Evaluation of AMG510 Therapy on KRAS-Mutant Non–Small Cell Lung Cancer and Colorectal Cancer Cell Using a 3D Invasive Tumor Spheroid System under Normoxia and Hypoxia
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Generation of Multicellular Spheroids
2.3. Drug Treatment
2.4. Imaging
2.5. Spheroid Viability Testing
2.6. Tumor Growth Inhibition Calculation
2.7. Statistical Analysis
3. Results
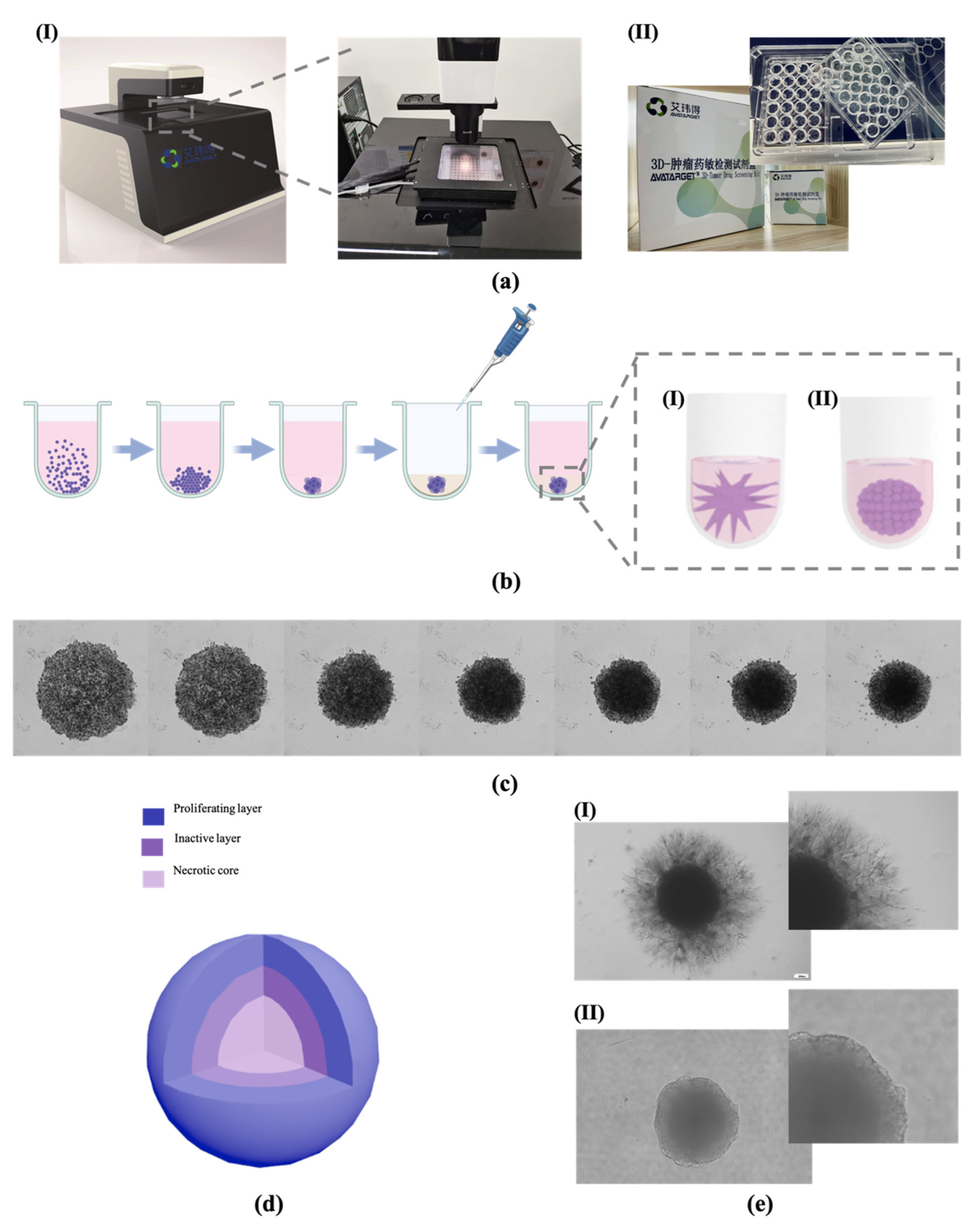
3.1. Tumor Spheroid Construction
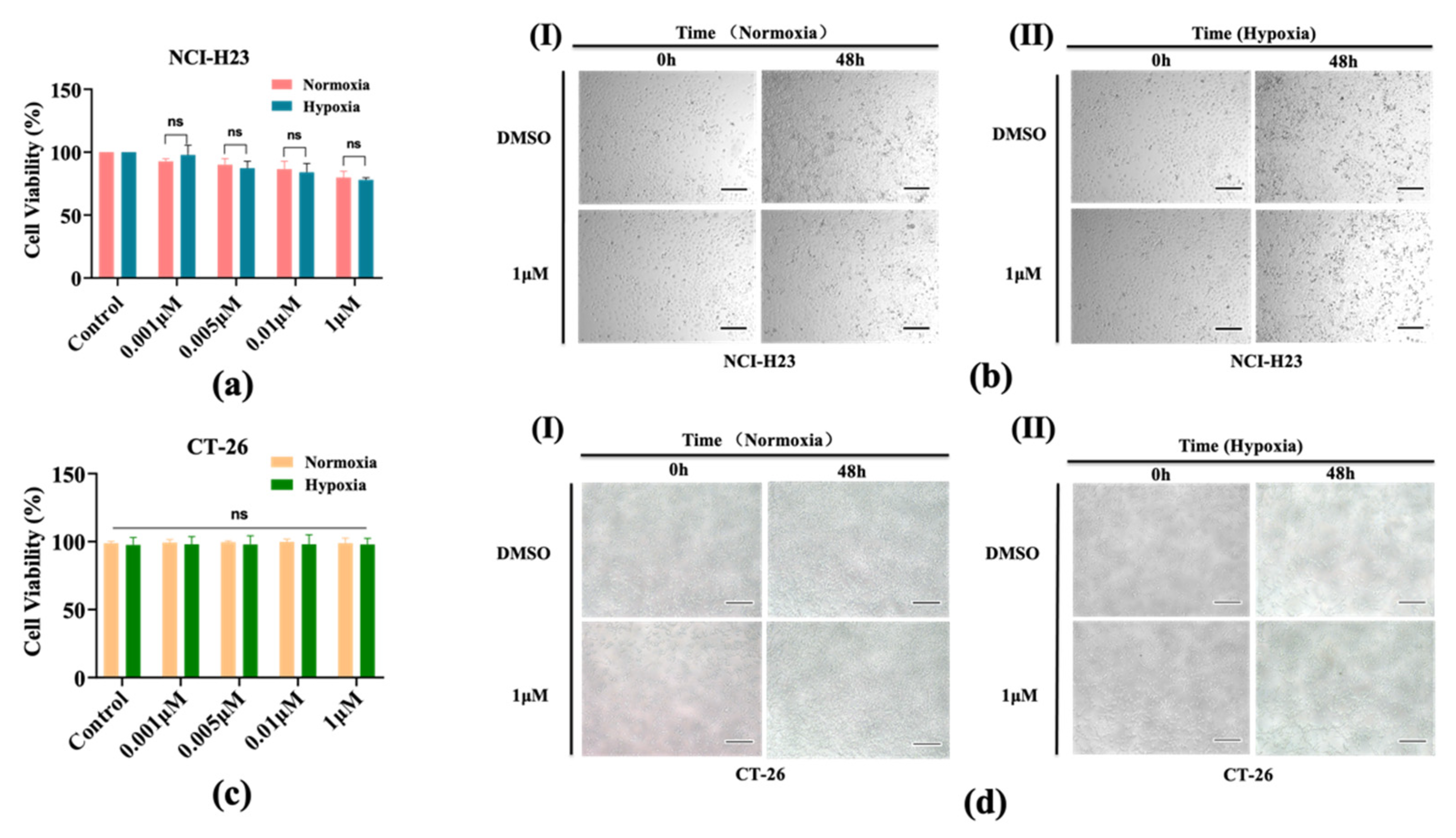
3.2. AMG510 Treatment to 2D Cultured NCI-H23 Cells
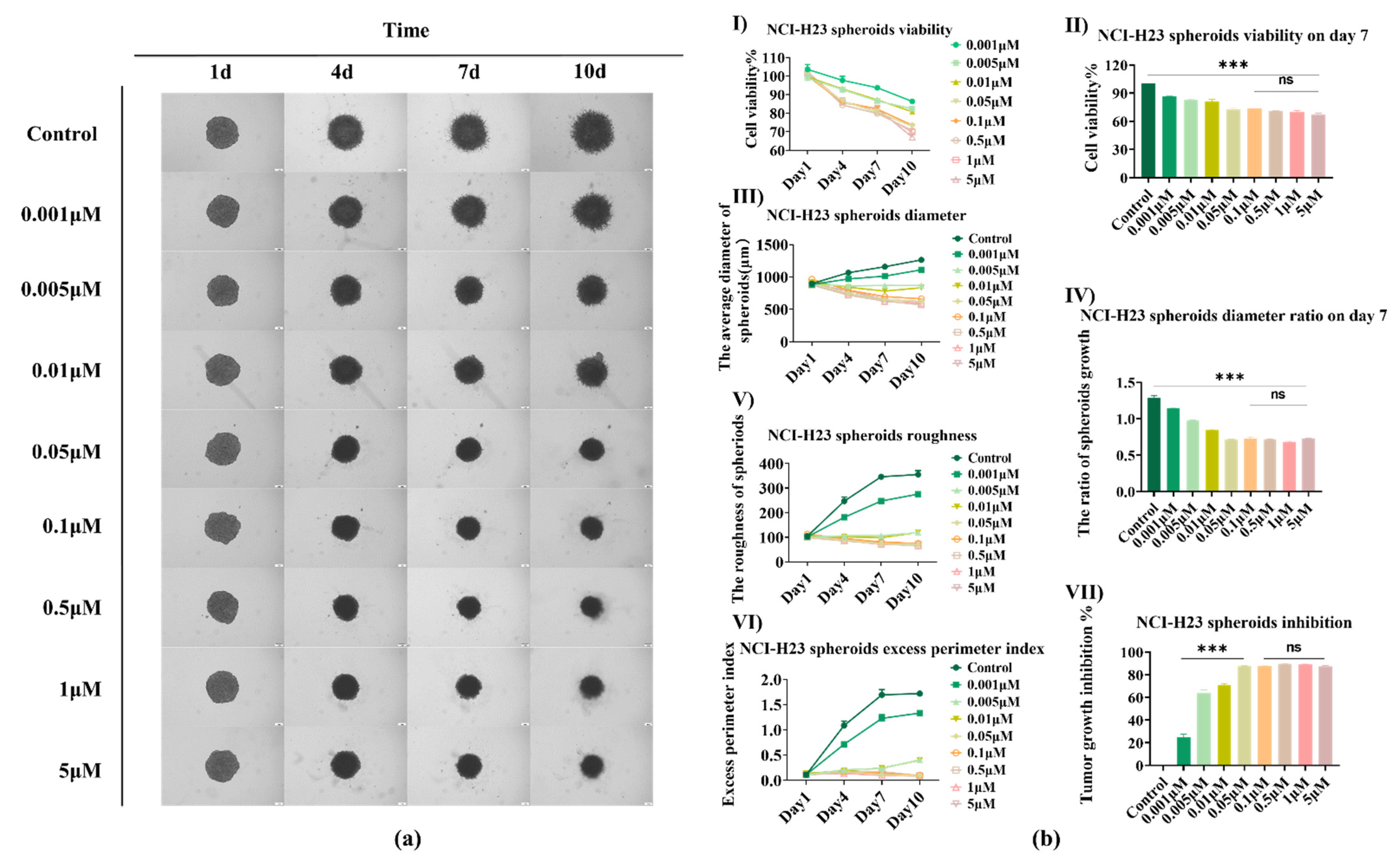
3.3. AMG510 Treatment to 3D NCI-H23 Spheroids
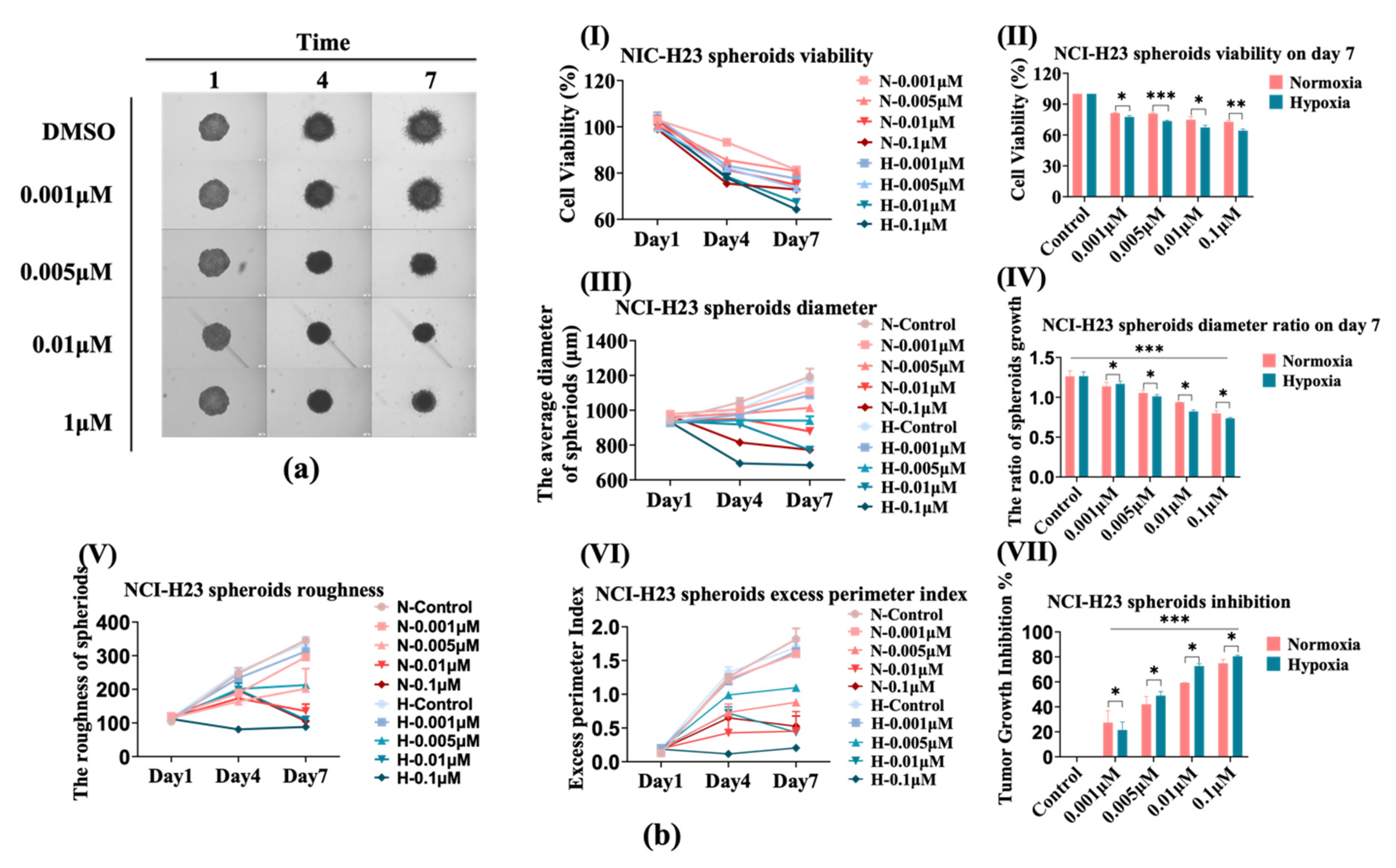
3.4. The Sensitivity of NCI-H23 Spheroids to AMG510 Increases under Hypoxia
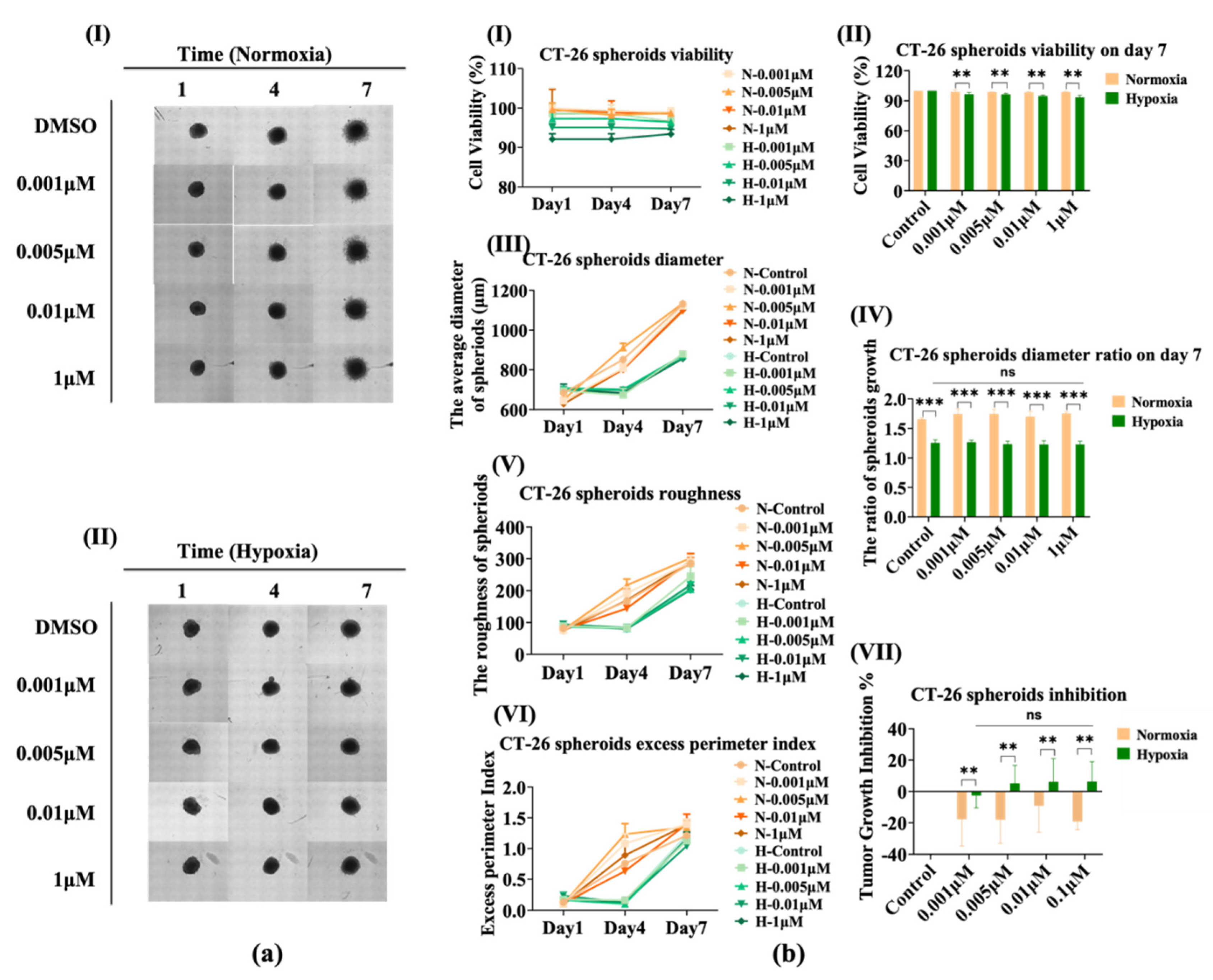
3.5. The Sensitivity of CT-26 Spheroids Does Not Change Significantly under Hypoxia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| KRAS | Kirsten ratsarcoma viral oncogene homolog |
| ECM | Extracellular matrix |
| TME | Tumor microenvironment |
| TSE | 3D tumor-spheroid-ECM |
| NSCLC | Non–small cell lung cancer |
| CRC | Colorectal cancer |
| EPI | Excess perimeter index |
| TGI | Tumor growth inhibition |
| RTV | Relative spheroids volumes |
| HIF | Hypoxia inducible factor |
References
- Costa, E.C.; Moreira, A.F.; De Melo-Diogo, D.M.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Selby, M.; Delosh, R.; Laudeman, J.; Ogle, C.; Reinhart, R.; Silvers, T.; Lawrence, S.; Kinders, R.; Parchment, R.; Teicher, B.A.; et al. 3D models of the nci60 cell lines for screening oncology compounds. SLAS Discov. 2017, 22, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Weiswald, L.B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Todd, W.; Ridky Chow, J.M.; Wong, D.J.; Khavari, P.A. Invasive 3-dimensional organotypic neoplasia from multiple normal human epithelia. Nat. Med. 2010, 16, 1450–1455. [Google Scholar]
- Johnson, J.I.; Decker, S.; Zaharevitz, D.; Rubinstein, L.V.; Venditti, J.M.; Schepartz, S.; Christian, M.; Arbuck, S.; Hollingshead, M.; Sausville, E.A. Relationships between drug activity in nci preclinical in vitro and in vivo models and early clinical trials. Br. J. Cancer 2001, 84, 1424–1431. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Friedrich, J.; Ebner, R.; Kunz-Schughart, L.A. Experimental anti-tumor therapy in 3-d: Spheroidsold hat or new challenge? Int. J. Radiat. Biol. 2007, 83, 849–871. [Google Scholar] [CrossRef]
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-based drug screen considerations and practical approach. Nat. Protoc. 2009, 4, 309–324. [Google Scholar] [CrossRef]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of threedimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K. Role of tumor microenvironment in tumorigenesi. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Pengfei, L.; Valerie, M.; Weaver, W.Z. The extracellular matrix: A dynamic nichein cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J.; Schwartz, M.A.; Burridge, K.; Firtel, R.A.; Ginsberg, M.H.; Borisy, G.; Parsons, J.T.; Horwitz, A.R. Cell migrationintegrating: Signals from front to back. Science 2003, 302, 1704–1709. [Google Scholar] [CrossRef]
- Adachi, Y.; Ito, K.; Hayashi, Y.; Kimura, R.; Tan, T.Z.; Yamaguchi, R.; Ebi, H. Epithelial-to-mesenchymal transition is a cause of bothintrinsic and acquired resistance to kras g12c inhibitorin kras g12c–mutant non–small cell lung cancer. Clin. Cancer Res. 2020, 26, 5962–5973. [Google Scholar] [CrossRef] [PubMed]
- Amodio, V.; Yaeger, R.; Arcella, P.; Cancelliere, C.; Lamba, S.; Lorenzato, A.; Arena, S.; Montone, M.; Mussolin, B.; Bian, Y.; et al. Egfr blockade reverts resistance to krasg12c inhibition in colorectal cancer. Cancer Discovery 2020, 10, 1129–1139. [Google Scholar] [CrossRef]
- Castle, J.C.; Loewer, M.; Boegel, S.; de Graaf, J.; Bender, C.; Tadmor, A.D.; Boisguerin, V.; Bukur, T.; Sorn, P.; Paret, P.; et al. Immunomic, genomic and transcriptomic characterization of ct26 colorectal carcinoma. BMC Genom. 2014, 15, 190. [Google Scholar] [CrossRef]
- Mazumdar, J.; Hickey, M.M.; Pant, D.K.; Durham, A.C.; Sweet-Cordero, A.; Vachani, A.; Jacks, T.; Chodosh, L.A.; Kissil, J.L.; Simon, M.C.; et al. Hif-2 alpha deletion promotes kras-driven lung tumor development. Proc. Natl. Acad. Sci. USA 2010, 107, 14182–14187. [Google Scholar] [CrossRef]
- Kang, R.; Hou, W.; Zhang, Q.; Chen, R.; Lee, Y.J.; Bartlett, D.L.; Lotze, M.T.; Tang, D.; Zeh, H.J. RAGE is essential for oncogenic KRAS-mediated hypoxic signaling in pancreatic cancer. Cell Death Dis. 2014, 5, e1480. [Google Scholar] [CrossRef]
- Saliakoura, M.; Sebastiano, M.R.; Pozzato, C.; Heidel, F.H.; Schnöder, T.M.; Prince, S.P.; Bubendorf, L.; Pinton, L.; Schmid, R.; Baumgartner, J.; et al. Plcγ1 suppression promotes the adaptation of kras-mutant lung adenocarcinomas to hypoxia. Nat. Cell Biol. 2020, 22, 1382–1395. [Google Scholar] [CrossRef]
- Chen, Z.; Ma, N.; Sun, X.; Li, Q.; Zeng, Y.; Chen, F.; Sun, S.; Xu, J.; Zhang, J.; Ye, H.; et al. Automated evaluation of tumor spheroid behavior in 3d culture using deep learning-based recognition. Biomaterials 2021, 272, 120770. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.M.; Dumas, J.; Adnane, L.; Lynch, M.; Carter, C.A.; Schütz, G.; Thierauch, K.-H.; Zopf, D. Regorafenib (bay 73-4506)a new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int. J. Cancer 2011, 129, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Nazari, S.S. Generation of 3d tumor spheroids with encapsulating basement membranes for invasion studies. Curr. Protoc. Cell Biol. 2020, 87, e105. [Google Scholar] [CrossRef] [PubMed]
- Tuomainen, K.A.-S.A.; Potdar, S.; Turunen, L.; Turunen, M.; Karhemo, P.-R.; Bergman, P.; Risteli, M.; Åström, P.; Tiikkaja, R.; Grenman, R.; et al. Human tumor–derived matrix improves the predictability of head and neck cancer drug testing. Cancers 2020, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Guan, X.; Zhang, X.; Luan, X.; Song, Z.; Cheng, X.; Qin, J.J. Targeting kras mutant cancers: From druggable therapy to drug resistance. Mol. Cancer 2022, 21, 159. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. Kras mutation: From undruggable to druggable in cancer. Signal Transduct. Target Ther. 2021, 6, 386. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Li, M.; Fang, M. The research progress of direct kras g12c mutation inhibitors. Pathol. Oncol. Res. 2021, 27, 67. [Google Scholar] [CrossRef] [PubMed]
- Lanman, B.A.; Allen, J.R.; Allen, J.G.; Amegadzie, A.K.; Ashton, K.S.; Booker, S.K.; Chen, J.J.; Chen, N.; Frohn, M.J.; Goodman, G.; et al. Discovery of a covalent inhibitor of krasg12c (amg 510) for the treatment of solid tumors. J. Med. Chem. 2020, 63, 52–65. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Su, P.; Miao, Z.; Hu, L.; Li, R.; Yin, C.; Li, D.; Chen, Z.; Zhao, F.; Qian, A. Methods of studying mammalian cell migration and invasion in vitro. In Proceedings of the 2017 14th International Bhurban Conference on Applied Sciences and Technology (IBCAST), Islamabad, Pakistan, 10–14 January 2017. [Google Scholar]
- Martín-Bernabé, A.; Tarragó-Celada, J.; Cunin, V.; Michelland, S.; Cortés, R.; Poignant, J.; Boyault, C.; Rachidi, W.; Bourgoin-Voillard, S.; Cascante, M.; et al. Quantitative proteomic approach reveals altered metabolic pathways in response to the inhibition of lysine deacetylases in a549 cells under normoxia and hypoxia. Int. J. Mol. Sci. 2021, 22, 3378. [Google Scholar] [CrossRef]
- Overcoming a Cancer Nemesis? Kras Inhibitor Shows Promise in Early Trial. Available online: https://www.cancer.gov/news-events/cancer-currents-blog/2019/kras-inhibitor-amg-510-clinical-trial (accessed on 5 December 2019).
- Minakata, K.; Takahashi, F.; Nara, T.; Hashimoto, M.; Tajima, K.; Murakami, A.; Nurwidya, F.; Yae, S.K.F.; Moriyama, H. Hypoxia induces gefitinib resistance in non-small-cell lung cancer with both mutant and wild-type epidermal growth factor receptors. Cancer Sci. 2012, 103, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Liu, X.; Chi, W.; Liu, Y.; Wei, L.; Wang, X.; Yu, J. Hypoxia-induced resistance to cisplatin and doxorubicin in non-small cell lung cancer is inhibited by silencing of hif-1α gene. Cancer Chemother. Pharm. 2006, 58, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wang, Y.; Lei, F.; Rong, W.; Zeng, Q. Positive feedback between oncogenic kras and hif-1α confers drug resistance in colorectal cancer. OncoTargets Ther. 2015, 8, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, J.; Yang, Z.; Zhao, H.; Deng, B.; Ren, Y.; Mai, R.; Huang, J.; Chen, J. Discovery of thieno[2,3-d]pyrimidine-based kras g12d inhibitors as potential anticancer agents via combinatorial virtual screening. Eur. J. Med. Chem. 2022, 233, 114243. [Google Scholar] [CrossRef]
- Fan, G.; Lou, L.; Song, Z.; Zhang, X.; Xiong, X.F. Targeting mutated gtpase kras in tumor therapies. Eur. J. Med. Chem. 2021, 226, 113816. [Google Scholar] [CrossRef]
- Ji, J.; Wang, C.; Fakih, M. Targeting krasg12c-mutated advanced colorectal cancer: Research and clinical developments. OncoTargets Ther. 2022, 15, 747–756. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.; Hou, W.; Zhang, J.; Li, M.; Zhang, Z.; Li, X.; Chen, Z.; Wang, C.; Yang, L. Evaluation of AMG510 Therapy on KRAS-Mutant Non–Small Cell Lung Cancer and Colorectal Cancer Cell Using a 3D Invasive Tumor Spheroid System under Normoxia and Hypoxia. Bioengineering 2022, 9, 792. https://doi.org/10.3390/bioengineering9120792
Huang M, Hou W, Zhang J, Li M, Zhang Z, Li X, Chen Z, Wang C, Yang L. Evaluation of AMG510 Therapy on KRAS-Mutant Non–Small Cell Lung Cancer and Colorectal Cancer Cell Using a 3D Invasive Tumor Spheroid System under Normoxia and Hypoxia. Bioengineering. 2022; 9(12):792. https://doi.org/10.3390/bioengineering9120792
Chicago/Turabian StyleHuang, Meng, Wei Hou, Jing Zhang, Menglan Li, Zilin Zhang, Xiaoran Li, Zaozao Chen, Cailian Wang, and Lihua Yang. 2022. "Evaluation of AMG510 Therapy on KRAS-Mutant Non–Small Cell Lung Cancer and Colorectal Cancer Cell Using a 3D Invasive Tumor Spheroid System under Normoxia and Hypoxia" Bioengineering 9, no. 12: 792. https://doi.org/10.3390/bioengineering9120792
APA StyleHuang, M., Hou, W., Zhang, J., Li, M., Zhang, Z., Li, X., Chen, Z., Wang, C., & Yang, L. (2022). Evaluation of AMG510 Therapy on KRAS-Mutant Non–Small Cell Lung Cancer and Colorectal Cancer Cell Using a 3D Invasive Tumor Spheroid System under Normoxia and Hypoxia. Bioengineering, 9(12), 792. https://doi.org/10.3390/bioengineering9120792





