Current and Perspective Sensing Methods for Monkeypox Virus
Abstract
:1. Introduction
2. Overview of Monkeypox Virus
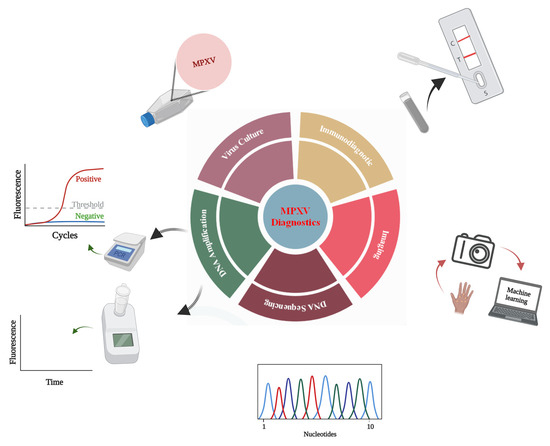
3. Monkeypox Diagnosis Approaches
3.1. Indirect Detection
3.1.1. Monkeypox Diagnosis Based on Virus Culture
3.1.2. Diagnosis Based on Image Analysis
3.2. Direct Detection
3.2.1. Monkeypox Immunodiagnostics
3.2.2. Whole-Particle Detection
3.2.3. Detection by Genome Sequencing
3.2.4. Monkeypox Virus Detection Based on PCR
3.2.5. Detection Based on Isothermal Amplification
| Sr. No | Assay Name | Target Gene | Primers’ Sequences | Probes’ Sequences | Detection Limit | Real-Sample Analysis | References |
|---|---|---|---|---|---|---|---|
| 1 | HA-PCR | HA gene | Forward: 5′-CTGATAATGTAGAAG AC -3′ Reverse: 5′-TTGTATTTACGTGGGTG-3′ | NA | Not reported | Yes | [52] |
| 2 | ATI-PCR | ATI-gene | Forward: 5′-AATACAAGGAGGATCT-3′ Reverse: 5′-CTTAACTTTTTCTTTTTCTTTCTC-3′ | NA | Not reported | Yes | [53] |
| 3 | MPXV PCR assay | ATI-gene | Forward: 5′-GAGAGAATCTCTTGATAT-3′ Reverse: 5′-ATTCTAGATTGTAATC-3′ | NA | Not reported | Yes | [55] |
| 4 | Real-time PCR | B6R | Forward: 5′-ATTGGTCATTATTTTTGTCACAGGAACA-3′ Reverse: 5′-AATGGCGTTGACAATTATGGGTG-3′ | 5′-MGB/DarkQuencher-AGAGATTAGAAATA-3′-FAM | ∼10 viral copies (2 fg) | Yes | [56] |
| 5 | Real-time PCR | G2R | Forward: 5′-CACACCGTCTCTTCCACAGA -3′ Reverse: 5′-GATACAGGTTAATTTCCACATCG -3′ | 5′-FAM AACCCGTCGTAACCAGCAATACATTT-3′-BHQ1 | ∼8.2 genome copies (1.7 fg) | Yes | [62] |
| 6 | Real-time PCR | G2R | Forward: 5′-TGTCTACCTGGATACAGAAAGCAA-3′ Reverse: 5′-GGCATCTCCGTTTAATACATTGAT -3′ | 5′-FAM-CCCATATATGCTAAATGTACCGGTACCGGA-3′-BHQ1 | ∼40.4 copies (9.46 fg) | Yes | [62] |
| 7 | Real-time PCR | F3L | Forward: 5′-CTCATTGATTTTTCG CGGGAT A-3′ Reverse: 5′-GACGATACTCCTCCT CGTTGGT-3′ | 5′-6FAM-CATCAGAATCTGTAGGCCGT-MGBNFQ-3′ | 11–55 fg (50–250 copies) | Yes | [74] |
| 8 | Real-time PCR | N3R | Forward: 5′-AACAACCGT CCTACA ATTAAA CAACA-3′ Reverse: 5′-CGCTATCGAACCATT TTTGTAGTCT-3′ | 5′-6FAM-TAT AAC GGC GAA GAA TAT ACT-MGBNFQ-3′ | 11–55 fg (50–250 copies) | Rodents | [74] |
| 9 | Real-time PCR | B7R | Forward: 5′-ACGTGTTAAACAATGGGTGATG-3′ Reverse: 5′-AACATTTCCATGAATCGTAGTCC-3′ | 5′-TAMRA-TGAATGAATGCGATACTGTATGTGTGGG-3′-BHQ2 | 50 copies per reaction | Yes | [75] |
| 10 | C-LAMP | D14L | FIP-C: 5′-TGGGAGCATTGTAACTTATAGTTGCCCTCCTGAACACATGACA-3′ F3-C: 5′-TGGGTGGATTGGACCATT-3′ BIP-C: 5′-ATCCTCGTATCCGTTATGTCTTCCCACCTATTTGCGAATCTGTT-3′ B3-C: 5′-ATGGTATGGAATCCTGAGG-3′ LOOP-F-C: 5′-GATATTCGTTGATTGGTAACTCTGG-3′ LOOP-C-C: 5′-GTTGGATATAGATGGAGGTGATTGG-3′ | N/A | 102.4 copies per reaction | Yes | [69] |
| 11 | C-LAMP | ATI | FIP-W: 5′-CCGTTACCGTTTTTACAATCGTTAATCAATGCTGATATGGAAAAGAGA-3′ F3-W: 5′-TACAGTTGAACGACTGCG-3′ BIP-W: 5′-ATAGGCTAAAGACTAGAATCAGGGATTCTGATTCATCCTTTGAGAAG-3′ B3-W: 5′-AGTTCAGTTTTATATGCCGAAT-3′ LOOP-F-W: 5′-GATGTCTATCAAGATCCATGATTCT-3′ LOOP-C-W: 5′-TCTTGAACGATCGCTAGAGA-3′ | N/A | 103 copies per reaction | Yes | [69] |
| 12 | RPA | G2R | Forward: 5′-AATAAACGGAAGAGATATAGCACCACATGCAC-3′ Reverse: 5′-GTGAGATGTAAAGGTATCCGAACCACACG-3′ | 5′-ACAGAAGCCGTAATCTATGTTGTCTATCGQTFCCTCCGGGAACTTA-3′ | 16 DNA molecules/μL | Yes | [73] |
4. Wastewater-Based Epidemiology of MPXV
5. WHO’s Sample Collection Guidelines
6. Conclusions and Prospects
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zumla, A.; Valdoleiros, S.R.; Haider, N.; Asogun, D.; Ntoumi, F.; Petersen, E.; Kock, R. Monkeypox outbreaks outside endemic regions: Scientific and social priorities. Lancet Infect. Dis. 2022, 22, 929–931. [Google Scholar] [CrossRef]
- Karbalaei, M.; Keikha, M. Human monkeypox coinfections; lessons from available cases—Correspondence. Int. J. Surg. 2022, 104, 106734. [Google Scholar] [CrossRef] [PubMed]
- Sahebi, S.; Keikha, M. Clinical characteristics of human monkeypox laboratory confirmed cases: Lessons from observational studies. Int. J. Surg. 2022, 104, 106795. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.; Abubakar, I.; Ihekweazu, C.; Heymann, D.; Ntoumi, F.; Blumberg, L.; Asogun, D.; Mukonka, V.; Lule, S.A.; Bates, M.; et al. Monkeypox—Enhancing public health preparedness for an emerging lethal human zoonotic epidemic threat in the wake of the smallpox post-eradication era. Int. J. Infect. Dis. 2018, 78, 78–84. [Google Scholar] [CrossRef] [PubMed]
- De Baetselier, I.; Van Dijck, C.; Kenyon, C.; Coppens, J.; Michiels, J.; de Block, T.; Smet, H.; Coppens, S.; Vanroye, F.; Bugert, J.J.; et al. Retrospective detection of asymptomatic monkeypox virus infections among male sexual health clinic attendees in Belgium. Nat. Med. 2022. [Google Scholar] [CrossRef]
- Kyaw, W.M.; Vasoo, S.; Ho, H.J.A.; Chan, M.; Yeo, T.W.; Manauis, C.M.; Ang, H.; De, P.P.; Ang, B.S.P.; Chow, A.L.P. Monitoring healthcare professionals after monkeypox exposure: Experience from the first case imported to Asia. Infect. Control. Hosp. Epidemiol. 2020, 41, 373–375. [Google Scholar] [CrossRef] [Green Version]
- WHO. 2022 Monkeypox Outbreak: Global Trends; World Health Organization: Geneva, Switzerland, 2022; Available online: https://worldhealthorg.shinyapps.io/mpx_global/ (accessed on 14 September 2022).
- Isidro, J.; Borges, V.; Pinto, M.; Sobral, D.; Santos, J.D.; Nunes, A.; Mixão, V.; Ferreira, R.; Santos, D.; Duarte, S.; et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat. Med. 2022, 28, 1569–1572. [Google Scholar] [CrossRef]
- Frey, S.E.; Belshe, R.B. Poxvirus zoonoses—Putting pocks into context. New. Engl. J. Med. 2004, 350, 324–327. [Google Scholar] [CrossRef]
- Thornhill, J.P.; Barkati, S.; Walmsley, S.; Rockstroh, J.; Antinori, A.; Harrison, L.B.; Palich, R.; Nori, A.; Reeves, I.; Habibi, M.S.; et al. Monkeypox virus infection in humans across 16 countries—April–June 2022. New. Engl. J. Med. 2022, 387, 679–691. [Google Scholar] [CrossRef]
- WHO Monkeypox Declared a Global Health Emergency by the World Health Organization. Available online: https://news.un.org/en/story/2022/07/1123152 (accessed on 23 July 2022).
- Jiang, Z.; Sun, J.; Zhang, L.; Yan, S.; Li, D.; Zhang, C.; Lai, A.; Su, S. Laboratory diagnostics for monkeypox: An overview of sensitivities from various published tests. Travel Med. Infect. Dis. 2022, 49, 102425. [Google Scholar] [CrossRef]
- Glökler, J.; Lim, T.S.; Ida, J.; Frohme, M. Isothermal amplifications—A comprehensive review on current methods. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 543–586. [Google Scholar] [CrossRef] [PubMed]
- Al-Tawfiq, J.A.; Barry, M.; Memish, Z.A. International outbreaks of monkeypox virus infection with no established travel: A public health concern with significant knowledge gap. Travel Med. Infect. Dis. 2022, 49, 102364. [Google Scholar] [CrossRef] [PubMed]
- Chadha, J.; Khullar, L.; Gulati, P.; Chhibber, S.; Harjai, K. Insights into the monkeypox virus: Making of another pandemic within the pandemic? Environ. Microbiol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, D.B.; Eckburg, P.B. Human monkeypox: An emerging zoonosis. Lancet Infect. Dis. 2004, 4, 15–25. [Google Scholar] [CrossRef]
- Alakunle, E.; Moens, U.; Nchinda, G.; Okeke, M. Monkeypox virus in Nigeria: Infection biology, epidemiology, and evolution. Viruses 2020, 12, 1257. [Google Scholar] [CrossRef] [PubMed]
- Gigante, C.M.; Smole, S.; Seabolt, M.H.; Wilkins, K.; McCollum, A.; Hutson, C.; Davidson, W.; Rao, A.; Brown, C.; Li, Y. Draft monkeypox virus genome from confirmed monkeypox case in Massachusetts, United States, May 2022. Available online: https://www.ncbi.nlm.nih.gov/nuccore/ON563414 (accessed on 12 September 2022).
- Saxena, S.K.; Ansari, S.; Maurya, V.K.; Kumar, S.; Jain, A.; Paweska, J.T.; Tripathi, A.K.; Abdel-Moneim, A.S. Re-emerging human monkeypox: A major public-health debacle. J. Med. Virol. 2022. [Google Scholar] [CrossRef]
- Shchelkunov, S.N.; Totmenin, A.; Safronov, P.F.; Mikheev, M.; Gutorov, V.; Ryazankina, O.I.; Petrov, N.A.; Babkin, I.; Uvarova, E.A.; Sandakhchiev, L.S.; et al. Analysis of the monkeypox virus genome. Virology 2002, 297, 172–194. [Google Scholar] [CrossRef] [Green Version]
- Okyay, R.A.; Bayrak, E.; Kaya, E.; Sahin, A.R.; Kocyigit, B.F.; Tasdogan, A.M.; Avci, A.; Erdem, S.H. Another epidemic in the shadow of COVID-19 pandemic: A review of monkeypox. Eurasian J. Med. Oncol. 2022, 6, 95–99. [Google Scholar] [CrossRef]
- Weaver, J.R.; Isaacs, S.N. Monkeypox virus and insights into its immunomodulatory proteins. Immunol. Rev. 2008, 225, 96–113. [Google Scholar] [CrossRef] [Green Version]
- WHO Monkeypox: Experts Give Virus Variants New Names. Available online: https://www.who.int/news/item/12-08-2022-monkeypox--experts-give-virus-variants-new-names (accessed on 13 September 2022).
- Christian, H.; Ifedayo, A.; Placide, M.; Richard, N.; Emmanuel, N.; Anise, H.; Nnaemeka, N.; Oyeronke, A.; Mboowa, G.; Trevor, B.; et al. Urgent Need for a Non-Discriminatory and Non-Stigmatizing Nomenclature for Monkeypox Virus. Available online: https://virological.org/t/urgent-need-for-a-non-discriminatory-and-non-stigmatizing-nomenclature-for-monkeypox-virus/853 (accessed on 30 July 2022).
- WHO Multi-Country Monkeypox Outbreak: Situation Update. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON393 (accessed on 23 June 2022).
- WHO. 2022 Monkeypox Outbreak: Global Trends; World Health Organization: Geneva, Switzerland, 2022; Available online: https://worldhealthorg.shinyapps.io/mpx_global/#section-global (accessed on 13 October 2022).
- Von Magnus, P.; Andersen, E.K.; Petersen, K.B.; Birch-Andersen, A. A pox-like disease in cynomolgus monkeys. Acta Pathol. Microbiol. Scand. 2009, 46, 156–176. [Google Scholar] [CrossRef]
- Rondle, C.J.; Sayeed, K.A. Studies on monkeypox virus. Bull World Health Organ 1972, 46, 577–583. [Google Scholar] [PubMed]
- Prier, J.E.; Sauer, R.M. A pox disease of monkeys. Ann. N. Y. Acad. Sci. 1960, 85, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Fohlerová, Z.; Pekárek, J.; Basova, E.; Neužil, P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020, 153, 112041. [Google Scholar] [CrossRef] [PubMed]
- Artika, I.M.; Ma’Roef, C.N. Laboratory biosafety for handling emerging viruses. Asian Pac. J. Trop. Biomed. 2017, 7, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Jayakeerthi, R.S.; Potula, R.V.; Srinivasan, S.; Badrinath, S. Shell vial culture assay for the rapid diagnosis of Japanese encephalitis, west Nile and dengue-2 viral encephalitis. Virol. J. 2006, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Battineni, G.; Chintalapudi, N.; Amenta, F. AI chatbot design during an epidemic like the novel coronavirus. Healthcare 2020, 8, 20154. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Uddin, M.R.; Luna, S.A. Monkeypox image data collection. arXiv 2022, arXiv:2206.01774. [Google Scholar]
- Ahsan, M.M.; Uddin, M.R.; Farjana, M.; Sakib, A.N.; Momin, K.; Luna, S.A. Image data collection and implementation of deep learning-based model in detecting monkeypox disease using modified VGG16. arXiv 2022, arXiv:2206.01862. [Google Scholar]
- Portela-Dias, J.; Sereno, S.; Falcão-Reis, I.; Rasteiro, C. Female monkeypox infection with localized genital lesions. Am. J. Obstet. Gynecol. 2022. [Google Scholar] [CrossRef]
- Mailhe, M.; Beaumont, A.-L.; Thy, M.; Le Pluart, D.; Perrineau, S.; Houhou-Fidouh, N.; Deconinck, L.; Bertin, C.; Ferré, V.M.; Cortier, M.; et al. Clinical characteristics of ambulatory and hospitalized patients with monkeypox virus infection: An observational cohort study. Clin. Microbiol. Infect. 2022. [Google Scholar] [CrossRef]
- Hierholzer, J.C.; Suggs, M.T.; Hall, E.C. Standardized viral hemagglutination and hemagglutination-inhibition tests II. Description and statistical evaluation. Appl. Microbiol. 1969, 18, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, P.; Sohrabi, H.; Hejazi, M.; Jahanban-Esfahlan, A.; Baradaran, B.; Tohidast, M.; Majidi, M.R.; Mokhtarzadeh, A.; Tavangar, S.M.; de la Guardia, M. Lateral flow assays (LFA) as an alternative medical diagnosis method for detection of virus species: The intertwine of nanotechnology with sensing strategies. TrAC Trends Anal. Chem. 2021, 145, 116460. [Google Scholar] [CrossRef] [PubMed]
- Townsend, M.B.; MacNeil, A.; Reynolds, M.G.; Hughes, C.M.; Olson, V.A.; Damon, I.K.; Karem, K.L. Evaluation of the tetracore orthopox biothreat® antigen detection assay using laboratory grown orthopoxviruses and rash illness clinical specimens. J. Virol. Methods 2013, 187, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.; Pauly, D.; Zydek, M.; Miller, L.; Piesker, J.; Laue, M.; Lisdat, F.; Dorner, M.B.; Dorner, B.G.; Nitsche, A. Development of a genus-specific antigen capture ELISA for orthopoxviruses—Target selection and optimized screening. PLoS ONE 2016, 11, e0150110. [Google Scholar] [CrossRef]
- Stern, D.; Olson, V.A.; Smith, S.K.; Pietraszczyk, M.; Miller, L.; Miethe, P.; Dorner, B.G.; Nitsche, A. Rapid and sensitive point-of-care detection of orthopoxviruses by ABICAP immunofiltration. Virol. J. 2016, 13, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Poltavchenko, A.G.; Ersh, A.V.; Filatov, P.V.; Yakubitskiy, S.N. Rapid protocol of dot-immunnoassay for orthopoxviruses detection. J. Virol. Methods 2020, 279, 113859. [Google Scholar] [CrossRef]
- Ulaeto, D.O.; Lonsdale, S.G.; Laidlaw, S.M.; Clark, G.C.; Horby, P.; Carroll, M.W. A prototype lateral flow assay for detection of orthopoxviruses. Lancet Infect. Dis. 2022, 22, 1279–1280. [Google Scholar] [CrossRef]
- Hughes, L.; Wilkins, K.; Goldsmith, C.S.; Smith, S.; Hudson, P.; Patel, N.; Karem, K.; Damon, I.; Li, Y.; Olson, V.A.; et al. A rapid orthopoxvirus purification protocol suitable for high-containment laboratories. J. Virol. Methods 2017, 243, 68–73. [Google Scholar] [CrossRef]
- Richert-Pöggeler, K.R.; Franzke, K.; Hipp, K.; Kleespies, R.G. Electron microscopy methods for virus diagnosis and high resolution analysis of viruses. Front. Microbiol. 2019, 9, 3255. [Google Scholar] [CrossRef] [Green Version]
- Lavazza, A.; Tittarelli, C.; Cerioli, M. The use of convalescent sera in immune-electron microscopy to detect non-suspected/new viral agents. Viruses 2015, 7, 2683–2703. [Google Scholar] [CrossRef] [Green Version]
- Gelderblom, H.R.; Madeley, D. Rapid viral diagnosis of orthopoxviruses by electron microscopy: Optional or a must? Viruses 2018, 10, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumont, C.; Irenge, L.M.; Magazani, E.K.; Garin, D.; Muyembe, J.-J.T.; Bentahir, M.; Gala, J.-L. Simple technique for in field samples collection in the cases of skin rash illness and subsequent PCR detection of orthopoxviruses and varicella zoster virus. PLoS ONE 2014, 9, e96930. [Google Scholar] [CrossRef]
- NCBI Monkeypox Genome Sequences. Available online: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?SeqType_s=Nucleotide&VirusLineage_ss=Monkeypoxvirus,taxid:10244&CreateDate_dt=2022-04-01T00:00:00.00ZTO2022-12-31T23:59:59.00Z (accessed on 9 August 2022).
- WHO Laboratory Testing for the Monkeypox Virus. Available online: https://www.who.int/publications/i/item/WHO-MPX-laboratory-2022.1 (accessed on 16 June 2022).
- Ropp, S.L.; Jin, Q.; Knight, J.C.; Massung, R.F.; Esposito, J.J. PCR strategy for identification and differentiation of small pox and other orthopoxviruses. J. Clin. Microbiol. 1995, 33, 2069–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, H.; Ropp, S.L.; Esposito, J.J. Gene for a-type inclusion body protein is useful for a polymerase chain reaction assay to differentiate orthopoxviruses. J. Virol. Methods 1997, 64, 217–221. [Google Scholar] [CrossRef]
- Meyer, H.; Damon, I.K.; Esposito, J.J. Orthopoxvirus diagnostics. In Vaccinia Virus and Poxvirology; Humana Press: Totowa, NJ, USA, 2004; pp. 119–133. [Google Scholar]
- Neubauer, H.; Reischl, U.; Ropp, S.; Esposito, J.; Wolf, H.; Meyer, H. Specific detection of monkeypox virus by polymerase chain reaction. J. Virol. Methods 1998, 74, 201–207. [Google Scholar] [CrossRef]
- Li, Y.; Olson, V.A.; Laue, T.; Laker, M.T.; Damon, I.K. Detection of monkeypox virus with real-time PCR assays. J. Clin. Virol. 2006, 36, 194–203. [Google Scholar] [CrossRef]
- Afonina, I.; Reed, M.; Lusby, E.; Shishkina, I.; Belousov, Y. Minor groove binder-conjugated DNA probes for quantitative DNA detection by hybridization-triggered fluorescence. BioTechniques 2002, 32, 940–949. [Google Scholar] [CrossRef] [Green Version]
- Gelaye, E.; Mach, L.; Kolodziejek, J.; Grabherr, R.; Loitsch, A.; Achenbach, J.E.; Nowotny, N.; Diallo, A.; Lamien, C.E. A novel HRM assay for the simultaneous detection and differentiation of eight poxviruses of medical and veterinary importance. Sci. Rep. 2017, 7, srep42892. [Google Scholar] [CrossRef] [Green Version]
- Nitsche, A.; Ellerbrok, H.; Pauli, G. Detection of orthopoxvirus DNA by real-time PCR and identification of variola virus DNA by melting analysis. J. Clin. Microbiol. 2004, 42, 1207–1213. [Google Scholar] [CrossRef] [Green Version]
- Olson, V.A.; Laue, T.; Laker, M.T.; Babkin, I.V.; Drosten, C.; Shchelkunov, S.; Niedrig, M.; Damon, I.K.; Meyer, H. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. J. Clin. Microbiol. 2004, 42, 1940–1946. [Google Scholar] [CrossRef] [Green Version]
- Simpson, K.; Heymann, D.; Brown, C.S.; Edmunds, W.J.; Elsgaard, J.; Fine, P.; Hochrein, H.; Hoff, N.A.; Green, A.; Ihekweazu, C.; et al. Human monkeypox—After 40 years, an unintended consequence of smallpox eradication. Vaccine 2020, 38, 5077–5081. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, H.; Wilkins, K.; Hughes, C.; Damon, I.K. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J. Virol. Methods 2010, 169, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.M.; Liu, L.; Davidson, W.B.; Radford, K.W.; Wilkins, K.; Monroe, B.; Metcalfe, M.G.; Likafi, T.; Lushima, R.S.; Kabamba, J.; et al. A tale of two viruses: Coinfections of monkeypox and varicella zoster virus in the Democratic Republic of Congo. Am. J. Trop. Med. Hyg. 2021, 104, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Luciani, L.; Inchauste, L.; Ferraris, O.; Charrel, R.; Nougairède, A.; Piorkowski, G.; Peyrefitte, C.; Bertagnoli, S.; de Lamballerie, X.; Priet, S. A novel and sensitive real-time PCR system for universal detection of poxviruses. Sci. Rep. 2021, 11, 1798. [Google Scholar] [CrossRef] [PubMed]
- Maksyutov, R.A.; Gavrilova, E.V.; Shchelkunov, S.N. Species-specific differentiation of variola, monkeypox, and varicella-zoster viruses by multiplex real-time PCR assay. J. Virol. Methods 2016, 236, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Meyer, H.; Zhao, H.; Damon, I.K. GC Content-based pan-pox universal PCR assays for poxvirus detection. J. Clin. Microbiol. 2010, 48, 268–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becherer, L.; Borst, N.; Bakheit, M.; Frischmann, S.; Zengerle, R.; von Stetten, F. Loop-mediated isothermal amplification (LAMP)—Review and classification of methods for sequence-specific detection. Anal. Methods 2020, 12, 717–746. [Google Scholar] [CrossRef] [Green Version]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Iizuka, I.; Saijo, M.; Shiota, T.; Ami, Y.; Suzaki, Y.; Nagata, N.; Hasegawa, H.; Sakai, K.; Fukushi, S.; Mizutani, T.; et al. Loop-mediated isothermal amplification-based diagnostic assay for monkeypox virus infections. J. Med. Virol. 2009, 81, 1102–1108. [Google Scholar] [CrossRef]
- Bhadra, S.; Ellington, A.D. Portable nucleic acid tests for rapid detection of monkeypox virus. medRxiv 2022. [Google Scholar] [CrossRef]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends Anal. Chem. 2017, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Li, L.; Zhao, Y.; Liu, Y.; Liu, C.; Wang, Q.; Dong, Y.; Wang, S.; Chi, T.; Song, F.; et al. Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front. Microbiol. 2020, 11, 1696. [Google Scholar] [CrossRef] [PubMed]
- Davi, S.D.; Kissenkötter, J.; Faye, M.; Böhlken-Fascher, S.; Stahl-Hennig, C.; Faye, O.; Faye, O.; Sall, A.A.; Weidmann, M.; Ademowo, O.G.; et al. Recombinase polymerase amplification assay for rapid detection of monkeypox virus. Diagn. Microbiol. Infect. Dis. 2019, 95, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Kulesh, D.A.; Loveless, B.M.; Norwood, D.; Garrison, J.; Whitehouse, C.A.; Hartmann, C.; Mucker, E.; Miller, D.; Wasieloski, L.P.; Huggins, J.; et al. Monkeypox virus detection in rodents using real-time 3′-minor groove binder TaqMan® assays on the roche light cycler. Lab. Investig. 2004, 84, 1200–1208. [Google Scholar] [CrossRef] [Green Version]
- Shchelkunov, S.N.; Shcherbakov, D.N.; Maksyutov, R.A.; Gavrilova, E.V. Species-specific identification of variola, monkeypox, cowpox, and vaccinia viruses by multiplex real-time PCR assay. J. Virol. Methods 2011, 175, 163–169. [Google Scholar] [CrossRef]
- Lorenzo, M.; Picó, Y. Wastewater-based epidemiology: Current status and future prospects. Curr. Opin. Environ. Sci. Health 2019, 9, 77–84. [Google Scholar] [CrossRef]
- Mahmoudi, T.; Naghdi, T.; Morales-Narváez, E.; Golmohammadi, H. Toward smart diagnosis of pandemic infectious diseases using wastewater-based epidemiology. TrAC Trends Anal. Chem. 2022, 153, 116635. [Google Scholar] [CrossRef]
- Antinori, A.; Mazzotta, V.; Vita, S.; Carletti, F.; Tacconi, D.; Lapini, L.E.; D’Abramo, A.; Cicalini, S.; Lapa, D.; Pittalis, S.; et al. Epidemiological, clinical and virological characteristics of four cases of monkeypox support transmission through sexual contact, Italy, May 2022. Eurosurveillance 2022, 27, 2200421. [Google Scholar] [CrossRef]
- Wolfe, M.K.; Duong, D.; Hughes, B.; Chan-Herur, V.; White, B.J.; Boehm, A.B. Detection of monkeypox viral DNA in a routine wastewater monitoring program. medRxiv 2022. [Google Scholar] [CrossRef]
- Jain, S.; Nehra, M.; Kumar, R.; Dilbaghi, N.; Hu, T.; Kumar, S.; Kaushik, A.; Li, C. Internet of medical things (IoMT)-integrated biosensors for point-of-care testing of infectious diseases. Biosens. Bioelectron. 2021, 179, 113074. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Bui, H.K.; Phan, V.M.; Seo, T.S. An internet of things-based point-of-care device for direct reverse-transcription-loop mediated isothermal amplification to identify SARS-CoV-2. Biosens. Bioelectron. 2022, 195, 113655. [Google Scholar] [CrossRef] [PubMed]
- Ge, A.; Liu, F.; Teng, X.; Cui, C.; Wu, F.; Liu, W.; Liu, Y.; Chen, X.; Xu, J.; Ma, B. A palm germ-radar (PaGeR) for rapid and simple COVID-19 detection by reverse transcription loop-mediated isothermal amplification (RT-LAMP). Biosens. Bioelectron. 2021, 200, 113925. [Google Scholar] [CrossRef] [PubMed]
- Rohrman, B.; Richards-Kortum, R. Inhibition of recombinase polymerase amplification by background DNA: A lateral flow-based method for enriching target DNA. Anal. Chem. 2015, 87, 1963–1967. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.; Batule, B.S.; Seok, Y.; Kim, M.-G. Single-step recombinase polymerase amplification assay based on a paper chip for simultaneous detection of multiple foodborne pathogens. Anal. Chem. 2018, 90, 10211–10216. [Google Scholar] [CrossRef]
- Li, J.; Pollak, N.M.; Macdonald, J. Multiplex detection of nucleic acids using recombinase polymerase amplification and a molecular colorimetric 7-segment display. ACS Omega 2019, 4, 11388–11396. [Google Scholar] [CrossRef] [Green Version]
- del Río, J.S.; Adly, N.Y.; Acero-Sánchez, J.L.; Henry, O.Y.; O’Sullivan, C.K. Electrochemical detection of francisella tularensis genomic DNA using solid-phase recombinase polymerase amplification. Biosens. Bioelectron. 2014, 54, 674–678. [Google Scholar] [CrossRef]
- Bao, M.; Zhang, S.; Pas, C.T.; Dollery, S.J.; Bushnell, R.V.; Yuqing, F.N.U.; Liu, R.; Lu, G.; Tobin, G.J.; Du, K. Computer vision enabled funnel adapted sensing tube (FAST) for power-free and pipette-free nucleic acid detection. Lab A Chip 2022. [Google Scholar] [CrossRef]
- Chaudhury, S.; Yu, C.; Liu, R.; Kumar, K.; Hornby, S.; Duplessis, C.; Sklar, J.M.; Epstein, J.E.; Reifman, J. Wearables detect malaria early in a controlled human-infection study. IEEE Trans. Biomed. Eng. 2021, 69, 2119–2129. [Google Scholar] [CrossRef]
- Frew, J.W. Monkeypox: Cutaneous clues to clinical diagnosis. J. Am. Acad. Dermatol. 2022. [Google Scholar] [CrossRef]
- Mishra, T.; Wang, M.; Metwally, A.A.; Bogu, G.K.; Brooks, A.W.; Bahmani, A.; Alavi, A.; Celli, A.; Higgs, E.; Dagan-Rosenfeld, O.; et al. Pre-symptomatic detection of COVID-19 from smartwatch data. Nat. Biomed. Eng. 2020, 4, 1208–1220. [Google Scholar] [CrossRef]
- Cho, H.R.; Kim, J.H.; Yoon, H.R.; Han, Y.S.; Kang, T.S.; Choi, H.; Lee, S. Machine learning-based optimization of pre-symptomatic COVID-19 detection through smartwatch. Sci. Rep. 2022, 12, 7886. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.; Marinsek, N.; Clay, I.; Bradshaw, B.; Ramirez, E.; Min, J.; Trister, A.; Wang, Y.; Althoff, T.; Foschini, L. Characterizing COVID-19 and influenza illnesses in the real world via person-generated health data. Patterns 2021, 2, 100188. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.; Gao, S. In silico identification of non-cross-reactive epitopes for monkeypox cell surface-binding protein. Researchsquare 2022, 1–11. [Google Scholar] [CrossRef]
- Guo, K.; Wustoni, S.; Koklu, A.; Díaz-Galicia, E.; Moser, M.; Hama, A.; Alqahtani, A.A.; Ahmad, A.N.; Alhamlan, F.S.; Shuaib, M.; et al. Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat. Biomed. Eng. 2021, 5, 666–677. [Google Scholar] [CrossRef]
- Shin, Y.; Perera, P.; Tang, W.Y.; Fu, D.L.; Liu, Q.; Sheng, J.K.; Gu, Z.; Lee, T.Y.; Barkham, T.; Park, M.K. A rapid amplification/detection assay for analysis of Mycobacterium tuberculosis using an isothermal and silicon bio-photonic sensor complex. Biosens. Bioelectron. 2015, 68, 390–396. [Google Scholar] [CrossRef]
- Ming, K.; Kim, J.; Biondi, M.J.; Syed, A.; Chen, K.; Lam, A.; Ostrowski, M.; Rebbapragada, A.; Feld, J.J.; Chan, W.C.W. Integrated quantum dot barcode smartphone optical device for wireless multiplexed diagnosis of infected patients. ACS Nano 2015, 9, 3060–3074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolaou, P.; Sciuto, E.L.; Zanut, A.; Petralia, S.; Valenti, G.; Paolucci, F.; Prodi, L.; Conoci, S. Ultrasensitive PCR-Free detection of whole virus genome by electrochemiluminescence. Biosens. Bioelectron. 2022, 209, 114165. [Google Scholar] [CrossRef]
- Yuan, X.; Yang, C.; He, Q.; Chen, J.; Yu, D.; Li, J.; Zhai, S.; Qin, Z.; Du, K.; Chu, Z.; et al. Current and perspective diagnostic techniques for COVID-19. ACS Infect. Dis. 2020, 6, 1998–2016. [Google Scholar] [CrossRef]
- Bao, M.; Chen, Q.; Xu, Z.; Jensen, E.C.; Liu, C.; Waitkus, J.T.; Yuan, X.; He, Q.; Qin, P.; Du, K. Challenges and opportunities for clustered regularly interspaced short palindromic repeats based molecular biosensing. ACS Sensors 2021, 6, 2497–2522. [Google Scholar] [CrossRef]
- Gul, I.; Aer, L.; Zhang, M.; Jiang, H.; Khan, A.A.; Bilal, M.; Fang, R.; Feng, J.; Zeng, H.; Tang, L. Multifunctional 3D-printed platform integrated with a smartphone ambient light sensor for halocarbon contaminants monitoring. Environ. Technol. Innov. 2021, 24, 101883. [Google Scholar] [CrossRef]
- Gul, I.; Zhai, S.; Zhong, X.; Chen, Q.; Yuan, X.; Du, Z.; Qin, P. Angiotensin-converting enzyme 2-based biosensing modalities and devices for coronavirus detection. OSF 2022. [Google Scholar] [CrossRef]
- Gul, I.; Le, W.; Jie, Z.; Ruiqin, F.; Bilal, M.; Tang, L. Recent advances on engineered enzyme-conjugated biosensing modalities and devices for halogenated compounds. TrAC Trends Anal. Chem. 2021, 134, 116145. [Google Scholar] [CrossRef]
- Gul, I.; Wang, Q.; Jiang, Q.; Fang, R.; Tang, L. Enzyme immobilization on glass fiber membrane for detection of halogenated compounds. Anal. Biochem. 2020, 609, 113971. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gul, I.; Liu, C.; Yuan, X.; Du, Z.; Zhai, S.; Lei, Z.; Chen, Q.; Raheem, M.A.; He, Q.; Hu, Q.; et al. Current and Perspective Sensing Methods for Monkeypox Virus. Bioengineering 2022, 9, 571. https://doi.org/10.3390/bioengineering9100571
Gul I, Liu C, Yuan X, Du Z, Zhai S, Lei Z, Chen Q, Raheem MA, He Q, Hu Q, et al. Current and Perspective Sensing Methods for Monkeypox Virus. Bioengineering. 2022; 9(10):571. https://doi.org/10.3390/bioengineering9100571
Chicago/Turabian StyleGul, Ijaz, Changyue Liu, Xi Yuan, Zhicheng Du, Shiyao Zhai, Zhengyang Lei, Qun Chen, Muhammad Akmal Raheem, Qian He, Qiuyue Hu, and et al. 2022. "Current and Perspective Sensing Methods for Monkeypox Virus" Bioengineering 9, no. 10: 571. https://doi.org/10.3390/bioengineering9100571
APA StyleGul, I., Liu, C., Yuan, X., Du, Z., Zhai, S., Lei, Z., Chen, Q., Raheem, M. A., He, Q., Hu, Q., Xiao, C., Haihui, Z., Wang, R., Han, S., Du, K., Yu, D., Zhang, C. Y., & Qin, P. (2022). Current and Perspective Sensing Methods for Monkeypox Virus. Bioengineering, 9(10), 571. https://doi.org/10.3390/bioengineering9100571