, J.; Xia, N.; Zhao, Q. Escherichia Coli-Derived Virus-like Particles in Vaccine Development. npj Vaccines 2017, 2, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owczarek, B.; Gerszberg, A.; Hnatuszko-Konka, K. A Brief Reminder of Systems of Production and Chromatography-Based Recovery of Recombinant Protein Biopharmaceuticals. Biomed Res. Int. 2019, 2019, 4216060. [Google Scholar] [CrossRef]
- Tripathi, N.K.; Shrivastava, A. Recent Developments in Bioprocessing of Recombinant Proteins: Expression Hosts and Process Development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Kumar, P. Yeast-Based Vaccines: New Perspective in Vaccine Development and Application. FEMS Yeast Res. 2019, 19, foz007. [Google Scholar] [PubMed] [Green Version]
- Roohvand, F.; Shokri, M.; Abdollahpour-Alitappeh, M.; Ehsani, P. Biomedical Applications of Yeast-a Patent View, Part One: Yeasts as Workhorses for the Production of Therapeutics and Vaccines. Expert Opin. Ther. Pat. 2017, 27, 929–951. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Farajnia, S.; Rajabibazl, M.; Ghasemi, Y.; Mafi, A.A.; Hoseinpoor, R.; Rahbarnia, L.; Aria, M. Yeast Expression Systems: Overview and Recent Advances. Mol. Biotechnol. 2019, 61, 365–384. [Google Scholar] [CrossRef] [PubMed]
- Roohvand, F.; Ehsani, P.; Abdollahpour-Alitappeh, M.; Shokri, M.; Kossari, N. Biomedical Applications of Yeasts—A Patent View, Part Two: Era of Humanized Yeasts and Expanded Applications. Expert Opin. Ther. Pat. 2020, 30, 609–631. [Google Scholar] [CrossRef]
- Haller, A.A.; Lauer, G.M.; King, T.H.; Kemmler, C.; Fiolkoski, V.; Lu, Y.; Bellgrau, D.; Rodell, T.C.; Apelian, D.; Franzusoff, A.; et al. Whole Recombinant Yeast-Based Immunotherapy Induces Potent T Cell Responses Targeting HCV NS3 and Core Proteins. Vaccine 2007, 25, 1452–1463. [Google Scholar] [CrossRef]
- Zeng, Y.; Cai, D.; Peng, M.; Ren, H. The primary study on the anti-HBV effect of whole recombinant yeast. Zhonghua Gan Zang Bing Za Zhi Zhonghua Ganzangbing Zazhi Chin. J. Hepatol. 2003, 11, 728–730. [Google Scholar]
- Hadiji-Abbes, N.; Martin, M.; Benzina, W.; Karray-Hakim, H.; Gergely, C.; Gargouri, A.; Mokdad-Gargouri, R. Extraction and Purification of Hepatitis B Virus-like M Particles from a Recombinant Saccharomyces Cerevisiae Strain Using Alumina Powder. J. Virol. Methods 2013, 187, 132–137. [Google Scholar]
- Ramasamy, V.; Arora, U.; Shukla, R.; Poddar, A.; Shanmugam, R.K.; White, L.J.; Mattocks, M.M.; Raut, R.; Perween, A.; Tyagi, P.; et al. A Tetravalent Virus-like Particle Vaccine Designed to Display Domain III of Dengue Envelope Proteins Induces Multi-Serotype Neutralizing Antibodies in Mice and Macaques Which Confer Protection against Antibody Dependent Enhancement in AG129 Mice. PLoS Negl. Trop. Dis. 2018, 12, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Yao, Y.; Yang, X.; Bai, H.; Huang, W.; Xia, Y.; Ma, Y. Production of Recombinant Human Papillomavirus Type 52 L1 Protein in Hansenula Polymorpha Formed Virus-like Particles. J. Microbiol. Biotechnol. 2015, 25, 936–940. [Google Scholar] [CrossRef]
- Wang, H.; Chen, X.; Wang, D.; Yao, C.; Wang, Q.; Xie, J.; Shi, X.; Xiang, Y.; Liu, W.; Zhang, L. Epitope-Focused Immunogens against the CD4-Binding Site of HIV-1 Envelope Protein Induce Neutralizing Antibodies against Auto- and Heterologous Viruses. J. Biol. Chem. 2018, 293, 830–846. [Google Scholar] [CrossRef] [Green Version]
- Wasilenko, J.L.; Sarmento, L.; Spatz, S.; Pantin-Jackwood, M. Cell Surface Display of Highly Pathogenic Avian Influenza Virus Hemagglutinin on the Surface of Pichia Pastoris Cells Using α-Agglutinin for Production of Oral Vaccines. Biotechnol. Prog. 2010, 26, 542–547. [Google Scholar]
- Khulape, S.A.; Maity, H.K.; Pathak, D.C.; Mohan, C.M.; Dey, S. Antigenic Validation of Recombinant Hemagglutinin-Neuraminidase Protein of Newcastle Disease Virus Expressed in Saccharomyces Cerevisiae. Acta Virol. 2015, 59, 240–246. [Google Scholar] [CrossRef] [Green Version]
- Fazlalipour, M.; Keyvani, H.; Reza Monavari, S.H.; Mollaie, H.R. Expression, Purification and Immunogenic Description of a Hepatitis c Virus Recombinant CoreE1E2 Protein Expressed by Yeast Pichia Pastoris. Jundishapur J. Microbiol. 2015, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Lorent, E.; Bierau, H.; Engelborghs, Y.; Verheyden, G.; Bosman, F. Structural Characterisation of the Hepatitis C Envelope Glycoprotein E1 Ectodomain Derived from a Mammalian and a Yeast Expression System. Vaccine 2008, 26, 399–410. [Google Scholar] [CrossRef]
- Ahmad, M.; Hirz, M.; Pichler, H.; Schwab, H. Protein Expression in Pichia Pastoris: Recent Achievements and Perspectives for Heterologous Protein Production. Appl. Microbiol. Biotechnol. 2014, 98, 5301–5317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia Pastoris: A Highly Successful Expression System for Optimal Synthesis of Heterologous Proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, V.; Reddy, G.R.; Suryanarayana, V.V.S. Pichia Pastoris: A Notable Heterologous Expression System for the Production of Foreign Proteins—Vaccines. Indian J. Biotechnol. 2007, 6, 175–186. [Google Scholar]
- Charoenrat, T.; Khumruaengsri, N.; Promdonkoy, P.; Rattanaphan, N.; Eurwilaichitr, L.; Tanapongpipat, S.; Roongsawang, N. Improvement of Recombinant Endoglucanase Produced in Pichia Pastoris KM71 through the Use of Synthetic Medium for Inoculum and PH Control of Proteolysis. J. Biosci. Bioeng. 2013, 116, 193–198. [Google Scholar] [CrossRef]
- Velez-Suberbie, M.L.; Betts, J.P.J.; Walker, K.L.; Robinson, C.; Zoro, B.; Keshavarz-Moore, E. High Throughput Automated Microbial Bioreactor System Used for Clone Selection and Rapid Scale-down Process Optimization. Biotechnol. Prog. 2018, 34, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Werten, M.W.T.; Eggink, G.; Cohen Stuart, M.A.; de Wolf, F.A. Production of Protein-Based Polymers in Pichia Pastoris. Biotechnol. Adv. 2019, 37, 642–666. [Google Scholar] [CrossRef]
- Vogl, T.; Sturmberger, L.; Fauland, P.C.; Hyden, P.; Fischer, J.E.; Schmid, C.; Thallinger, G.G.; Geier, M.; Glieder, A. Methanol Independent Induction in Pichia Pastoris by Simple Derepressed Overexpression of Single Transcription Factors. Biotechnol. Bioeng. 2018, 115, 1037–1050. [Google Scholar] [CrossRef]
- Fischer, J.E.; Glieder, A. Current Advances in Engineering Tools for Pichia Pastoris. Curr. Opin. Biotechnol. 2019, 59, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Gong, T.; Wang, Q.H.; Liang, X.; Chen, J.J.; Zhu, P. Scaling-up Fermentation of Pichia Pastoris to Demonstration-Scale Using New Methanol-Feeding Strategy and Increased Air Pressure Instead of Pure Oxygen Supplement. Sci. Rep. 2016, 6, 1–12. [Google Scholar]
- Jacobs, P.P.; Geysens, S.; Vervecken, W.; Contreras, R.; Callewaert, N. Engineering Complex-Type N-Glycosylation in Pichia Pastoris Using GlycoSwitch Technology. Nat. Protoc. 2009, 4, 58–70. [Google Scholar] [CrossRef]
- Li, S.; Sun, P.; Gong, X.; Chang, S.; Li, E.; Xu, Y.; Wu, J.; Liu, B. Engineering O-Glycosylation in Modified N-Linked Oligosaccharide (Man12GlcNAc2∼Man16GlcNAc2) Pichia Pastoris Strains. RSC Adv. 2019, 9, 8246–8252. [Google Scholar] [CrossRef] [Green Version]
- Cregg, J.M.; Cereghino, J.L.; Shi, J.; Higgins, D.R. Recombinant Protein Expression in Pichia Pastoris. Appl. Biochem. Biotechnol. Part B Mol. Biotechnol. 2000, 16, 23–52. [Google Scholar] [CrossRef]
- Barrero, J.J.; Casler, J.C.; Valero, F.; Ferrer, P.; Glick, B.S. An Improved Secretion Signal Enhances the Secretion of Model Proteins from Pichia Pastoris. Microb. Cell Fact. 2018, 17, 1–13. [Google Scholar] [CrossRef]
- Looser, V.; Bruhlmann, B.; Bumbak, F.; Stenger, C.; Costa, M.; Camattari, A.; Fotiadis, D.; Kovar, K. Cultivation Strategies to Enhance Productivity of Pichia Pastoris: A Review. Biotechnol. Adv. 2014, 33, 1177–1193. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zhang, Z. Engineering Strategies for Enhanced Production of Protein and Bio-Products in Pichia Pastoris: A Review. Biotechnol. Adv. 2018, 36, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P.; Hubbuch, J. Downstream Process Development Strategies for Effective Bioprocesses: Trends, Progress, and Combinatorial Approaches. Eng. Life Sci. 2017, 17, 1142–1158. [Google Scholar] [CrossRef]
- Crowell, L.E.; Lu, A.E.; Love, K.R.; Stockdale, A.; Timmick, S.M.; Wu, D.; Wang, Y.A.; Doherty, W.; Bonnyman, A.; Vecchiarello, N.; et al. On-Demand Manufacturing of Clinical-Quality Biopharmaceuticals. Nat. Biotechnol. 2018, 36, 988. [Google Scholar] [CrossRef]
- Zhou, W.J.; Yang, J.K.; Mao, L.; Miao, L.H. Codon Optimization, Promoter and Expression System Selection That Achieved High-Level Production of Yarrowia Lipolytica Lipase in Pichia Pastoris. Enzym. Microb. Technol. 2015, 71, 66–72. [Google Scholar] [CrossRef]
- Cai, H.; Chen, L.; Wan, L.; Zeng, L.; Yang, H.; Li, S.; Li, Y.; Cheng, J.; Lu, X. High-Level Expression of a Functional Humanized Anti-CTLA4 Single-Chain Variable Fragment Antibody in Pichia Pastoris. Appl. Microbiol. Biotechnol. 2009, 82, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Baeshen, M.N.; Bouback, T.A.F.; Alzubaidi, M.A.; Bora, R.S.; Alotaibi, M.A.T.; Alabbas, O.T.O.; Alshahrani, S.M.; Aljohani, A.A.M.; Munshi, R.A.A.; Al-Hejin, A.; et al. Expression and Purification of C-Peptide Containing Insulin Using Pichia Pastoris Expression System. Biomed Res. Int. 2016, 2016, 3423685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, W.; Su, L.; Liao, Q.; Ye, L.; Wu, Y.; Wu, Z.; She, Y. Expression, Purification and Immunogenic Characterization of Hepatitis C Virus Recombinant E1E2 Protein Expressed by Pichia Pastoris Yeast. Antivir. Res. 2010, 88, 80–85. [Google Scholar] [CrossRef]
- Zahrl, R.J.; Peña, D.A.; Mattanovich, D.; Gasser, B. Systems Biotechnology for Protein Production in Pichia Pastoris. FEMS Yeast Res. 2017, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komor, A.C.; Badran, A.H.; Liu, D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017, 168, 20–36. [Google Scholar] [CrossRef] [Green Version]
- Gassler, T.; Heistinger, L.; Mattanovich, D.; Gasser, B.; Prielhofer, R. CRISPR/Cas9-Mediated Homology-Directed Genome Editing in Pichia Pastoris. Methods Mol. Biol. 2019, 1923, 211–225. [Google Scholar]
- Weninger, A.; Fischer, J.E.; Raschmanová, H.; Kniely, C.; Vogl, T.; Glieder, A. Expanding the CRISPR/Cas9 Toolkit for Pichia Pastoris with Efficient Donor Integration and Alternative Resistance Markers. J. Cell Biochem. 2018, 119, 3183–3198. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Shi, X.; Song, L.; Liu, H.; Zhou, X.; Wang, Q.; Zhang, Y.; Cai, M. CRISPR-Cas9-Mediated Genomic Multiloci Integration in Pichia Pastoris. Microb. Cell Fact. 2019, 18, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Liu, G.; Chen, X.; Liu, M.; Zhan, C.; Liu, X.; Bai, Z. High Efficiency CRISPR/Cas9 Genome Editing System with an Eliminable Episomal SgRNA Plasmid in Pichia Pastoris. Enzym. Microb. Technol. 2020, 138, 109556. [Google Scholar] [CrossRef]
- Obst, U.; Lu, T.K.; Sieber, V. A Modular Toolkit for Generating Pichia Pastoris Secretion Libraries. ACS Synth. Biol. 2017, 6, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Lin-Cereghino, G.P.; Stark, C.M.; Kim, D.; Chang, J.; Shaheen, N.; Poerwanto, H.; Agari, K.; Moua, P.; Low, L.K.; Tran, N.; et al. The Effect of α-Mating Factor Secretion Signal Mutations on Recombinant Protein Expression in Pichia Pastoris. Gene 2013, 519, 311–317. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Kuang, Y.; Li, H.; Liu, Y.; Hui, X.; Li, P.; Jiang, Z.; Zhou, Y.; Wang, Y.; Xu, A.; et al. Enhanced Production of Recombinant Secretory Proteins in Pichia Pastoris by Optimizing Kex2 P1′ Site. PLoS ONE 2013, 8, e75347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartner, F.S.; Glieder, A. Regulation of Methanol Utilisation Pathway Genes in Yeasts. Microb. Cell Fact. 2006, 5, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Krainer, F.W.; Dietzsch, C.; Hajek, T.; Herwig, C.; Spadiut, O.; Glieder, A. Krainer 2012 Recombinant Protein. Microb. Cell Fact. 2012, 22, 1–14. [Google Scholar]
- Qin, X.; Qian, J.; Yao, G.; Zhuang, Y.; Zhang, S.; Chu, J. GAP Promoter Library for Fine-Tuning of Gene Expression in Pichia Pastoris. Appl. Environ. Microbiol. 2011, 77, 3600–3608. [Google Scholar] [CrossRef] [Green Version]
- Hartner, F.S.; Ruth, C.; Langenegger, D.; Johnson, S.N.; Hyka, P.; Lin-Cereghino, G.P.; Lin-Cereghino, J.; Kovar, K.; Cregg, J.M.; Glieder, A. Promoter Library Designed for Fine-Tuned Gene Expression in Pichia Pastoris. Nucleic Acids Res. 2008, 36, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Várnai, A.; Tang, C.; Bengtsson, O.; Atterton, A.; Mathiesen, G.; Eijsink, V.G.H. Expression of Endoglucanases in Pichia Pastoris under Control of the GAP Promoter. Microb. Cell Fact. 2014, 13, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.H.; Hsiung, H.A.; Hong, K.L.; Huang, C.T. Enhancing the Efficiency of the Pichia Pastoris AOX1 Promoter via the Synthetic Positive Feedback Circuit of Transcription Factor Mxr1. BMC Biotechnol. 2018, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mombeni, M.; Arjmand, S.; Siadat, S.O.R.; Alizadeh, H.; Abbasi, A. PMOX: A New Powerful Promoter for Recombinant Protein Production in Yeast Pichia Pastoris. Enzym. Microb. Technol. 2020, 139, 109582. [Google Scholar] [CrossRef] [PubMed]
- Rajamanickam, V.; Metzger, K.; Schmid, C.; Spadiut, O. A Novel Bi-Directional Promoter System Allows Tunable Recombinant Protein Production in Pichia Pastoris. Microb. Cell Fact. 2017, 16, 1–7. [Google Scholar] [CrossRef]
- Vogl, T.; Glieder, A. Regulation of Pichia Pastoris Promoters and Its Consequences for Protein Production. N. Biotechnol. 2013, 30, 385–404. [Google Scholar] [CrossRef]
- Vogl, T.; Ruth, C.; Pitzer, J.; Kickenweiz, T.; Glieder, A. Synthetic Core Promoters for Pichia Pastoris. ACS Synth. Biol. 2014, 3, 188–191. [Google Scholar] [CrossRef]
- Brady, J.R.; Whittaker, C.A.; Tan, M.C.; Kristensen, D.L.; Ma, D.; Dalvie, N.C.; Love, K.R.; Love, J.C. Comparative Genome-Scale Analysis of Pichia Pastoris Variants Informs Selection of an Optimal Base Strain. Biotechnol. Bioeng. 2020, 117, 543–555. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, S.R.; Gerngross, T.U. Glycosylation Engineering in Yeast: The Advent of Fully Humanized Yeast. Curr. Opin. Biotechnol. 2007, 18, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Bollok, M.; Resina, D.; Valero, F.; Ferrer, P. Recent Patents on the Pichia Pastoris Expression System: Expanding the Toolbox for Recombinant Protein Production. Recent Pat. Biotechnol. 2009, 3, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Peña, D.A.; Gasser, B.; Zanghellini, J.; Steiger, M.G.; Mattanovich, D. Metabolic Engineering of Pichia Pastoris. Metab. Eng. 2018, 50, 2–15. [Google Scholar] [CrossRef]
- Wang, S.; Rong, Y.; Wang, Y.; Kong, D.; Wang, P.G.; Chen, M.; Kong, Y. Homogeneous Production and Characterization of Recombinant N-GlcNAc-Protein in Pichia Pastoris. Microb. Cell Fact. 2020, 19, 1–11. [Google Scholar] [CrossRef]
- Claes, K.; Vandewalle, K.; Laukens, B.; Laeremans, T.; Vosters, O.; Langer, I.; Parmentier, M.; Steyaert, J.; Callewaert, N. Modular Integrated Secretory System Engineering in Pichia Pastoris to Enhance G-Protein Coupled Receptor Expression. ACS Synth. Biol. 2016, 5, 1070–1075. [Google Scholar] [CrossRef]
- Liu, C.P.; Tsai, T.I.; Cheng, T.; Shivatare, V.S.; Wu, C.Y.; Wu, C.Y.; Wong, C.H. Glycoengineering of Antibody (Herceptin) through Yeast Expression and in Vitro Enzymatic Glycosylation. Proc. Natl. Acad. Sci. USA 2018, 115, 720–725. [Google Scholar] [CrossRef] [Green Version]
- Kaushik, N.; Lamminmäki, U.; Khanna, N.; Batra, G. Enhanced Cell Density Cultivation and Rapid Expression-Screening of Recombinant Pichia Pastoris Clones in Microscale. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Katla, S.; Karmakar, B.; Tadi, S.R.R.; Mohan, N.; Anand, B.; Pal, U.; Sivaprakasam, S. High Level Extracellular Production of Recombinant Human Interferon Alpha 2b in Glycoengineered Pichia Pastoris: Culture Medium Optimization, High Cell Density Cultivation and Biological Characterization. J. Appl. Microbiol. 2019, 126, 1438–1453. [Google Scholar] [CrossRef]
- Matthews, C.B.; Kuo, A.; Love, K.R.; Love, J.C. Development of a General Defined Medium for Pichia Pastoris. Biotechnol. Bioeng. 2018, 115, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Velez-Suberbie, M.L.; Morris, S.A.; Kaur, K.; Hickey, J.M.; Joshi, S.B.; Volkin, D.B.; Bracewell, D.G.; Mukhopadhyay, T.K. Holistic Process Development to Mitigate Proteolysis of a Subunit Rotavirus Vaccine Candidate Produced in Pichia Pastoris by Means of an Acid PH Pulse during Fed-Batch Fermentation. Biotechnol. Prog. 2020, 36, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kastilan, R.; Boes, A.; Spiegel, H.; Voepel, N.; Chudobová, I.; Hellwig, S.; Buyel, J.F.; Reimann, A.; Fischer, R. Improvement of a Fermentation Process for the Production of Two PfAMA1-DiCo-Based Malaria Vaccine Candidates in Pichia Pastoris. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Nieto-Taype, M.A.; Garcia-Ortega, X.; Albiol, J.; Montesinos-Seguí, J.L.; Valero, F. Continuous Cultivation as a Tool Toward the Rational Bioprocess Development with Pichia Pastoris Cell Factory. Front. Bioeng. Biotechnol. 2020, 8, 1–21. [Google Scholar] [CrossRef]
- Rahimi, A.; Hosseini, S.N.; Javidanbardan, A.; Khatami, M. Continuous Fermentation of Recombinant Pichia Pastoris Mut+ Producing HBsAg: Optimizing Dilution Rate and Determining Strain-Specific Parameters. Food Bioprod. Process. 2019, 118, 248–257. [Google Scholar] [CrossRef]
- Tan, L.; Wang, H.; Tan, X.; Zou, J.; Yao, Z. Yeast Expressed Foldable Quadrivalent Aβ15 Elicited Strong Immune Response against Aβ without Aβ-Specific T Cell Response in Wild C57BL/6 Mice. Hum. Vaccines Immunother. 2012, 8, 1090–1098. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Jiang, S.; Liu, X.; Wang, Y. Expression, Purification, and Immunogenic Characterization of Epstein-Barr Virus Recombinant EBNA1 Protein in Pichia Pastoris. Appl. Microbiol. Biotechnol. 2013, 97, 6251–6262. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiang, S.; Han, Z.; Zhao, B.; Wang, L.; Zhou, Z.; Wang, Y. Expression and Immunogenic Characterization of Recombinant Gp350 for Developing a Subunit Vaccine against Epstein-Barr Virus. Appl. Microbiol. Biotechnol. 2016, 100, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiang, S.; Wang, Y. Recombinant VP1 Protein Expressed in Pichia Pastoris Induces Protective Immune Responses against EV71 in Mice. Biochem. Biophys. Res. Commun. 2013, 430, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ku, Z.; Liu, Q.; Wang, X.; Chen, T.; Ye, X.; Li, D.; Jin, X.; Huang, Z. High-Yield Production of Recombinant Virus-like Particles of Enterovirus 71 in Pichia Pastoris and Their Protective Efficacy against Oral Viral Challenge in Mice. Vaccine 2015, 33, 2335–2341. [Google Scholar] [CrossRef]
- O’Riordan, A.A.; Morales, V.A.; Mulligan, L.; Faheem, N.; Windle, H.J.; Kelleher, D.P. Alkyl Hydroperoxide Reductase: A Candidate Helicobacter Pylori Vaccine. Vaccine 2012, 30, 3876–3884. [Google Scholar] [CrossRef]
- Gurramkonda, C.; Zahid, M.; Nemani, S.K.; Adnan, A.; Gudi, S.K.; Khanna, N.; Ebensen, T.; Lünsdorf, H.; Guzmán, C.A.; Rinas, U. Purification of Hepatitis B Surface Antigen Virus-like Particles from Recombinant Pichia Pastoris and in Vivo Analysis of Their Immunogenic Properties. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 940, 104–111. [Google Scholar] [CrossRef]
- Curti, E.; Seid, C.A.; Hudspeth, E.; Center, L.; Rezende, W.; Pollet, J.; Kwityn, C.; Hammond, M.; Matsunami, R.K.; Engler, D.A.; et al. Optimization and Revision of the Production Process of the Necator Americanus Glutathione S-Transferase 1 (Na-GST-1), the Lead Hookworm Vaccine Recombinant Protein Candidate. Hum. Vaccines Immunother. 2014, 10, 1914–1925. [Google Scholar] [CrossRef] [Green Version]
- Curti, E.; Kwityn, C.; Zhan, B.; Gillespie, P.; Brelsford, J.; Deumic, V.; Plieskatt, J.; Rezende, W.C.; Tsao, E.; Kalampanayil, B.; et al. Expression at a 20L Scale and Purification of the Extracellular Domain of the Schistosoma Mansoni TSP-2 Recombinant Protein: A Vaccine Candidate for Human Intestinal Schistosomiasis. Hum. Vaccines Immunother. 2013, 9, 2342–2350. [Google Scholar] [CrossRef] [Green Version]
- Coimbra, E.C.; Gomes, F.B.; Campos, J.F.; D’arc, M.; Carvalho, J.C.; Mariz, F.C.; Jesus, A.L.S.; Stocco, R.C.; Beçak, W.; Freitas, A.C. Production of L1 Protein from Different Types of HPV in Pichia Pastoris Using an Integrative Vector. Braz. J. Med. Biol. Res. 2011, 44, 1209–1214. [Google Scholar] [CrossRef] [Green Version]
- Sanchooli, A.; Aghayipour, K.; Naghlani, S.K.; Samiee, Z.; Kiasari, B.A.; Makvandi, M. Production of Human Papillomavirus Type-16 L1 VLP in Pichia Pastoris. Appl. Biochem. Microbiol. 2020, 56, 51–57. [Google Scholar] [CrossRef]
- Sherry, L.; Grehan, K.; Snowden, J.S.; Rowlands, D.J.; Stonehouse, J. Comparative Molecular Biology Approaches for the Production of Poliovirus Virus-Like Particles Using Pichia Pastoris. Msphere 2020, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Batra, G.; Gurramkonda, C.; Nemani, S.K.; Jain, S.K.; Swaminathan, S.; Khanna, N. Optimization of Conditions for Secretion of Dengue Virus Type 2 Envelope Domain III Using Pichia Pastoris. J. Biosci. Bioeng. 2010, 110, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, L.; Mani, S.; Raut, R.; Poddar, A.; Tyagi, P.; Arora, U.; de Silva, A.; Swaminathan, S.; Khanna, N. Pichia Pastoris-Expressed Dengue 3 Envelope-Based Virus-like Particles Elicit Predominantly Domain III-Focused High Titer Neutralizing Antibodies. Front. Microbiol. 2015, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Rajpoot, R.K.; Shukla, R.; Arora, U.; Swaminathan, S.; Khanna, N. Dengue Envelope-Based “four-in-One” Virus-like Particles Produced Using Pichia Pastoris Induce Enhancement-Lacking, Domain III-Directed Tetravalent Neutralising Antibodies in Mice. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraswat, S.; Athmaram, T.N.; Parida, M.; Agarwal, A.; Saha, A.; Dash, P.K. Expression and Characterization of Yeast Derived Chikungunya Virus Like Particles (CHIK-VLPs) and Its Evaluation as a Potential Vaccine Candidate. PLoS Negl. Trop. Dis. 2016, 10, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Matos, M.N.; Sánchez Alberti, A.; Morales, C.; Cazorla, S.I.; Malchiodi, E.L. A Prime-Boost Immunization with Tc52 N-Terminal Domain DNA and the Recombinant Protein Expressed in Pichia Pastoris Protects against Trypanosoma Cruzi Infection. Vaccine 2016, 34, 3243–3251. [Google Scholar] [CrossRef]
- Jacob, D.; Ruffie, C.; Dubois, M.; Combredet, C.; Amino, R.; Formaglio, P.; Gorgette, O.; Pehau-Arnaudet, G.; Guery, C.; Puijalon, O.; et al. Whole Pichia Pastoris Yeast Expressing Measles Virus Nucleoprotein as a Production and Delivery System to Multimerize Plasmodium Antigens. PLoS ONE 2014, 9, e86658. [Google Scholar] [CrossRef] [Green Version]
- Spiegel, H.; Schinkel, H.; Kastilan, R.; Dahm, P.; Boes, A.; Scheuermayer, M.; Chudobová, I.; Maskus, D.; Fendel, R.; Schillberg, S.; et al. Optimization of a Multi-Stage, Multi-Subunit Malaria Vaccine Candidate for the Production in Pichia Pastoris by the Identification and Removal of Protease Cleavage Sites. Biotechnol. Bioeng. 2015, 112, 659–667. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; O’Brien, K.L.; Madhi, S.A.; Widdowson, M.A.; Byass, P.; Omer, S.B.; Abbas, Q.; Ali, A.; Amu, A.; et al. Global Burden of Respiratory Infections Associated with Seasonal Influenza in Children under 5 Years in 2018: A Systematic Review and Modelling Study. Lancet Glob. Heal. 2020, 8, e497–e510. [Google Scholar] [CrossRef] [Green Version]
- Ameli, J. Communicable Diseases and Outbreak Control. Turk. J. Emerg. Med. 2015, 15, 20–26. [Google Scholar]
- Aw, R.; McKay, P.F.; Shattock, R.J.; Polizzi, K.M. A Systematic Analysis of the Expression of the Anti-HIV VRC01 Antibody in Pichia Pastoris through Signal Peptide Optimization. Protein Expr. Purif. 2018, 149, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Liao, H.Y.; Zhang, J.Y.; Cheng, T.J.R.; Wong, C.H. Development of a Universal Influenza Vaccine Using Hemagglutinin Stem Protein Produced from Pichia Pastoris. Virology 2019, 526, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, M.; Macioła, A.; Zdanowski, K.; Protas-Klukowska, A.M.; Olszewska, M.; Śmietanka, K.; Minta, Z.; Szewczyk, B.; Kopera, E. An Avian Influenza H5N1 Virus Vaccine Candidate Based on the Extracellular Domain Produced in Yeast System as Subviral Particles Protects Chickens from Lethal Challenge. Antivir. Res. 2016, 133, 242–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.H.; Hotez, P.J.; Bottazzi, M.E. Potential for Developing a SARS-CoV Receptor-Binding Domain (RBD) Recombinant Protein as a Heterologous Human Vaccine against Coronavirus Infectious Disease (COVID)-19. Hum. Vaccines Immunother. 2020, 16, 1239–1242. [Google Scholar] [CrossRef] [Green Version]
- Purcell, O.; Opdensteinen, P.; Chen, W.; Lowenhaupt, K.; Brown, A.; Hermann, M.; Cao, J.; Tenhaef, N.; Kallweit, E.; Kastilan, R.; et al. Production of Functional Anti-Ebola Antibodies in Pichia Pastoris. ACS Synth. Biol. 2017, 6, 2183–2190. [Google Scholar] [CrossRef]
- Osman, E.E.A.; Toogood, P.L.; Neamati, N. COVID-19: Living through Another Pandemic. ACS Infect. Dis. 2020, 6, 1548–1552. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J.; Bottazzi, M.E. Developing a Low-Cost and Accessible Covid-19 Vaccine for Global Health. PLoS Negl. Trop. Dis. 2020, 14, 1–6. [Google Scholar] [CrossRef]
- Nkengasong, J.N.; Ndembi, N.; Tshangela, A.; Raji, T. COVID-19 Vaccines: How to Ensure Africa Has Access. Nature 2020, 586, 197–199. [Google Scholar] [CrossRef]
- WHO. COVID-19 Vaccines. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines (accessed on 5 February 2021).
- Chen, W.-H.; Tao, X.; Agrawal, A.; Algaissi, A.; Peng, B.-H.; Pollet, J.; Strych, U.; Bottazzi, M.E.; Hotez, P.; Lustigman, S.; et al. Yeast-Expressed SARS-CoV Recombinant Receptor-Binding Domain (RBD219-N1) Formulated with Aluminum Hydroxide Induces Protective Immunity and Reduces Immune Enhancement. bioRxiv Prepr. Serv. Biol. 2020. [Google Scholar] [CrossRef]
- Hayman, B.; Bowles, A.; Evans, B.; Eyermann, E.; Nepomnyashchiy, L.; Pagliusi, S. Advancing Innovation for Vaccine Manufacturers from Developing Countries: Prioritization, Barriers, Opportunities. Vaccine 2021, 39, 1190–1194. [Google Scholar] [CrossRef] [PubMed]
- Kraigsley, A.M.; Moore, K.A.; Bolster, A.; Peters, M.; Richardson, D.; Arpey, M.; Sonnenberger, M.; McCarron, M.; Lambach, P.; Maltezou, H.C.; et al. Barriers and Activities to Implementing or Expanding Influenza Vaccination Programs in Low- and Middle-Income Countries: A Global Survey. Vaccine 2021, 39, 3419–3427. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.E.; Dieleman, J.L.; Lim, S.S.; Shearer, J. Determinants of Effective Vaccine Coverage in Low and Middle-Income Countries: A Systematic Review and Interpretive Synthesis. BMC Health Serv. Res. 2017, 17, 1–17. [Google Scholar] [CrossRef] [PubMed]
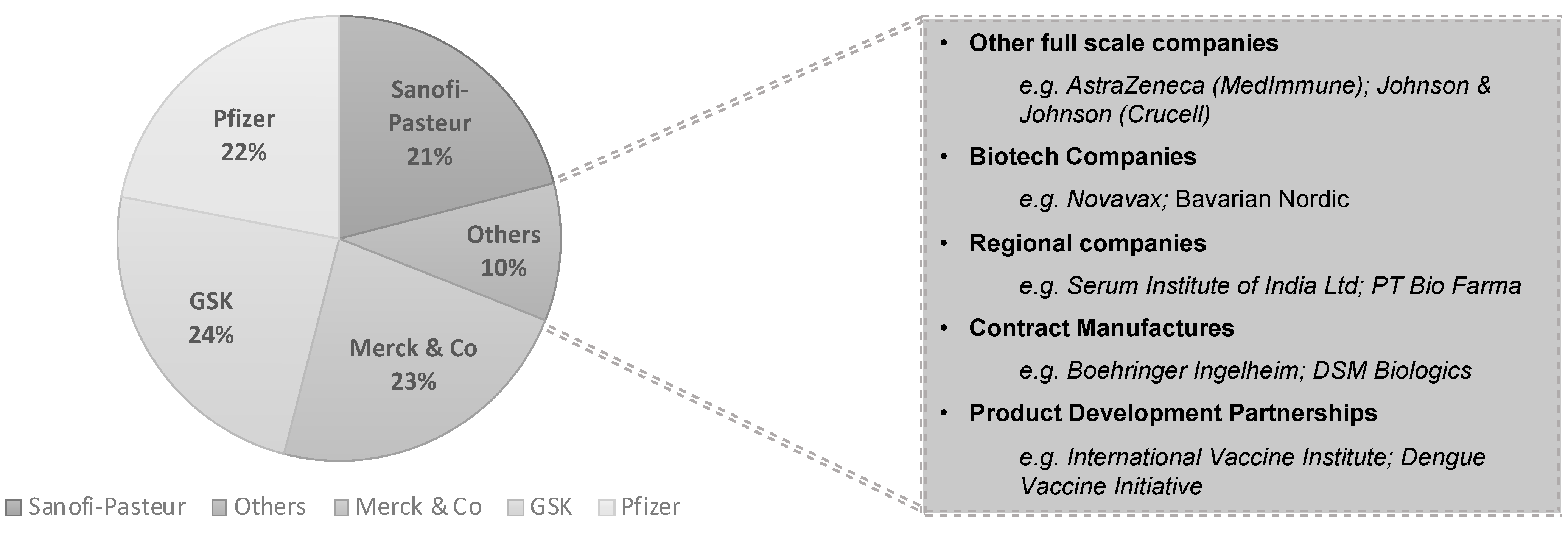
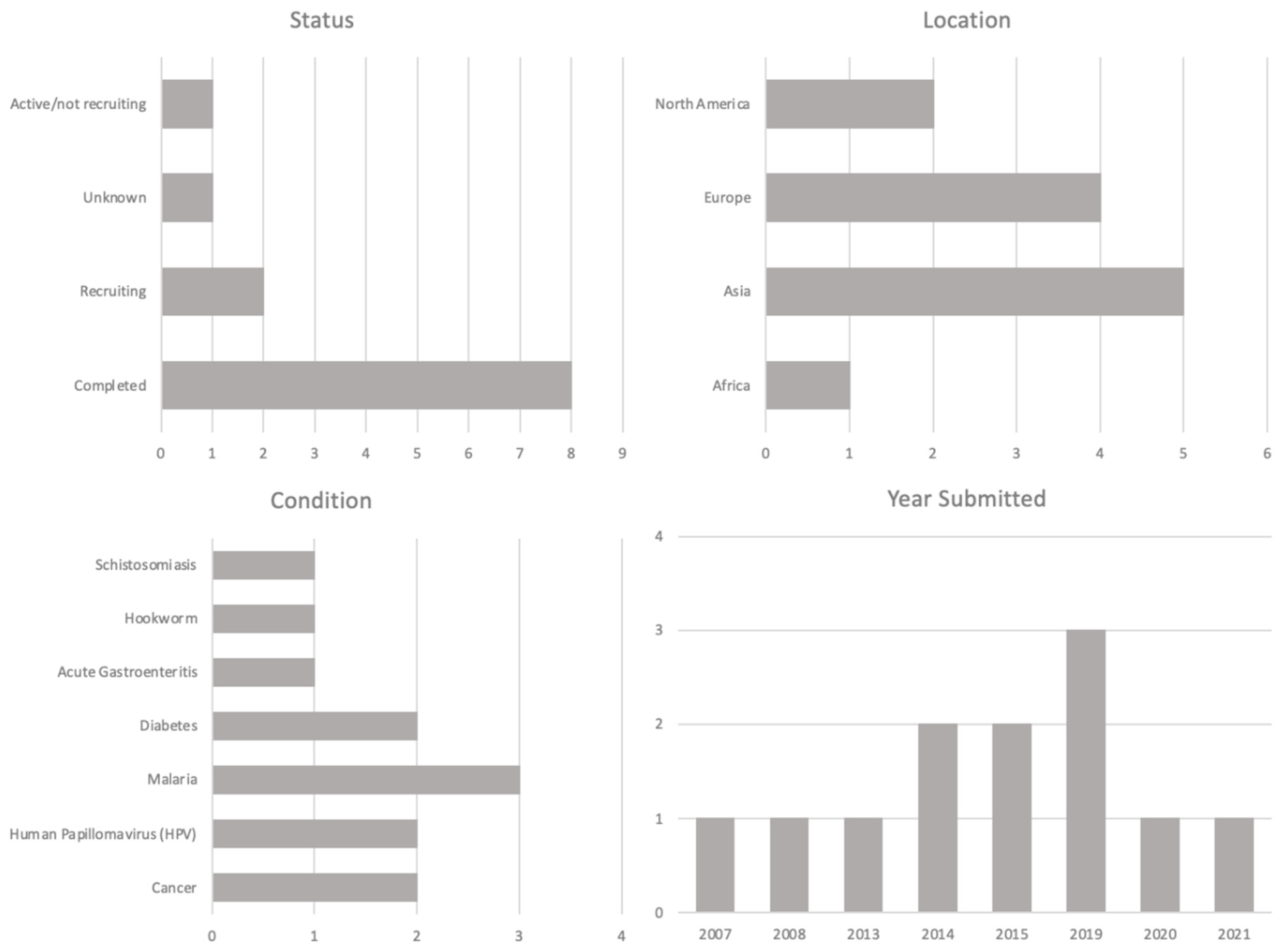
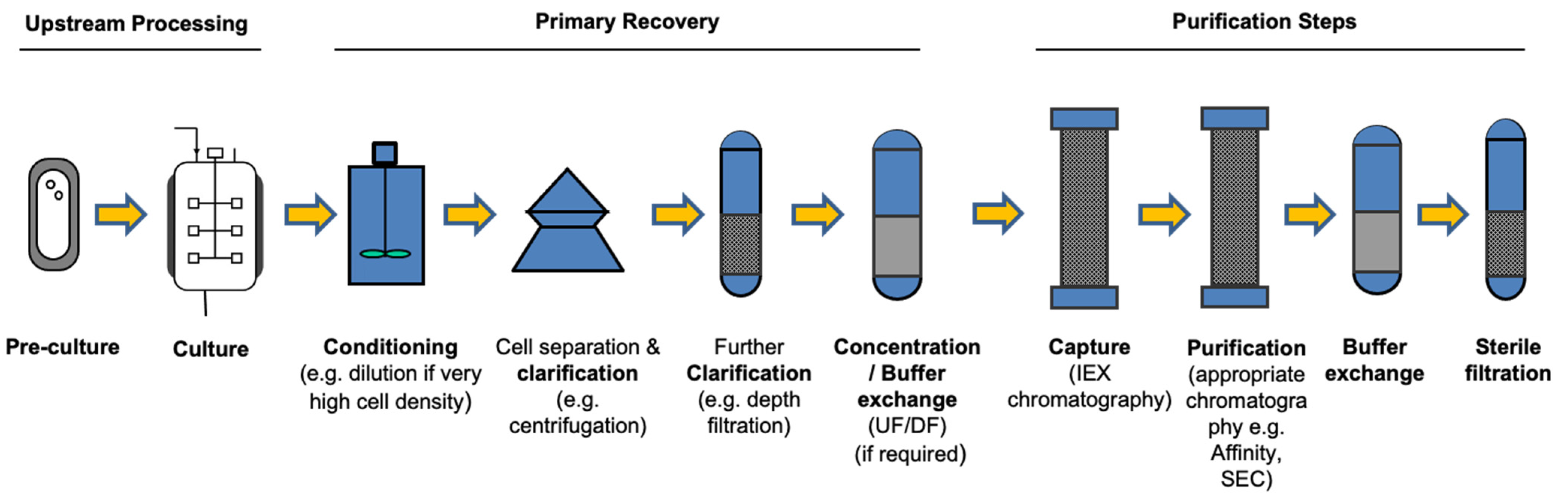
| Host/Platform | Characteristics | Examples of Expressed Products | |||||
|---|---|---|---|---|---|---|---|
| Overall cost | Production time | Scale-up capacity | Propagation | Product yield | Contamination risk | Vaccine Candidates | |
| Bacteria | low | low | high | easy | high | medium (e.g., endotoxins) | Hepatitis E virus (HEV) capsid polypeptide |
| Mammalian cells | high | high | low | hard | medium | very high (e.g., virus, DNA) | Recombinant varicella-zoster virus (rVZV) |
| Yeast | medium | medium | high | easy | high | low | Hepatitis B surface antigen (HBsAg) |
| Insect cells | medium | medium | high | feasible | high | low | Truncated dengue envelope proteins (DEN-80E) |
| Transgenic Plants | medium | medium | high | easy | high | low | Papillomavirus major capsidProtein L1 |
| Transgenic Animals | high | high | low | feasible | high | very high (e.g., virus, DNA) | Malaria major surface protein (MSP-1) antigen |
| Strengths | Weaknesses and Threats | Opportunities |
|---|---|---|
|
|
|
| Strategy | Yeast Species | Antigen Immunogen | Disease | Ref |
|---|---|---|---|---|
| Whole Recombinant Yeast (WRY) | S. cerevisiae | HCV NS3-core fusion | Hepatitis C | [17] |
| P. pastoris | HBsAg, HSP70 | Hepatitis B | [18] | |
| Virus Like Particles (VLPs) | S. cerevisiae | HBsAg | Hepatitis B | [19] |
| P. pastoris | DENV envelope protein domain III (EDIII) | Dengue | [20] | |
| H. polymorpha | HPV52L1 | Human papillomavirus | [21] | |
| Yeast Display (YD) | S. cerevisiae | HIV-1 envelope (Env) glycoprotein | AIDS | [22] |
| P. pastoris | α-aggulutinin | Avian Influenza virus | [23] | |
| Purified Protein | S. cerevisiae | Hemagglutinin-neuramidase | Newcastle disease | [24] |
| P. pastoris | HCV core E1E2 | Hepatitis C | [25] | |
| H. polymorpha | Envelope glycoprotein-E1 ectodomain | Hepatitis C | [26] |
| Pros | Cons | References |
|---|---|---|
| High Expression | Inefficient secretion of larger proteins (>30kDa) | [27,28,29] |
| Cost-effective | Proteolysis of secreted proteins | [29,30,31] |
| Relatively rapid growth | Methanol safety requirements (Methanol is highly flammable chemical)) | [28,32,33,34] |
| Scalability | Some glycosylation patterns different from mammalian | [27,28,35,36,37] |
| Efficient Secretion & Simple purification | [28,38] | |
| Choice of secreted/intracellular expression | [27,28,39] | |
| Efficient protein folding | [27,32] | |
| N-glycosylation close to higher eukaryotes (e.g., glycoengineered, GlycoSwitch® (Carlsbad, CA, USA)) | [36,37] |
| Strain Engineering | Process Engineering | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene Optimisation & Synthesis | Plasmid Construction | Host Strain | Fermentation Mode | |||||||
| Codon Optimisation | Signal Sequence | Promoter | Selectable Marker | Genomic Integration | Type | Mut Form | Selection | Batch | Fed-Batch | Continuous |
| Codon usage database | S.c. α-MF | Constitutive | Drug resistance | Single multicopy | Wild | + | Microscale cultivation | Carbon source | Induction temperature | Dilution rate (D) |
| Bioinformatic tools | P.p. PHO1 | Inducible | Auxotrophy | Homologous recombination | Protease-deficient | - | EasySelect™ | Medium composition | Substrate feed rate | |
| Kex2p | Ectopic integration | Auxotrophic | s | pH | Specific growth rate (µ) | |||||
| Ste13p | Glyco-engineered | DO | Mixed substrate | |||||||
| Process Development Methodologies | Application | Examples |
|---|---|---|
| High throughput (HT) approaches | Cell lysis | 24-well-HT sonication, high pressure homogenisation |
| Refolding | Circular dichroism, refolding kits | |
| Purification | Aqueous two-phase systems, pre-packed Predictor filter plates | |
| Solubility | Multiscreen assay system, Chromafil® (Loughborough, UK) Multi96 filter plates | |
| Single-use systems | Harvesting | Ksep® (centrifugation) (Surrey, UK), ARTeSYN’s® (TFF) (Kilbarry, Ireland) |
| Design of Experiments (DoE) | Screen critical process parameters (CPPs) based on critical quality attributes (CQAs) | Jmp, Design-Expert, Modde software’s |
| Process Analytical Technology (PAT) | Analysis of protein concentration, purity, host cell protein (HCP), host cell DNA (HCD), etc. | Spectroscopy, HPLC, circular dichroism |
| Continuous DSP processing | Purification | chromatography |
| Formulation | Freeze-drying | |
| Integrative system | Upstream and Downstream coupling | InSCyT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Sá Magalhães, S.; Keshavarz-Moore, E. Pichia pastoris (Komagataella phaffii) as a Cost-Effective Tool for Vaccine Production for Low- and Middle-Income Countries (LMICs). Bioengineering 2021, 8, 119. https://doi.org/10.3390/bioengineering8090119
de Sá Magalhães S, Keshavarz-Moore E. Pichia pastoris (Komagataella phaffii) as a Cost-Effective Tool for Vaccine Production for Low- and Middle-Income Countries (LMICs). Bioengineering. 2021; 8(9):119. https://doi.org/10.3390/bioengineering8090119
Chicago/Turabian Stylede Sá Magalhães, Salomé, and Eli Keshavarz-Moore. 2021. "Pichia pastoris (Komagataella phaffii) as a Cost-Effective Tool for Vaccine Production for Low- and Middle-Income Countries (LMICs)" Bioengineering 8, no. 9: 119. https://doi.org/10.3390/bioengineering8090119
APA Stylede Sá Magalhães, S., & Keshavarz-Moore, E. (2021). Pichia pastoris (Komagataella phaffii) as a Cost-Effective Tool for Vaccine Production for Low- and Middle-Income Countries (LMICs). Bioengineering, 8(9), 119. https://doi.org/10.3390/bioengineering8090119





