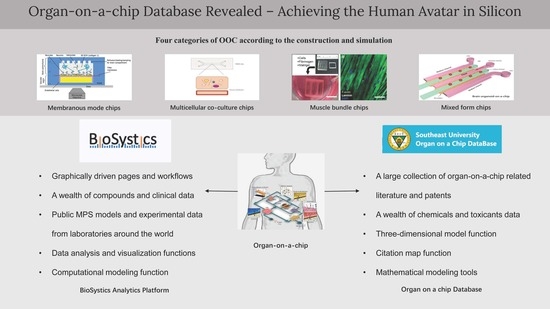
Organ-On-A-Chip Database Revealed—Achieving the Human Avatar in Silicon
Abstract
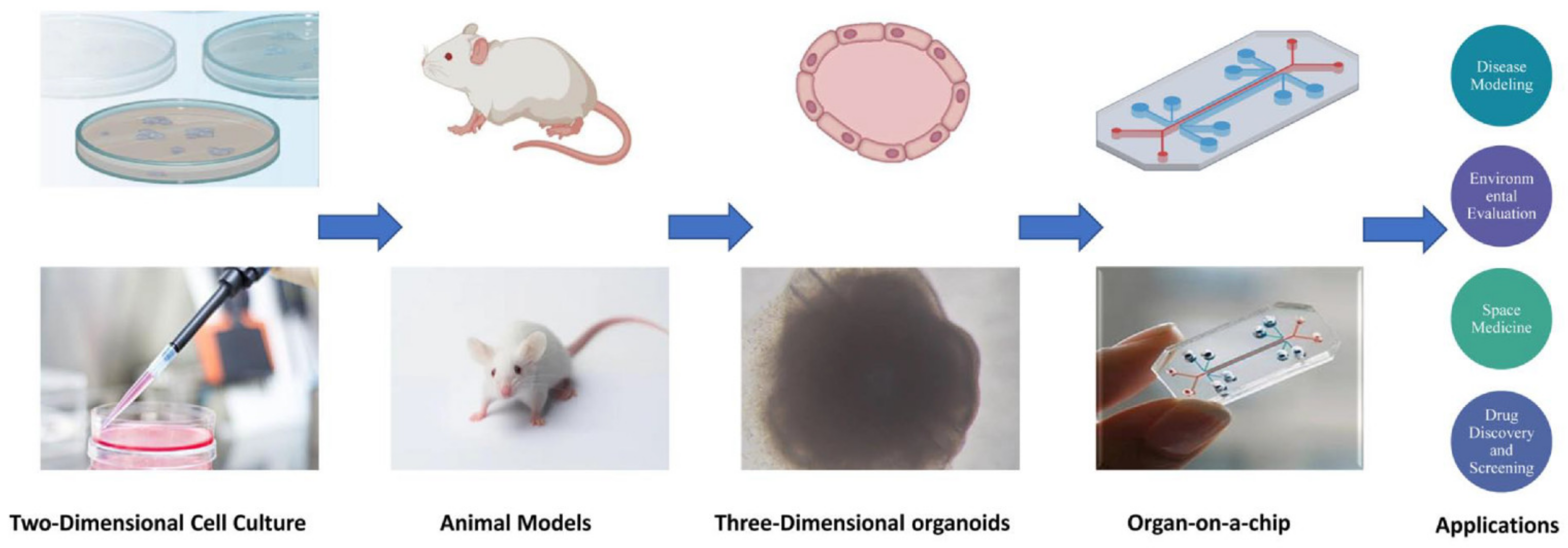
:1. Introduction
2. Classification and Structure of Organs-On-Chips
2.1. Simulation Mode of Organs-On-Chips for Organ Functions
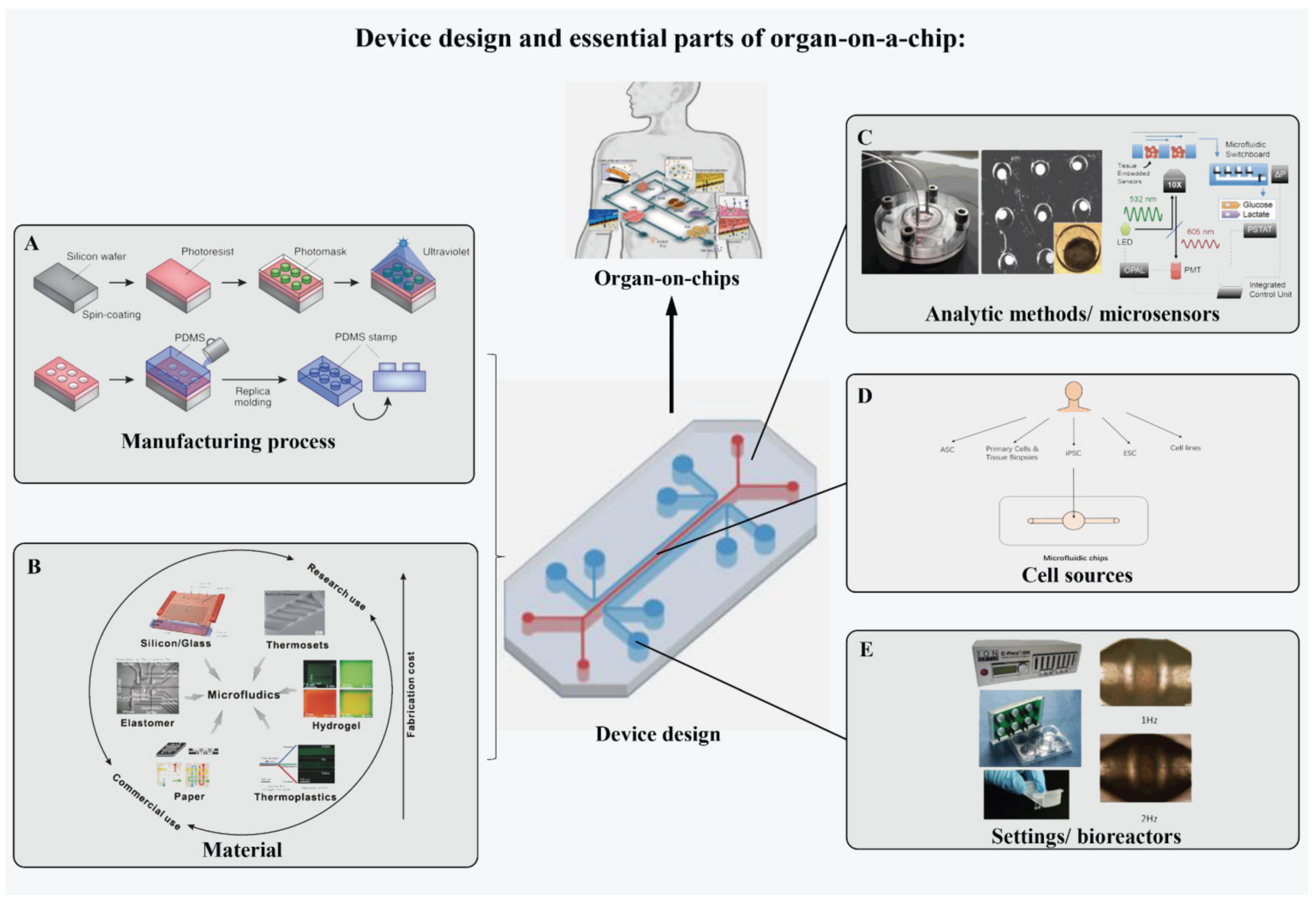
2.2. Device Design and Essential Parts of OOCs
2.2.1. Manufacturing Process of Device
2.2.2. Material of the Device
2.2.3. Cell Sources
2.2.4. Sensory Systems
2.2.5. Settings
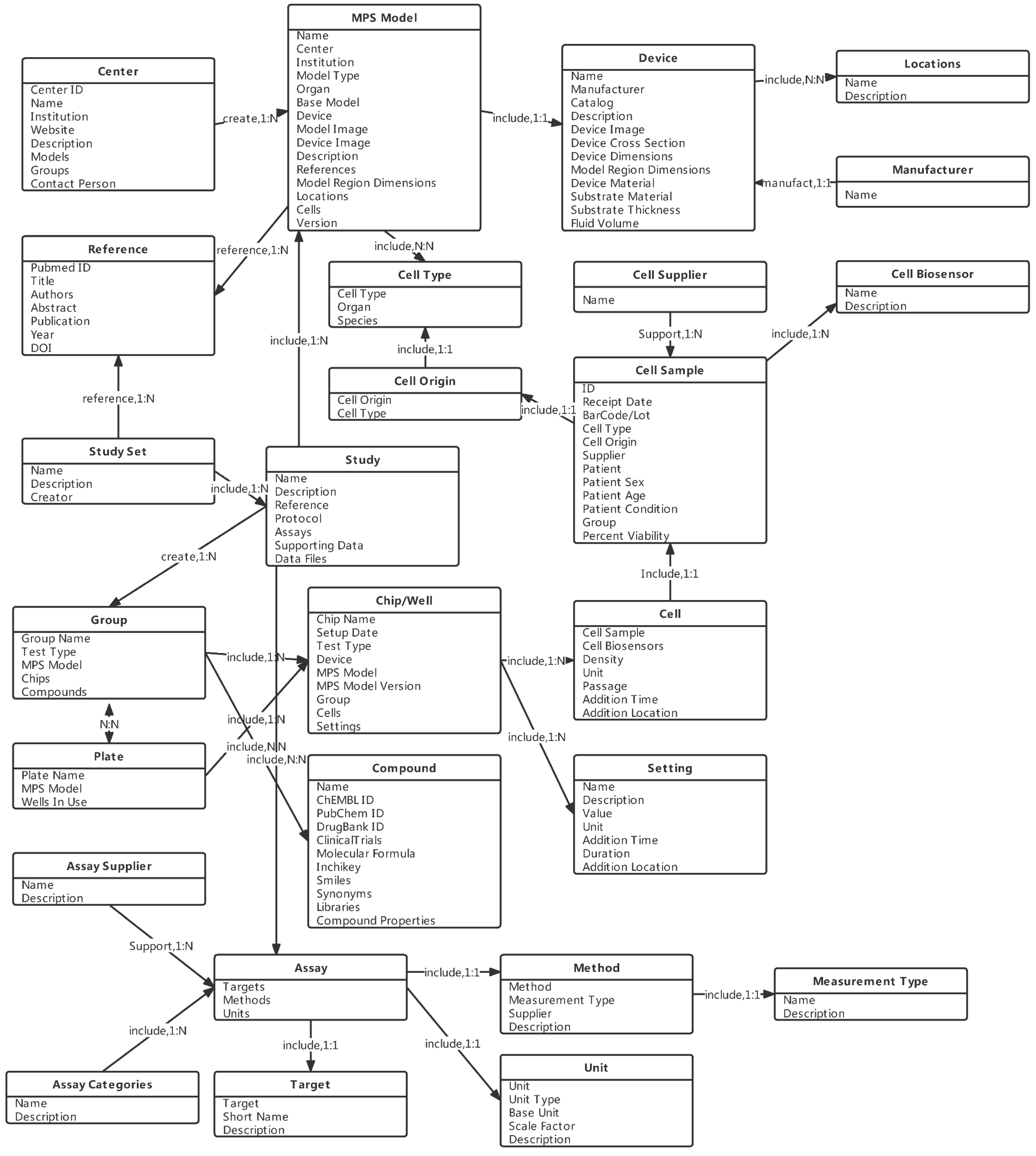
3. Database Design
3.1. Data Model
3.2. Entity and Relationship Design
4. Developed OOC Database in the World
4.1. BAP
4.1.1. Functions of BAP Website
- (i)
- Study design.
- (ii)
- Data analysis.
- (iii)
- Reproducibility analysis.
- (iv)
- Computational modeling.
4.1.2. Public Data Included in BAP
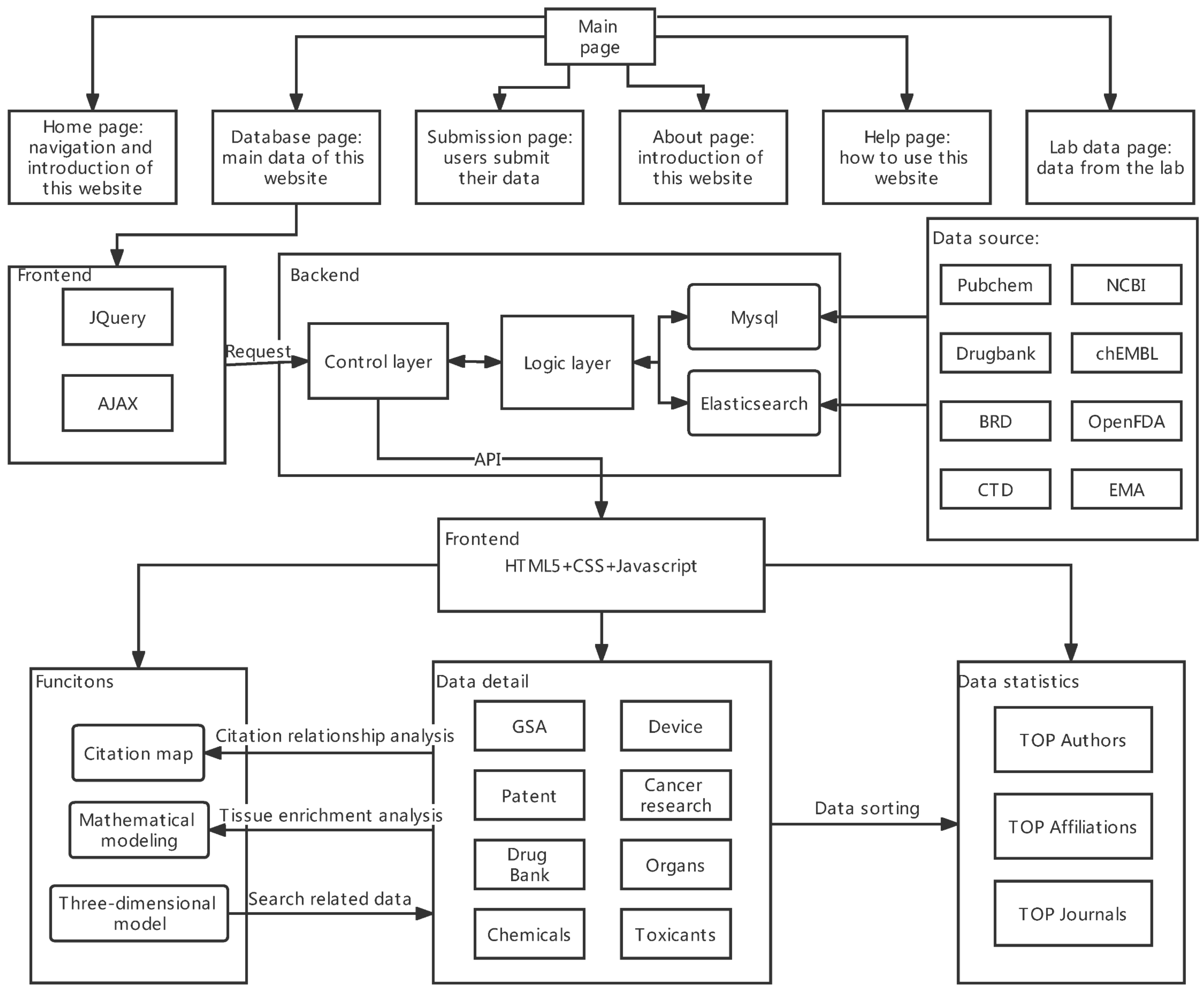
4.2. The Overview of Ocdb
4.3. Comparison of the Two Databases
5. Application Prospect and Future Development Direction of OOC and Its Database
5.1. Personalized Medicine
5.2. Pharmacology
5.3. Environmental Toxicology
5.4. Space Medicine
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Approval
References
- Top Ten Emerging Technologies. Available online: https://www.weforum.org/agenda/2016/06/top-10-emerging-technologies-2016/ (accessed on 23 September 2022).
- Ainslie, G.R.; Davis, M.; Ewart, L.; Lieberman, L.A.; Rowlands, D.J.; Thorley, A.J.; Yoder, G.; Ryan, A.M. Microphysiological lung models to evaluate the safety of new pharmaceutical modalities: A biopharmaceutical perspective. Lab Chip 2019, 19, 3152–3161. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-chip: Recent breakthroughs and future prospects. Biomed. Eng. Online 2020, 19, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Alepee, N.; Bahinski, A.; Daneshian, M.; De Wever, B.; Fritsche, E.; Goldberg, A.; Hansmann, J.; Hartung, T.; Haycock, J.; Hogberg, H.; et al. State-of-the-art of 3D cultures (organs-on-a-chip) in safety testing and pathophysiology. ALTEX 2014, 31, 441–477. [Google Scholar] [CrossRef] [PubMed]
- Low, L.A.; Sutherland, M.; Lumelsky, N.; Selimovic, S.; Lundberg, M.S.; Tagle, D.A. Organs-on-a-Chip. In Biomaterials- and Microfluidics-Based Tissue Engineered 3D Models; Oliveira, J.M., Reis, R.L., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 27–42. [Google Scholar]
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G.; Pennie, W.D.; Pickett, S.D.; Wang, J.; et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Suggitt, M.; Bibby, M.C. 50 years of preclinical anticancer drug screening: Empirical to target-driven approaches. Clin. Cancer Res. 2005, 11, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Parng, C.; Seng, W.L.; Semino, C.; McGrath, P. Zebrafish: A preclinical model for drug screening. Assay Drug Dev. Technol. 2002, 1, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, A.D.; van den Berg, A. Organs-on-chips: Breaking the in vitro impasse. Integr. Biol. 2012, 4, 461–470. [Google Scholar] [CrossRef]
- Schutgens, F.; Clevers, H. Human Organoids: Tools for Understanding Biology and Treating Diseases. Annu. Rev. Pathol. 2020, 15, 211–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Esch, M.B.; Prot, J.-M.; Long, C.J.; Smith, A.; Hickman, J.J.; Shuler, M.L. Microfabricated mammalian organ systems and their integration into models of whole animals and humans. Lab Chip 2013, 13, 1201–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, L. Creating databases for biological information: An introduction. Curr. Protoc. Bioinform. 2013, 42, 9.1.1–9.1.10. [Google Scholar] [CrossRef] [PubMed]
- De Silva, N.H.N.D. Relational Databases and Biomedical Big Data. In Bioinformatics in MicroRNA Research; Huang, J., Borchert, G.M., Dou, D., Huan, J., Lan, W., Tan, M., Wu, B., Eds.; Springer New York: New York, NY, USA, 2017; pp. 69–81. [Google Scholar]
- Organ on a Chip Database. Available online: http://www.organchip.cn/ (accessed on 23 September 2022).
- FDA. Organs-on-Chips Technology; FDA: Silver Spring, MA, USA, 2017.
- Lee-Montiel, F.T.; George, S.M.; Gough, A.H.; Sharma, A.D.; Wu, J.; DeBiasio, R.; Vernetti, L.A.; Taylor, D.L. Control of oxygen tension recapitulates zone-specific functions in human liver microphysiology systems. Exp. Biol. Med. 2017, 242, 1617–1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Li, N.; Long, M. Chapter 6—Liver sinusoid on a chip. In Methods in Cell Biology; Doh, J., Fletcher, D., Piel, M., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 146, pp. 105–134. [Google Scholar]
- Brown, J.A.; Pensabene, V.; Markov, D.A.; Allwardt, V.; Neely, M.D.; Shi, M.; Britt, C.M.; Hoilett, O.S.; Yang, Q.; Brewer, B.M.; et al. Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor. Biomicrofluidics 2015, 9, 054124. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, L.; Zhu, Y.; Qin, J. Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab Chip 2018, 18, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.P.; Hou, Z.; Propson, N.E.; Zhang, J.; Engstrom, C.J.; Santos Costa, V.; Jiang, P.; Nguyen, B.K.; Bolin, J.M.; Daly, W.; et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc. Natl. Acad. Sci. USA 2015, 112, 12516–12521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcendor, D.J.; Block, F.E., 3rd; Cliffel, D.E.; Daniels, J.S.; Ellacott, K.L.J.; Goodwin, C.R.; Hofmeister, L.H.; Li, D.; Markov, D.A.; May, J.C.; et al. Neurovascular unit on a chip: Implications for translational applications. Stem Cell Res. Ther. 2013, 4 (Suppl. S1), S18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, A.; Loskill, P.; Shao, K.; Huebsch, N.; Hong, S.; Marcus, S.G.; Marks, N.; Mandegar, M.; Conklin, B.R.; Lee, L.P.; et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 2015, 5, 8883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villasante, A.; Marturano-Kruik, A.; Vunjak-Novakovic, G. Bioengineered human tumor within a bone niche. Biomaterials 2014, 35, 5785–5794. [Google Scholar] [CrossRef]
- Torisawa, Y.-s.; Spina, C.S.; Mammoto, T.; Mammoto, A.; Weaver, J.C.; Tat, T.; Collins, J.J.; Ingber, D.E. Bone marrow–on–a–chip replicates hematopoietic niche physiology in vitro. Nat. Methods 2014, 11, 663–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, E.J.; Chapron, A.; Chapron, B.D.; Voellinger, J.L.; Lidberg, K.A.; Yeung, C.K.; Wang, Z.; Yamaura, Y.; Hailey, D.W.; Neumann, T.; et al. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. 2016, 90, 627–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmet Akif, K.; Tuğçe, P.; Gül Banu, A.; Ali, A. Lung on a Chip for Drug Screening and Design. Curr. Pharm. Des. 2018, 24, 5386–5396. [Google Scholar] [CrossRef]
- Madden, L.; Juhas, M.; Kraus, W.E.; Truskey, G.A.; Bursac, N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. Elife 2015, 4, e04885. [Google Scholar] [CrossRef] [Green Version]
- Blutt, S.E.; Broughman, J.R.; Zou, W.; Zeng, X.-L.; Karandikar, U.C.; In, J.; Zachos, N.C.; Kovbasnjuk, O.; Donowitz, M.; Estes, M.K. Gastrointestinal microphysiological systems. Exp. Biol. Med. 2017, 242, 1633–1642. [Google Scholar] [CrossRef] [Green Version]
- Trietsch, S.J.; Naumovska, E.; Kurek, D.; Setyawati, M.C.; Vormann, M.K.; Wilschut, K.J.; Lanz, H.L.; Nicolas, A.; Ng, C.P.; Joore, J.; et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun. 2017, 8, 262. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Higgins, C.A.; Gillette, B.M.; Itoh, M.; Umegaki, N.; Gledhill, K.; Sia, S.K.; Christiano, A.M. Building a microphysiological skin model from induced pluripotent stem cells. Stem Cell Res. Ther. 2013, 4 (Suppl. S1), S2. [Google Scholar] [CrossRef] [Green Version]
- Kwak, B.S.; Jin, S.-P.; Kim, S.J.; Kim, E.J.; Chung, J.H.; Sung, J.H. Microfluidic skin chip with vasculature for recapitulating the immune response of the skin tissue. Biotechnol. Bioeng. 2020, 117, 1853–1863. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Z.; Zhang, Y.; Wang, X.; Ouyang, J.; Zhu, J.; Yan, Y.; Sun, X.; Wang, F.; Li, X. Construction of a high fidelity epidermis-on-a-chip for scalable in vitro irritation evaluation. Lab Chip 2021, 21, 3804–3818. [Google Scholar] [CrossRef]
- McAleer, C.W.; Long, C.J.; Elbrecht, D.; Sasserath, T.; Bridges, L.R.; Rumsey, J.W.; Martin, C.; Schnepper, M.; Wang, Y.; Schuler, F.; et al. Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Sci. Transl. Med. 2019, 11, eaav1386. [Google Scholar] [CrossRef]
- Oleaga, C.; Bernabini, C.; Smith, A.S.; Srinivasan, B.; Jackson, M.; McLamb, W.; Platt, V.; Bridges, R.; Cai, Y.; Santhanam, N.; et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. 2016, 6, 20030. [Google Scholar] [CrossRef] [PubMed]
- Marturano-Kruik, A.; Nava, M.M.; Yeager, K.; Chramiec, A.; Hao, L.; Robinson, S.; Guo, E.; Raimondi, M.T.; Vunjak-Novakovic, G. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl. Acad. Sci. USA 2018, 115, 1256–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatherell, K.; Couraud, P.-O.; Romero, I.A.; Weksler, B.; Pilkington, G.J. Development of a three-dimensional, all-human in vitro model of the blood–brain barrier using mono-, co-, and tri-cultivation Transwell models. J. Neurosci. Methods 2011, 199, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J.; Patabendige, A.A.; Dolman, D.E.; Yusof, S.R.; Begley, D.J. Structure and function of the blood–brain barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Loskill, P.; Sezhian, T.; Tharp, K.M.; Lee-Montiel, F.T.; Jeeawoody, S.; Reese, W.M.; Zushin, P.H.; Stahl, A.; Healy, K.E. WAT-on-a-chip: A physiologically relevant microfluidic system incorporating white adipose tissue. Lab Chip 2017, 17, 1645–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edington, C.D.; Chen, W.L.K.; Geishecker, E.; Kassis, T.; Soenksen, L.R.; Bhushan, B.M.; Freake, D.; Kirschner, J.; Maass, C.; Tsamandouras, N.; et al. Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies. Sci. Rep. 2018, 8, 4530. [Google Scholar] [CrossRef] [Green Version]
- Chou, D.B.; Frismantas, V.; Milton, Y.; David, R.; Pop-Damkov, P.; Ferguson, D.; MacDonald, A.; Vargel Bölükbaşı, Ö.; Joyce, C.E.; Moreira Teixeira, L.S.; et al. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat. Biomed. Eng. 2020, 4, 394–406. [Google Scholar] [CrossRef]
- Miedel, M.T.; Gavlock, D.C.; Jia, S.; Gough, A.; Taylor, D.L.; Stern, A.M. Modeling the Effect of the Metastatic Microenvironment on Phenotypes Conferred by Estrogen Receptor Mutations Using a Human Liver Microphysiological System. Sci. Rep. 2019, 9, 8341. [Google Scholar] [CrossRef] [Green Version]
- MIMETAS. OrganoPlate® 3-Lane; MIMETAS: Leiden, The Netherlands, 2018. [Google Scholar]
- MIMETAS. OrganoPlate® 2-Lane; MIMETAS: Leiden, The Netherlands, 2018. [Google Scholar]
- Zhao, Y.; Rafatian, N.; Feric, N.T.; Cox, B.J.; Aschar-Sobbi, R.; Wang, E.Y.; Aggarwal, P.; Zhang, B.; Conant, G.; Ronaldson-Bouchard, K. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 2019, 176, 913–927.e918. [Google Scholar] [CrossRef] [Green Version]
- Park Sunghee, E.; Georgescu, A.; Huh, D. Organoids-on-a-chip. Science 2019, 364, 960–965. [Google Scholar] [CrossRef]
- Ren, K.; Zhou, J.; Wu, H. Materials for Microfluidic Chip Fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef] [PubMed]
- Bavli, D.; Prill, S.; Ezra, E.; Levy, G.; Cohen, M.; Vinken, M.; Vanfleteren, J.; Jaeger, M.; Nahmias, Y. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 2016, 113, E2231–E2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, D.C.; McDonald, J.C.; Schueller, O.J.; Whitesides, G.M. Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane). Anal. Chem. 1998, 70, 4974–4984. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Cai, D.; Chen, P. Micro-and nanotechnologies for study of cell secretion. Anal. Chem. 2011, 83, 4393–4406. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.H.; Stapleton, S.C.; Yang, M.T.; Cha, S.S.; Choi, C.K.; Galie, P.A.; Chen, C.S. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc. Natl. Acad. Sci. USA 2013, 110, 6712–6717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, D.T.T.; Wang, X.; Craver, B.M.; Sobrino, A.; Zhao, D.; Chen, J.C.; Lee, L.Y.N.; George, S.C.; Lee, A.P.; Hughes, C.C.W. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip 2017, 17, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Trietsch, S.J.; Israëls, G.D.; Joore, J.; Hankemeier, T.; Vulto, P. Microfluidic titer plate for stratified 3D cell culture. Lab Chip 2013, 13, 3548–3554. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.B.; Wu, Q.; Yazbeck, J.; Liu, C.; Okhovatian, S.; Radisic, M. Beyond Polydimethylsiloxane: Alternative Materials for Fabrication of Organ-on-a-Chip Devices and Microphysiological Systems. ACS Biomater. Sci. Eng. 2021, 7, 2880–2899. [Google Scholar] [CrossRef]
- Amer, S.; Badawy, W. An integrated platform for bio-analysis and drug delivery. Curr. Pharm. Biotechnol. 2005, 6, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Rubin, J.; Deng, Y.; Huang, C.; Demirci, U.; Karp, J.M.; Khademhosseini, A. A cell-laden microfluidic hydrogel. Lab Chip 2007, 7, 756–762. [Google Scholar] [CrossRef]
- Ziolkowska, K.; Jedrych, E.; Kwapiszewski, R.; Lopacinska, J.; Skolimowski, M.; Chudy, M. PDMS/glass microfluidic cell culture system for cytotoxicity tests and cells passage. Sens. Actuators B Chem. 2010, 145, 533–542. [Google Scholar] [CrossRef]
- Jang, K.; Sato, K.; Igawa, K.; Chung, U.-i.; Kitamori, T. Development of an osteoblast-based 3D continuous-perfusion microfluidic system for drug screening. Anal. Bioanal. Chem. 2008, 390, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Shirure, V.S.; George, S.C. Design considerations to minimize the impact of drug absorption in polymer-based organ-on-a-chip platforms. Lab Chip 2017, 17, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Davenport Huyer, L.; Bannerman, A.D.; Wang, Y.; Savoji, H.; Knee-Walden, E.J.; Brissenden, A.; Yee, B.; Shoaib, M.; Bobicki, E.; Amsden, B.G. One-Pot Synthesis of Unsaturated Polyester Bioelastomer with Controllable Material Curing for Microscale Designs. Adv. Healthc. Mater. 2019, 8, 1900245. [Google Scholar] [CrossRef] [PubMed]
- Lenguito, G.; Chaimov, D.; Weitz, J.R.; Rodriguez-Diaz, R.; Rawal, S.A.; Tamayo-Garcia, A.; Caicedo, A.; Stabler, C.L.; Buchwald, P.; Agarwal, A. Resealable, optically accessible, PDMS-free fluidic platform for ex vivo interrogation of pancreatic islets. Lab Chip 2017, 17, 772–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, R.; Korolj, A.; Liu, C.; Song, X.; Lu, R.X.Z.; Zhang, B.; Ramachandran, A.; Liang, Q.; Radisic, M. h-FIBER: Microfluidic topographical hollow fiber for studies of glomerular filtration barrier. ACS Cent. Sci. 2020, 6, 903–912. [Google Scholar] [CrossRef]
- Lu, Y.; Shi, W.; Qin, J.; Lin, B. Fabrication and characterization of paper-based microfluidics prepared in nitrocellulose membrane by wax printing. Anal. Chem. 2010, 82, 329–335. [Google Scholar] [CrossRef]
- Wang, Y.I.; Carmona, C.; Hickman, J.J.; Shuler, M.L. Multiorgan microphysiological systems for drug development: Strategies, advances, and challenges. Adv. Healthc. Mater. 2018, 7, 1701000. [Google Scholar] [CrossRef] [PubMed]
- Lind, J.U.; Busbee, T.A.; Valentine, A.D.; Pasqualini, F.S.; Yuan, H.; Yadid, M.; Park, S.-J.; Kotikian, A.; Nesmith, A.P.; Campbell, P.H. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 2017, 16, 303–308. [Google Scholar] [CrossRef]
- Ellis, B.W.; Acun, A.; Can, U.I.; Zorlutuna, P. Human iPSC-derived myocardium-on-chip with capillary-like flow for personalized medicine. Biomicrofluidics 2017, 11, 024105. [Google Scholar] [CrossRef]
- Kanamori, T.; Sugiura, S.; Sakai, Y. Technical aspects of microphysiological systems (MPS) as a promising wet human-in-vivo simulator. Drug Metab. Pharmacokinet. 2018, 33, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Jun, B.-H. Advances in dynamic microphysiological organ-on-a-chip: Design principle and its biomedical application. J. Ind. Eng. Chem. 2019, 71, 65–77. [Google Scholar] [CrossRef]
- Ahadian, S.; Civitarese, R.; Bannerman, D.; Mohammadi, M.H.; Lu, R.; Wang, E.; Davenport-Huyer, L.; Lai, B.; Zhang, B.; Zhao, Y. Organ-on-a-chip platforms: A convergence of advanced materials, cells, and microscale technologies. Adv. Healthc. Mater. 2018, 7, 1700506. [Google Scholar] [CrossRef] [PubMed]
- Matter, K.; Mellman, I. Mechamisms of cell polarity: Sorting and transport in epithelial cells. Curr. Opin. Cell Biol. 1994, 6, 545–554. [Google Scholar] [CrossRef]
- Encyclopedia of Database Systems; Springer: New York, NY, USA; London, UK, 2009.
- Codd, E.F. A relational model of data for large shared data banks. In Software Pioneers; Springer: Berlin/Heidelberg, Germany, 2002; pp. 263–294. [Google Scholar]
- Zhang, X.; Sun, X.F.; Cao, Y.; Ye, B.; Peng, Q.; Liu, X.; Shen, B.; Zhang, H. CBD: A biomarker database for colorectal cancer. Database 2018, 2018, bay046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giffen, C.A.; Carroll, L.E.; Adams, J.T.; Brennan, S.P.; Coady, S.A.; Wagner, E.L. Providing contemporary access to historical biospecimen collections: Development of the NHLBI Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC). Biopreservation Biobanking 2015, 13, 271–279. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liang, W.; Chen, Z.; Li, X.; Liu, A.; Chen, P.; Li, Q.; Mei, X.; Yang, J.; Liu, J.; et al. Organ on a Chip Database (Ocdb): A Comprehensive, Systematic and Real-time Organ-on-a-chip Database. bioRxiv 2022. [Google Scholar] [CrossRef]
- Cattell, R. Scalable SQL and NoSQL data stores. SIGMOD Rec. 2011, 39, 12–27. [Google Scholar] [CrossRef] [Green Version]
- Leavitt, N. Will NoSQL Databases Live Up to Their Promise? Computer 2010, 43, 12–14. [Google Scholar] [CrossRef]
- Schulz, W.L.; Nelson, B.G.; Felker, D.K.; Durant, T.J.S.; Torres, R. Evaluation of relational and NoSQL database architectures to manage genomic annotations. J. Biomed. Inform. 2016, 64, 288–295. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Brister, J.R.; Bolton, E.E.; Canese, K.; Comeau, D.C.; Funk, K.; Ketter, A.; Kim, S.; Kimchi, A.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020, 48, D9–D16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gough, A.; Vernetti, L.; Bergenthal, L.; Shun, T.Y.; Taylor, D.L. The Microphysiology Systems Database for Analyzing and Modeling Compound Interactions with Human and Animal Organ Models. Appl. In Vitro Toxicol. 2016, 2, 103–117. [Google Scholar] [CrossRef] [PubMed]
- The PostgreSQL Global Development Group. PostgreSQL; The PostgreSQL Global Development Group: Wolfville, NS, Canada, 2016. [Google Scholar]
- Chen, P.P.-S. The entity-relationship model—Toward a unified view of data. ACM Trans. Database Syst. (TODS) 1976, 1, 9–36. [Google Scholar] [CrossRef] [Green Version]
- Teorey, T.J.; Yang, D.; Fry, J.P. A logical design methodology for relational databases using the extended entity-relationship model. ACM Comput. Surv. (CSUR) 1986, 18, 197–222. [Google Scholar] [CrossRef] [Green Version]
- Markowitz, V.M.; Shoshani, A. Representing extended entity-relationship structures in relational databases: A modular approach. ACM Trans. Database Syst. (TODS) 1992, 17, 423–464. [Google Scholar] [CrossRef] [Green Version]
- Harrington, J.L.; Harrington, J.L. Relational Database Design and Implementation: Clearly Explained, 3rd ed.; Morgan Kaufmann/Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Microphysiology Systems Database Study List. Available online: https://mps.csb.pitt.edu/assays/assaystudy/ (accessed on 23 September 2022).
- Django; Django Software Foundation: Lawrence, KA, USA, 2016.
- Anaconda, Version 2-2.4.0; Anaconda Software Distribution: Austin, TX, USA, 2015.
- Duggan, M.A.; Anderson, W.F.; Altekruse, S.; Penberthy, L.; Sherman, M.E. The Surveillance, Epidemiology, and End Results (SEER) Program and Pathology: Toward Strengthening the Critical Relationship. Am. J. Surg. Pathol. 2016, 40, e94–e102. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.-w.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Anthony Celi, L.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DataTable, Version 1.10.4; SpryMedia Ltd.: Dunfermline, UK, 2016.
- Bartlett, J.; Frost, C. Reliability, repeatability and reproducibility: Analysis of measurement errors in continuous variables. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2008, 31, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Shrout, P.E.; Fleiss, J.L. Intraclass correlations: Uses in assessing rater reliability. Psychol. Bull. 1979, 86, 420. [Google Scholar] [CrossRef] [PubMed]
- Mcgraw, K.O.; Wong, S.P. Forming inferences about some intraclass correlation coefficients. Psychol. Methods 1996, 1, 390. [Google Scholar] [CrossRef]
- Cohen, J. Statistical power analysis. Curr. Dir. Psychol. Sci. 1992, 1, 98–101. [Google Scholar] [CrossRef]
- Glass, G.V. Primary, secondary, and meta-analysis of research. Educ. Res. 1976, 5, 3–8. [Google Scholar] [CrossRef]
- Hedges, L.V. Distribution theory for Glass’s estimator of effect size and related estimators. J. Educ. Stat. 1981, 6, 107–128. [Google Scholar] [CrossRef]
- Schurdak, M.; Vernetti, L.; Bergenthal, L.; Wolter, Q.K.; Shun, T.Y.; Karcher, S.; Taylor, D.L.; Gough, A. Applications of the microphysiology systems database for experimental ADME-Tox and disease models. Lab Chip 2020, 20, 1472–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, I.R.; Fuchs, O.; Hansen, G.; von Mutius, E.; Kopp, M.V. What is precision medicine? Eur. Respir. J. 2017, 50, 1700391. [Google Scholar] [CrossRef] [Green Version]
- Van den Berg, A.; Mummery, C.L.; Passier, R.; van der Meer, A.D. Personalised organs-on-chips: Functional testing for precision medicine. Lab chip 2019, 19, 198–205. [Google Scholar] [CrossRef] [Green Version]
- Pandian, N.K.; Mannino, R.G.; Lam, W.A.; Jain, A. Thrombosis-on-a-chip: Prospective impact of microphysiological models of vascular thrombosis. Curr. Opin. Biomed. Eng. 2018, 5, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Prantil-Baun, R.; Novak, R.; Das, D.; Somayaji, M.R.; Przekwas, A.; Ingber, D.E. Physiologically Based Pharmacokinetic and Pharmacodynamic Analysis Enabled by Microfluidically Linked Organs-on-Chips. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 37–64. [Google Scholar] [CrossRef] [PubMed]
- Kenny, J.R.; Mcginnity, D.F.; Grime, K.; Riley, R.J. Preclinical Development Handbook: ADME and Biopharmaceutical Properties; Gad Consulting Services: Raleigh, NC, USA, 2017. [Google Scholar]
- Sager, J.E.; Yu, J.; Ragueneau-Majlessi, I.; Isoherranen, N. Physiologically Based Pharmacokinetic (PBPK) Modeling and Simulation Approaches: A Systematic Review of Published Models, Applications, and Model Verification. Drug Metab. Dispos. Biol. Fate Chem. 2015, 43, 1823–1837. [Google Scholar] [CrossRef] [PubMed]
- Prot, J.M.; Maciel, L.; Bricks, T.; Merlier, F.; Cotton, J.; Paullier, P.; Bois, F.Y.; Leclerc, E. First pass intestinal and liver metabolism of paracetamol in a microfluidic platform coupled with a mathematical modeling as a means of evaluating ADME processes in humans. Biotechnol. Bioeng. 2015, 111, 2027–2040. [Google Scholar] [CrossRef] [PubMed]
- Esch, M.B.; Mahler, G.J.; Stokol, T.; Shuler, M.L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip 2014, 14, 3081–3092. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Chen, Z.; Cheng, Y.; Liu, T.; Lihong, Y.; Pu, Y.; Liang, G. Environmental toxicology wars: Organ-on-a-chip for assessing the toxicity of environmental pollutants. Environ. Pollut. 2021, 268, 115861. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Yoon, J.-Y. Organ-on-a-chip for assessing environmental toxicants. Curr. Opin. Biotechnol. 2017, 45, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.U.S. EPA to eliminate all mammal testing by 2035. Science 2019, 365, 1231. [Google Scholar] [CrossRef]
- Blaber, E.; Marçal, H.; Burns, B.P. Bioastronautics: The influence of microgravity on astronaut health. Astrobiology 2010, 10, 463–473. [Google Scholar] [CrossRef]
- Ilyina-Kakueva, E.; Burkovskaya, T. The microgravity effect on a repair process in M. soleus of the rats flown on Cosmos-2044. Physiologist 1991, 34, S141–S142. [Google Scholar]
- Kaplansky, A.; Durnova, G.; Burkovskaya, T.; Vorotnikova, E. The effect of microgravity on bone fracture healing in rats flown on Cosmos-2044. Physiologist 1991, 34, S196–S199. [Google Scholar]
- Blomqvist, C.; Buckey, J.; Gaffney, F.; Lane, L.; Levine, B.; Watenpaugh, D. Mechanisms of post-flight orthostatic intolerance. J. Gravit. Physiol. J. Int. Soc. Gravit. Physiol. 1994, 1, P122–P124. [Google Scholar]
- Fritsch-Yelle, J.M.; Charles, J.B.; Jones, M.M.; Wood, M.L. Microgravity decreases heart rate and arterial pressure in humans. J. Appl. Physiol. 1996, 80, 910–914. [Google Scholar] [CrossRef]
- Caillot-Augusseau, A.; Lafage-Proust, M.-H.; Soler, C.; Pernod, J.; Dubois, F.; Alexandre, C. Bone formation and resorption biological markers in cosmonauts during and after a 180-day space flight (Euromir 95). Clin. Chem. 1998, 44, 578–585. [Google Scholar] [CrossRef]
- Dai, Z.-Q.; Wang, R.; Ling, S.; Wan, Y.; Li, Y. Simulated microgravity inhibits the proliferation and osteogenesis of rat bone marrow mesenchymal stem cells. Cell Prolif. 2007, 40, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.H.; Trunkey, D.; Rebagliati, G.S. Emergency medicine in space. J. Emerg. Med. 2007, 32, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Tissue Chips in Space. Available online: https://ncats.nih.gov/tissuechip/projects/space (accessed on 23 September 2022).

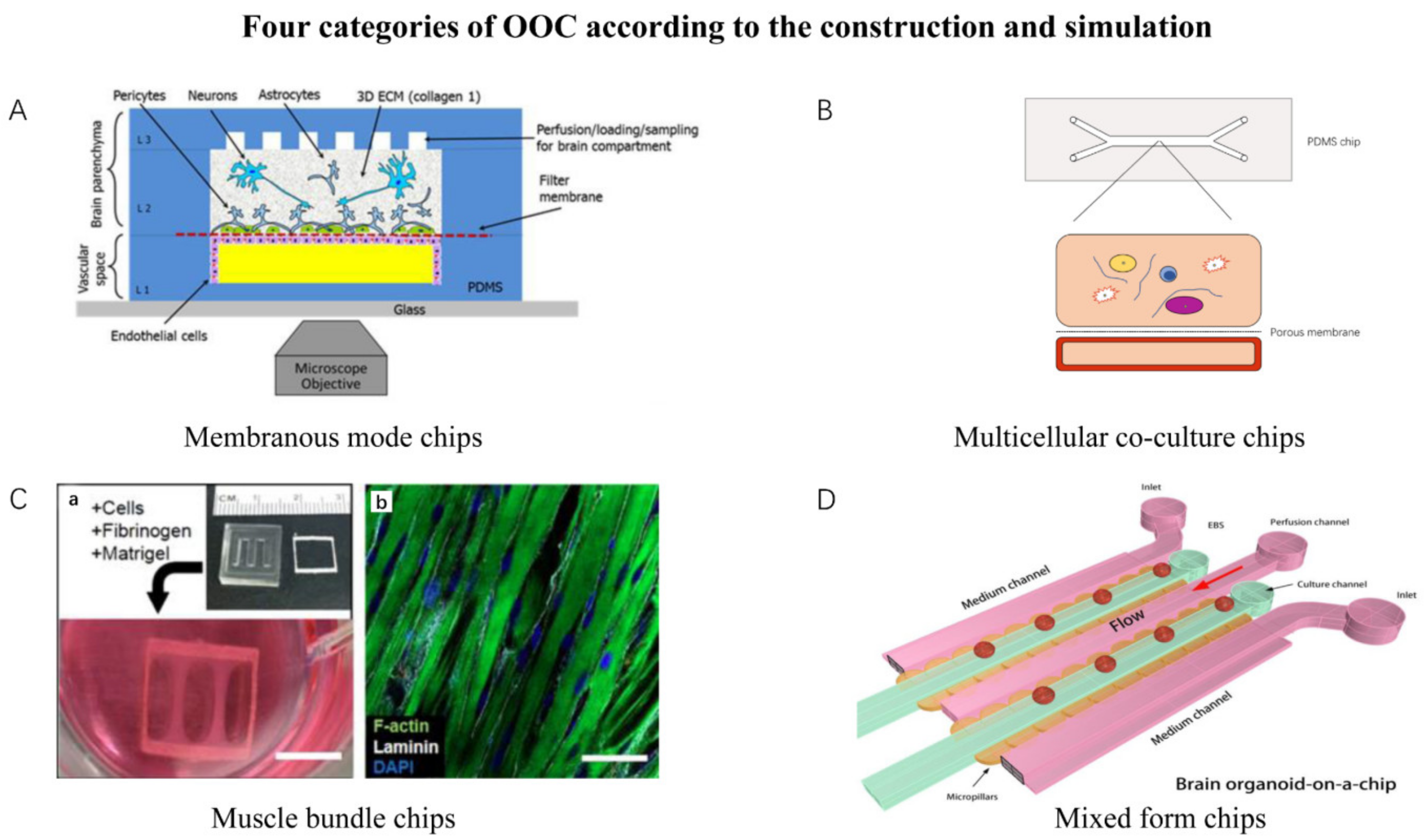
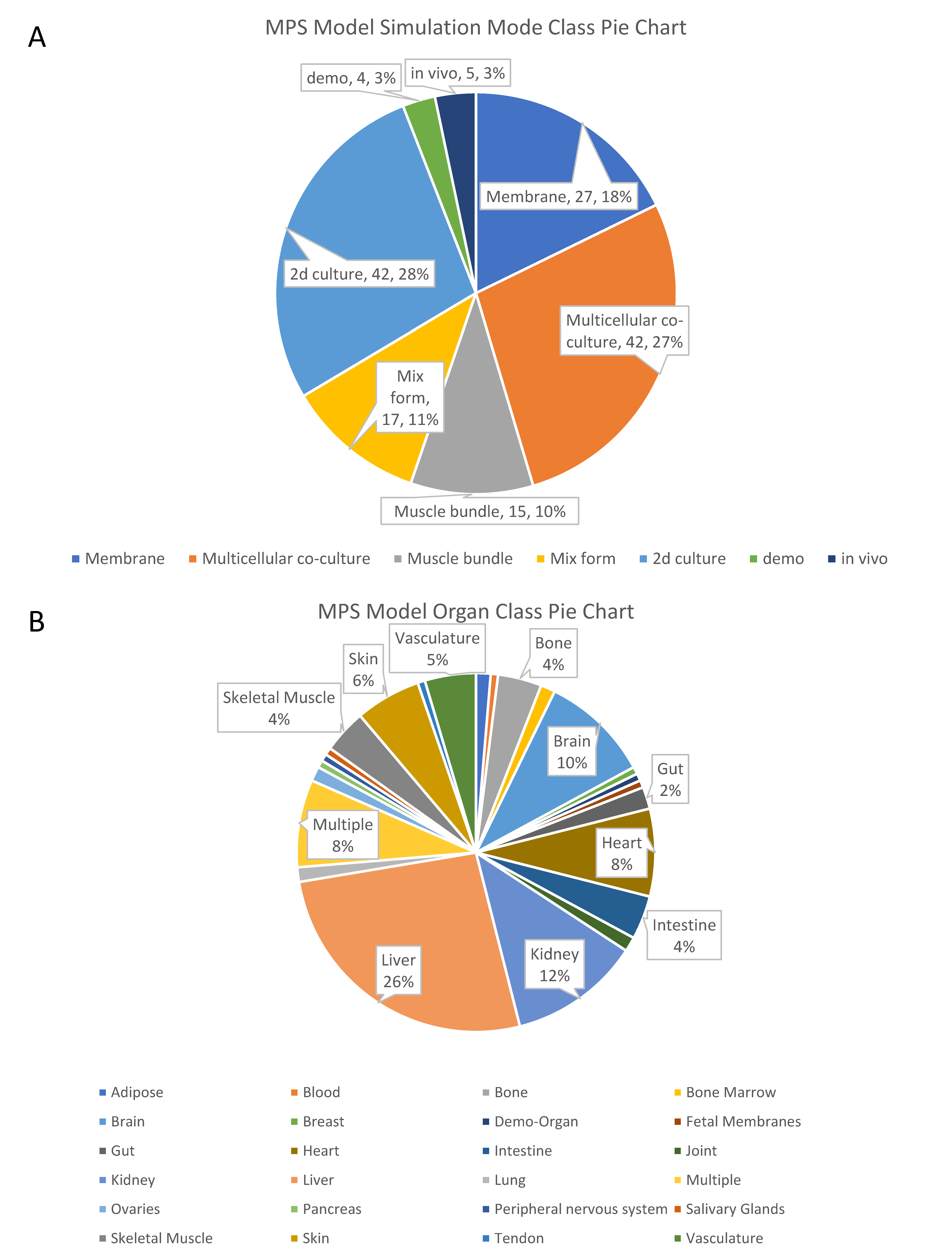
| Simulation Mode | Features | Representative Chips |
|---|---|---|
| Membranous mode chips | The chip has a layer of microporous membrane or semi-permeable membrane to connect two independent chambers. | Neurovascular unit on a chip. White adipose tissue on a chip. |
| Multicellular co-culture chips | The chip has one or multiple chambers for multicellular co-culture while allowing the cells’ spontaneous self-organization and interaction. | Liver Acinus Microphysiology System. Bone marrow on a chip. |
| Muscle bundle chips | Chips that culture muscle bundle. | Skeletal muscle culture system. Bioware II. |
| Mixed-form chips | Chips that involve multiple components together. Majority of organoids-on-chips could be classified into this category. | Brain organoid system on a chip. Tumor on a chip. Cardiac MPS. |
| Simulation Mode | MPS Model | Detection Target |
|---|---|---|
| Membrane Mode | Blood–brain barrier (NVU) | Dextran-FITC (10 kDa), Sigma-Aldrich: FD10S, Lactate Dehydrogenase, GM-CSF, IL-12/IL-23p40, IL-15, IL-16 |
| White Adipose Tissue | Bile Efflux, CYP3A4, Adiponectin, Lactate Dehydrogenase, Lipid Droplets, Lipid to Nuclei Ratio, Nuclei, PrestoBlue | |
| Intestinal Enteroid | FABP2/Human FABP2, Transepithelial Electrical Resistance (TEER), ATP, Dextran-FITC (10 kDa), Fexofenadine, Terfenadine | |
| Kidney Proximal Tubule | KIM-1 (human), Lactate Dehydrogenase (activity), 1α,25-Dihydroxyvitamin D, 24, 25-Dihydroxyvitamin D, 25-Hydroxyvitamin D, Gentamicin, Ammonium, Dead Cells, Human 1500+, PrestoBlue, Flowrate, Cadmium, Cisplatin | |
| Skin MPS | Corrosive, Non-Corrosive, Irritant, Non-Irritant, PrestoBlue, Lactate Dehydrogenase | |
| Multicellular polyculture | LAMPs (SQL-SAL) | Albumin, Bile Efflux, Blood Urea Nitrogen, Lactate Dehydrogenase, COL Ia1, Fexofenadine, ROS, Terfenadine, TNF-Alpha, chenodeoxycholic acid, glycochenodeoxycholic acid, taurocholate, Steatosis, α-SMA, Coumarin, Diclofenac, Phenacetin, Phenolphthalein, Terfenadine, Testosterone, Caffeine, Pioglitazone, Rosiglitazone, Tolcapone, Troglitazone, Trovafloxacin, 4-hydroxydiclofenac, 6beta-Hydroxytestosterone, 7-Hydroxycoumarin glucuronide, Acetaminophen, Pioglitazone, E-Cadherin, anti-E-Cadherin, PrestoBlue, Flowrate |
| Bone | Alkaline Phosphatase, Osteoprotegerin, Sclerostin, Osteocalcin, Osteopontin, Lactate Dehydrogenase, Luciferase Expression, Cisplatin, Dexamethasone, Methotrexate, Doxorubicin, Linsitinib | |
| Muscle bundle | Skeletal Myobundle | Maximum Elongation |
| Mix former | Cardiac MPS | Beat Interval, Beat Rate, Contraction Velocity, Relaxation Velocity |
| Brain | Park7-Recombinant (human), N-ACETYLASPARTIC ACID, Lactate Dehydrogenase | |
| Vascularized Tumor Model | Tumor Area, Tumor Growth, Tumor Integrated Intensity, Tumor Mean Intensity, Vessel Area, Vessel Junctions, Vessel Length, ATP |
| Search Keywords | Number of Studies in the Last 5 Years | Number of Studies in the Last 5–10 Years | Number of Studies from 10 Years Ago | Total |
|---|---|---|---|---|
| OOC | 782 | 112 | 11 | 905 |
| PBPK | 3711 | 2559 | 4102 | 10,372 |
| OOC AND PBPK | 15 | 2 | 0 | 17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Li, Q.; Liang, W.; Du, X.; Yang, Y.; Zhang, Z.; Xu, L.; Zhang, J.; Li, J.; Chen, Z.; et al. Organ-On-A-Chip Database Revealed—Achieving the Human Avatar in Silicon. Bioengineering 2022, 9, 685. https://doi.org/10.3390/bioengineering9110685
Jiang L, Li Q, Liang W, Du X, Yang Y, Zhang Z, Xu L, Zhang J, Li J, Chen Z, et al. Organ-On-A-Chip Database Revealed—Achieving the Human Avatar in Silicon. Bioengineering. 2022; 9(11):685. https://doi.org/10.3390/bioengineering9110685
Chicago/Turabian StyleJiang, Lincao, Qiwei Li, Weicheng Liang, Xuan Du, Yi Yang, Zilin Zhang, Lili Xu, Jing Zhang, Jian Li, Zaozao Chen, and et al. 2022. "Organ-On-A-Chip Database Revealed—Achieving the Human Avatar in Silicon" Bioengineering 9, no. 11: 685. https://doi.org/10.3390/bioengineering9110685
APA StyleJiang, L., Li, Q., Liang, W., Du, X., Yang, Y., Zhang, Z., Xu, L., Zhang, J., Li, J., Chen, Z., & Gu, Z. (2022). Organ-On-A-Chip Database Revealed—Achieving the Human Avatar in Silicon. Bioengineering, 9(11), 685. https://doi.org/10.3390/bioengineering9110685