Automatic Segmentation of Monofilament Testing Sites in Plantar Images for Diabetic Foot Management
Abstract
:1. Introduction
1.1. Diabetes and the Risk of Lower-Extremity Amputation
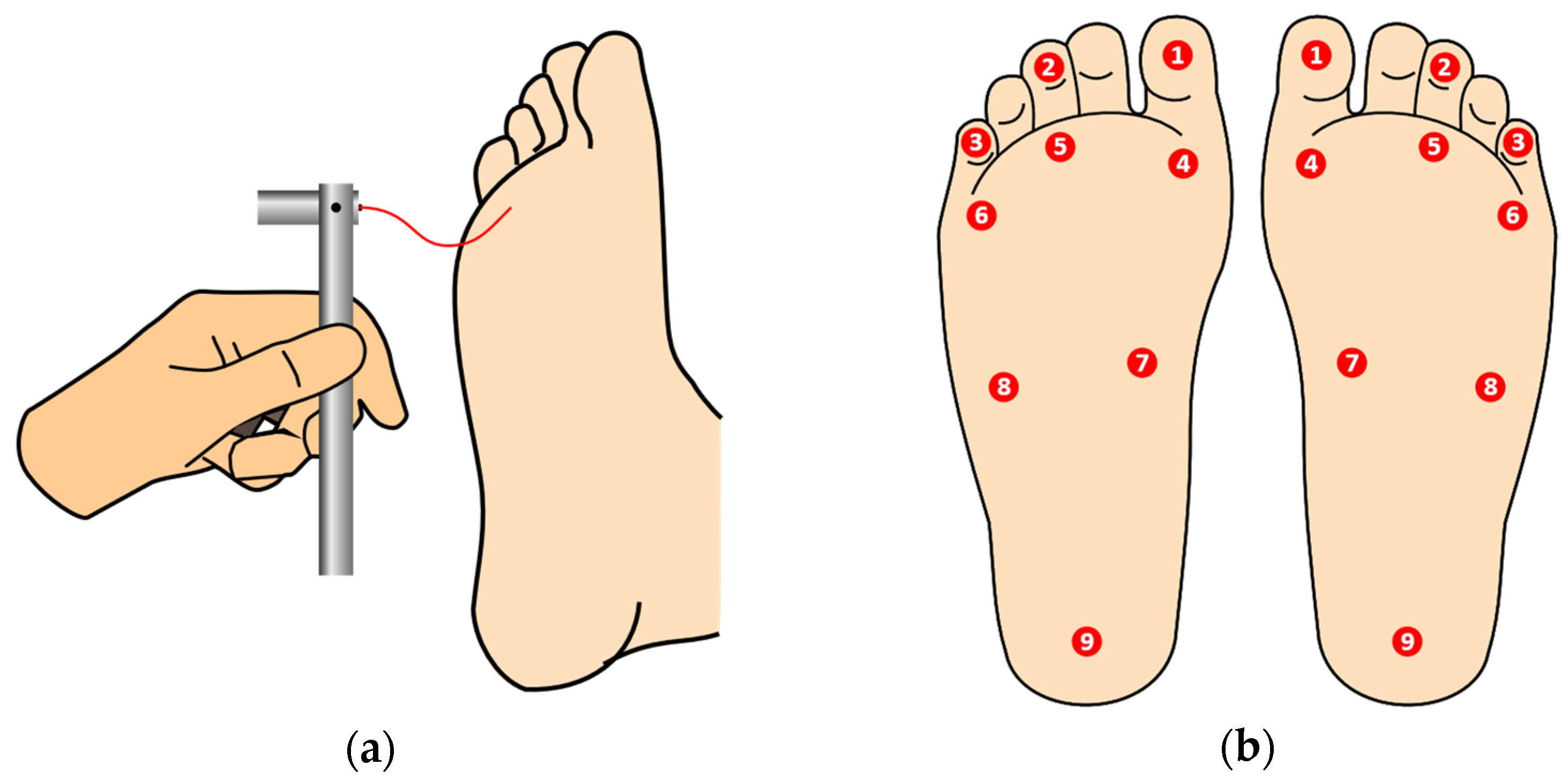
1.2. The Semmes–Weinstein Monofilament Examination
1.3. Automated and Semi/Automated Evaluation of DFN
1.4. Manuscript Organization
2. Photographic Plantar Image Segmentation for SWME
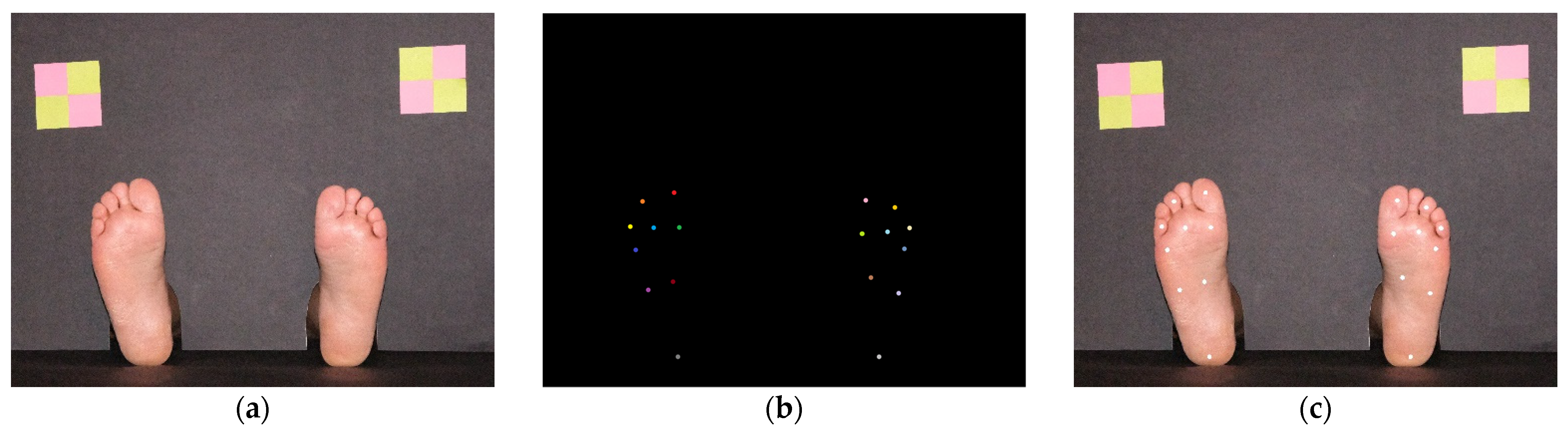
2.1. Plantar Photographic Images Database
2.2. System Overview
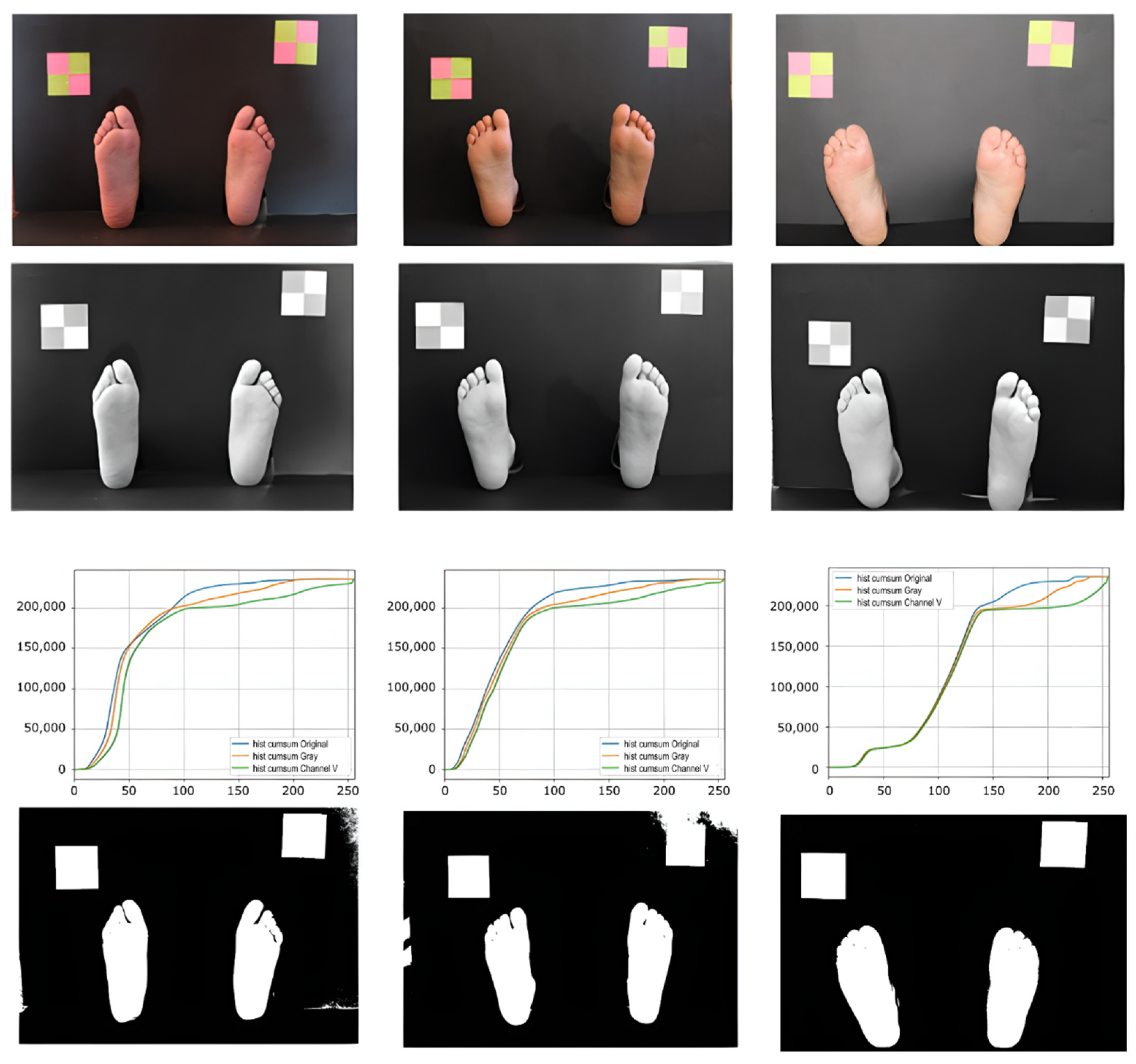
2.3. Background Segmentation
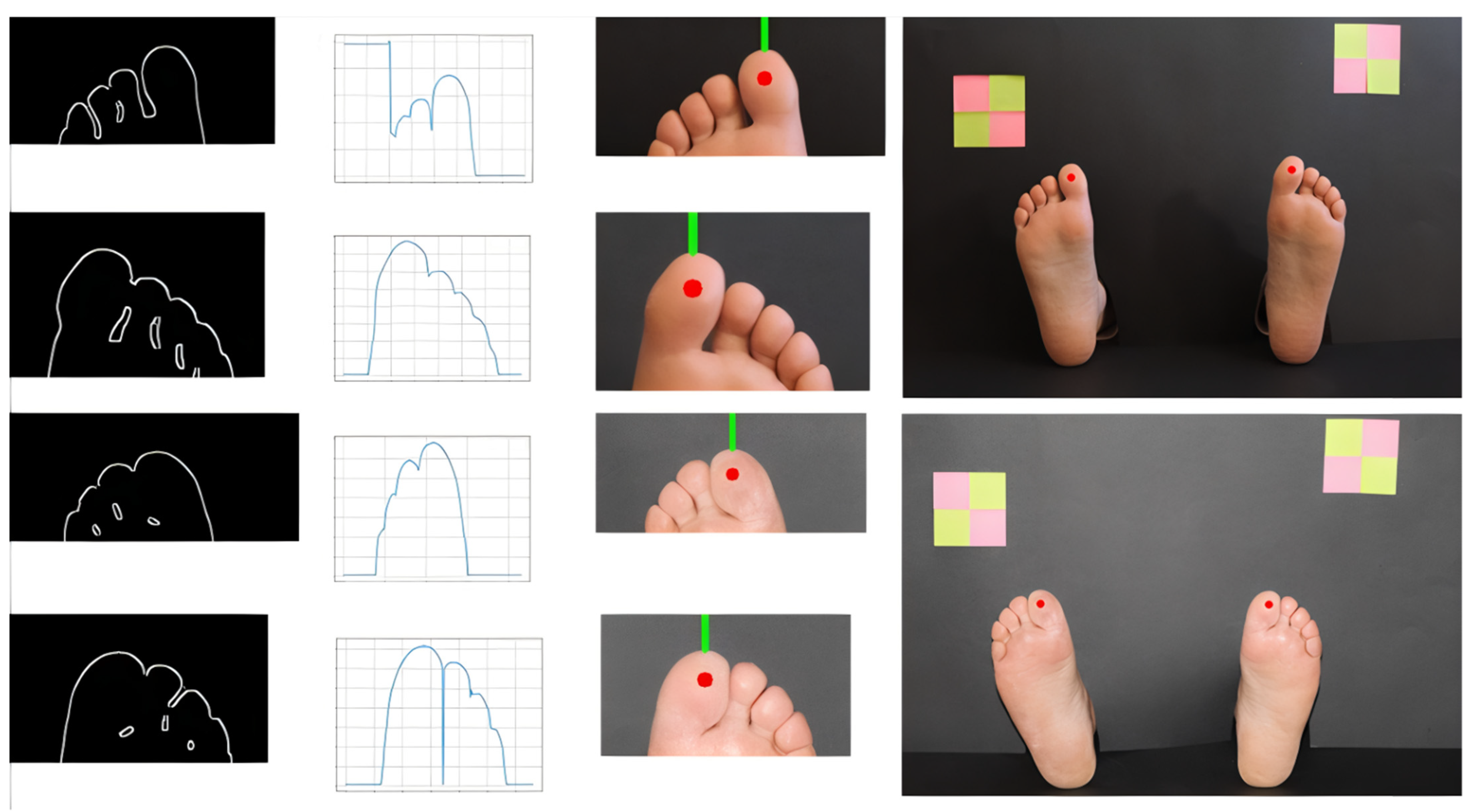
2.4. Hallux Segmentation
2.5. Segmentation of Third and Fifth Toes Sites
2.6. Segmentation of Heel, Central, and Metatarsophalangeal Sites
3. Results
3.1. Background Segmentation
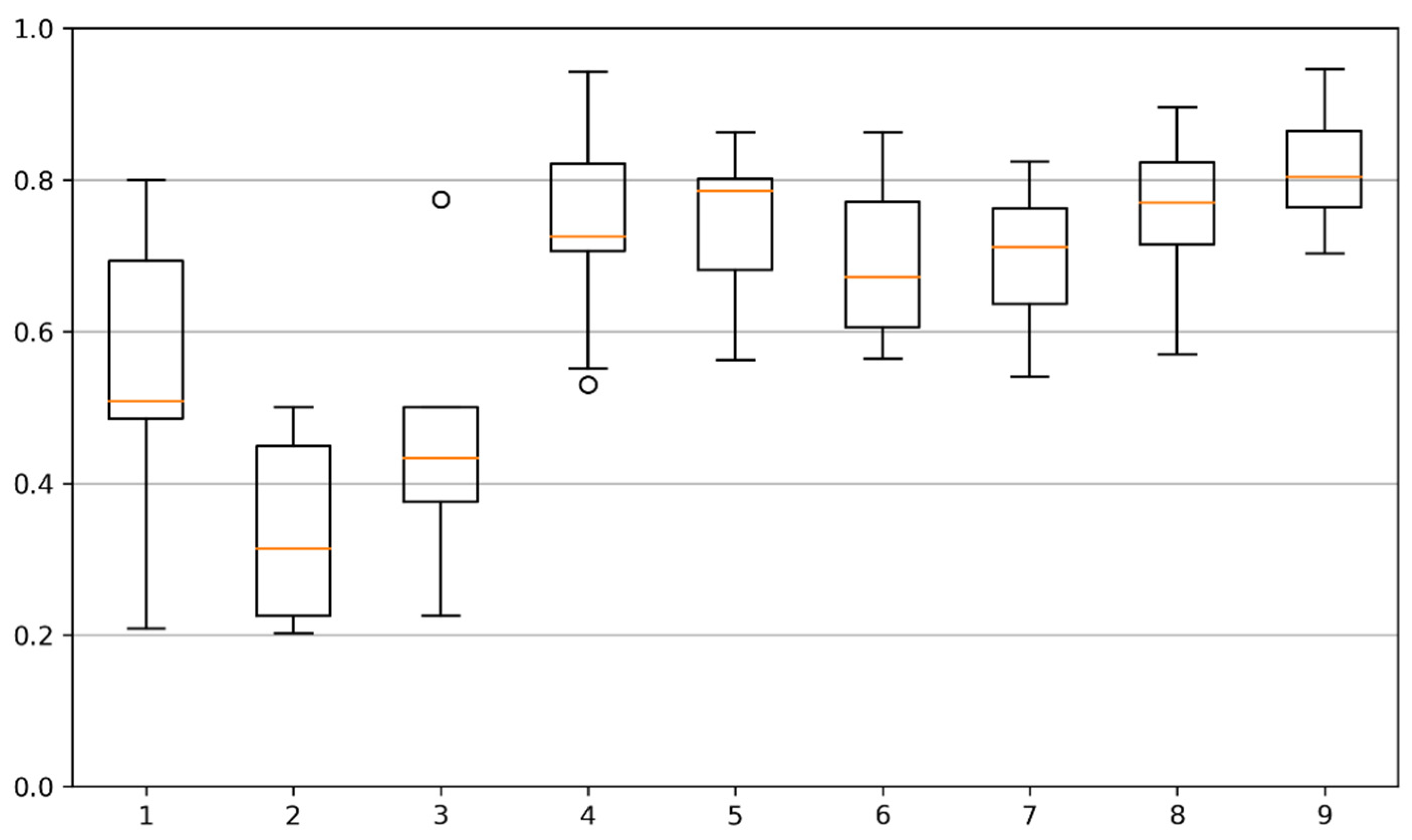
3.2. Testing Sites Segmentation
3.3. Time Analysis
4. Discussion
5. Conclusions
6. Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization Diabetes Fact Sheet. Available online: https://www.who.int/news-room/fact-sheets/detail/diabetes (accessed on 22 December 2021).
- Walicka, M.; Raczyńska, M.; Marcinkowska, K.; Lisicka, I.; Czaicki, A.; Wierzba, W.; Franek, E. Amputations of Lower Limb in Subjects with Diabetes Mellitus: Reasons and 30-Day Mortality. J. Diabetes Res. 2021, 2021, e8866126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lazzarini, P.A.; McPhail, S.M.; van Netten, J.J.; Armstrong, D.G.; Pacella, R.E. Global Disability Burdens of Diabetes-Related Lower-Extremity Complications in 1990 and 2016. Diabetes Care 2020, 43, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Boulton, A.J.M.; Armstrong, D.G.; Albert, S.F.; Frykberg, R.G.; Hellman, R.; Kirkman, M.S.; Lavery, L.A.; LeMaster, J.W.; Mills, J.L., Sr.; Mueller, M.J.; et al. Comprehensive Foot Examination and Risk Assessment: A Report of the Task Force of the Foot Care Interest Group of the American Diabetes Association, with Endorsement by the American Association of Clinical Endocrinologists. Diabetes Care 2008, 31, 1679–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aring, A.M.; Jones, D.E.; Falko, J.M. Evaluation and Prevention of Diabetic Neuropathy. Am. Fam. Physician 2005, 71, 2123–2128. [Google Scholar] [PubMed]
- Martins, P.; Coelho, L. Evaluation of the Semmes-Weinstein Monofilament (SWM) on the Diabetic Foot Assessment. In Advances and Current Trends in Biomechanics; CRC Press: Boca Raton, FL, USA, 2021; ISBN 978-1-00-321715-2. [Google Scholar]
- Monofilament Testing for Loss of Protective Sensation of Diabetic/Neuropathic Feet for Adults & Children; British Columbia Provincial Nursing Skin and Wound Committee: Victoria, BC, Canada, 2012.
- Wilasrusmee, C.; Suthakorn, J.; Guerineau, C.; Itsarachaiyot, Y.; Sa-Ing, V.; Proprom, N.; Lertsithichai, P.; Jirasisrithum, S.; Kittur, D. A Novel Robotic Monofilament Test for Diabetic Neuropathy. Asian J. Surg. 2010, 33, 193–198. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, H.-U.-R.; Alty, S.R.; Spruce, M.; Dudley, S.E. Automated Peripheral Neuropathy Assessment of Diabetic Patients Using Optical Imaging and Binary Processing Techniques. In Proceedings of the 2013 IEEE Point-of-Care Healthcare Technologies (PHT), Bangalore, India, 16–18 January 2013; pp. 200–203. [Google Scholar]
- Sun, P.-C.; Lin, H.-D.; Jao, S.-H.E.; Ku, Y.-C.; Chan, R.-C.; Cheng, C.-K. Relationship of Skin Temperature to Sympathetic Dysfunction in Diabetic At-Risk Feet. Diabetes Res. Clin. Pract. 2006, 73, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; van Netten, J.J.; Van Baal, J.G.; Bus, S.A.; van Der Heijden, F. Automatic Detection of Diabetic Foot Complications with Infrared Thermography by Asymmetric Analysis. J. Biomed. Opt. 2015, 20, 026003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majeed, A.; Chong, A.K. Forensic Perpetrator Identification Utilising Foot Pressure under Shoes during Gait. In Proceedings of the 2021 IEEE 11th IEEE Symposium on Computer Applications Industrial Electronics (ISCAIE), Penang, Malaysia, 3–4 April 2021; pp. 123–126. [Google Scholar]
- Wang, X.; Wang, H.; Cheng, Q.; Nankabirwa, N.L.; Zhang, T. Single 2D Pressure Footprint Based Person Identification. In Proceedings of the 2017 IEEE International Joint Conference on Biometrics (IJCB), Denver, CO, USA, 1–4 October 2017; pp. 413–419. [Google Scholar]
- Kulkarni, P.S.; Kulkarni, V.B. Human Footprint Classification Using Image Parameters. In Proceedings of the 2015 International Conference on Pervasive Computing (ICPC), Pune, India, 8–10 January 2015; pp. 1–5. [Google Scholar]
- Guerrero-Turrubiates, J.D.J.; Cruz-Aceves, I.; Ledesma, S.; Sierra-Hernandez, J.M.; Velasco, J.; Avina-Cervantes, J.G.; Avila-Garcia, M.S.; Rostro-Gonzalez, H.; Rojas-Laguna, R. Fast Parabola Detection Using Estimation of Distribution Algorithms. Comput. Math. Methods Med. 2017, 2017, 6494390. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, Y.; Xing, L.; Li, W. An Adaptive Foot-Image Segmentation Algorithm Based on Morphological Partition. In Proceedings of the 2018 IEEE International Conference on Progress in Informatics and Computing (PIC), Suzhou, China, 14–16 December 2018; pp. 231–235. [Google Scholar]
- Maestre-Rendon, J.R.; Rivera-Roman, T.A.; Sierra-Hernandez, J.M.; Cruz-Aceves, I.; Contreras-Medina, L.M.; Duarte-Galvan, C.; Fernandez-Jaramillo, A.A. Low Computational-Cost Footprint Deformities Diagnosis Sensor through Angles, Dimensions Analysis and Image Processing Techniques. Sensors 2017, 17, 2700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bougrine, A.; Harba, R.; Canals, R.; Ledee, R.; Jabloun, M. On the Segmentation of Plantar Foot Thermal Images with Deep Learning. In Proceedings of the 2019 27th European Signal Processing Conference (EUSIPCO), A Coruna, Spain, 2–6 September 2019; pp. 1–5. [Google Scholar]
- Kwolek, K.; Liszka, H.; Kwolek, B.; Gądek, A. Measuring the Angle of Hallux Valgus Using Segmentation of Bones on X-Ray Images. In Proceedings of the Artificial Neural Networks and Machine Learning—ICANN 2019: Workshop and Special Sessions, Munich, Germany, 17–19 September 2019; Tetko, I.V., Kůrková, V., Karpov, P., Theis, F., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 313–325. [Google Scholar]
- Dutta, A.; Dutta, A. SoleSCAN—Mobile Phone Based Monitoring of Foot Sole for Callus Formation and the Shoe Insole for Pressure “Hot Spots”. In Proceedings of the 2013 IEEE Point-of-Care Healthcare Technologies (PHT), Bangalore, India, 16–18 January 2013; pp. 339–342. [Google Scholar]
- Garrido-Jurado, S.; Muñoz-Salinas, R.; Madrid-Cuevas, F.J.; Marín-Jiménez, M.J. Automatic Generation and Detection of Highly Reliable Fiducial Markers under Occlusion. Pattern Recognit. 2014, 47, 2280–2292. [Google Scholar] [CrossRef]
- Olson, E. AprilTag: A Robust and Flexible Visual Fiducial System. In Proceedings of the 2011 IEEE International Conference on Robotics and Automation, Shanghai, China, 9–13 May 2011; pp. 3400–3407. [Google Scholar]
- Shabalina, K.; Sagitov, A.; Sabirova, L.; Li, H.; Magid, E. ARTag, AprilTag and CALTag Fiducial Systems Comparison in a Presence of Partial Rotation: Manual and Automated Approaches. In Proceedings of the Informatics in Control, Automation and Robotics, Prague, Czech Republic, 29–31 July 2019; Gusikhin, O., Madani, K., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 536–558. [Google Scholar]
- Shaik, K.B.; Ganesan, P.; Kalist, V.; Sathish, B.S.; Jenitha, J.M.M. Comparative Study of Skin Color Detection and Segmentation in HSV and YCbCr Color Space. Procedia Comput. Sci. 2015, 57, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Rahmat, R.F.; Chairunnisa, T.; Gunawan, D.; Sitompul, O.S. Skin Color Segmentation Using Multi-Color Space Threshold. In Proceedings of the 2016 3rd International Conference on Computer and Information Sciences (ICCOINS), Kuala Lumpur, Malaysia, 15–17 August 2016; pp. 391–396. [Google Scholar]
- Jurca, A.; Žabkar, J.; Džeroski, S. Analysis of 1.2 Million Foot Scans from North America, Europe and Asia. Sci. Rep. 2019, 9, 19155. [Google Scholar] [CrossRef] [PubMed]
- Chabrier, S.; Laurent, H.; Rosenberger, C.; Emile, B. Comparative Study of Contour Detection Evaluation Criteria Based on Dissimilarity Measures. EURASIP J. Image Video Process. 2008, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Hu, Z.; Zhang, Y. An Image Comparison Algorithm Based on Contour Similarity. In Proceedings of the 2020 International Conference on Computer Network, Electronic and Automation (ICCNEA), Xi’an China, 25–27 September 2020; pp. 111–116. [Google Scholar]
- Choi, S.; Cha, S. A Survey of Binary Similarity and Distance Measures. J. Syst. Cybern. Inform. 2010, 8, 43–48. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerlandm, 2015; pp. 234–241. [Google Scholar]
- Fysikopoulos, E.; Rouchota, M.; Eleftheriadis, V.; Gatsiou, C.-A.; Pilatis, I.; Sarpaki, S.; Loudos, G.; Kostopoulos, S.; Glotsos, D. Photograph to X-Ray Image Translation for Anatomical Mouse Mapping in Preclinical Nuclear Molecular Imaging. In Proceedings of 2021 International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD 2021); Su, R., Zhang, Y.-D., Liu, H., Eds.; Springer: Singapore, 2022; pp. 302–311. [Google Scholar]





| #TS | Overall | Foot | Gender | ||
|---|---|---|---|---|---|
| Left | Right | Male | Female | ||
| 1 | 99.4 | 98.9 | 100.0 | 100.0 | 98.8 |
| 2 | 35.6 | 36.7 | 34.4 | 19.2 | 53.5 |
| 3 | 57.8 | 62.2 | 53.3 | 56.4 | 59.3 |
| 4 | 88.9 | 86.7 | 91.1 | 96.8 | 80.2 |
| 5 | 85.9 | 82.2 | 88.9 | 95.7 | 74.4 |
| 6 | 86.1 | 83.3 | 88.9 | 96.8 | 74.4 |
| 7 | 99.4 | 98.9 | 100.0 | 100.0 | 98.8 |
| 8 | 98.3 | 97.8 | 98.9 | 100.0 | 96.5 |
| 9 | 98.3 | 97.8 | 98.9 | 100.0 | 96.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, T.; Coelho, L.; Silva, M.F. Automatic Segmentation of Monofilament Testing Sites in Plantar Images for Diabetic Foot Management. Bioengineering 2022, 9, 86. https://doi.org/10.3390/bioengineering9030086
Costa T, Coelho L, Silva MF. Automatic Segmentation of Monofilament Testing Sites in Plantar Images for Diabetic Foot Management. Bioengineering. 2022; 9(3):86. https://doi.org/10.3390/bioengineering9030086
Chicago/Turabian StyleCosta, Tatiana, Luis Coelho, and Manuel F. Silva. 2022. "Automatic Segmentation of Monofilament Testing Sites in Plantar Images for Diabetic Foot Management" Bioengineering 9, no. 3: 86. https://doi.org/10.3390/bioengineering9030086
APA StyleCosta, T., Coelho, L., & Silva, M. F. (2022). Automatic Segmentation of Monofilament Testing Sites in Plantar Images for Diabetic Foot Management. Bioengineering, 9(3), 86. https://doi.org/10.3390/bioengineering9030086








