Genome Editing Technology for Genetic Amelioration of Fruits and Vegetables for Alleviating Post-Harvest Loss
Abstract
:1. Introduction
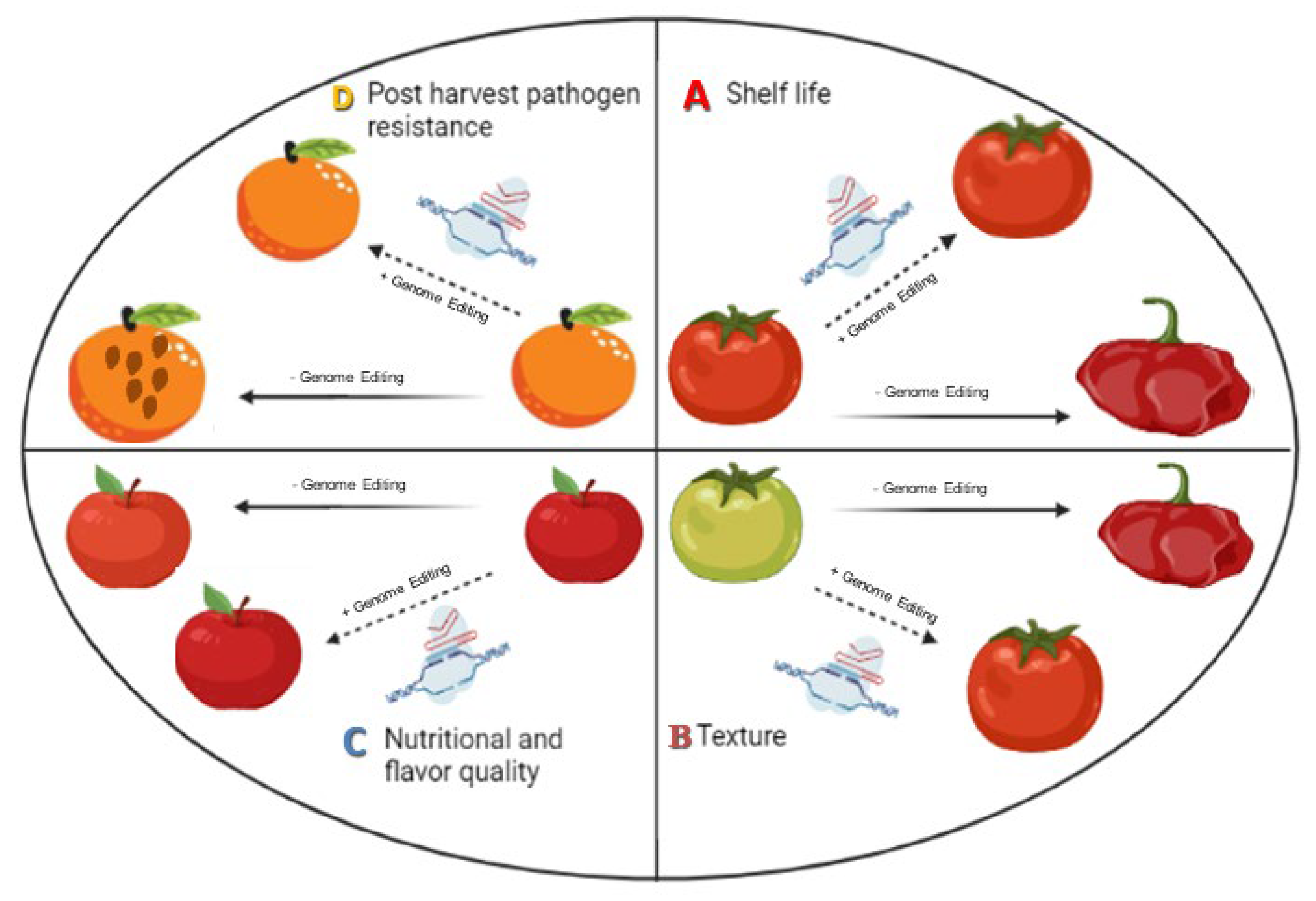
2. Genome Editing for Enhancing Post-Harvest Quality Attributes
2.1. Enhancement of Shelf Life
2.2. Fruit Texture Quality Improvement
2.3. Improving Post-Harvest Pathogen Resistance
2.4. Nutritional and Flavor Quality Enhancement
3. Obstacles, Challenges, and Solutions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hegazy, R. Post-Harvest Situation and Losses in India. J. Contrib. 2016, 2016, 584551. [Google Scholar] [CrossRef]
- Shipman, E.N.; Yu, J.; Zhou, J.; Albornoz, K.; Beckles, D.M. Can Gene Editing Reduce Postharvest Wasteand Loss of Fruit, Vegetables, and Ornamentals? Hortic. Res. 2021, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Meli, V.S.; Ghosh, S.; Prabha, T.N.; Chakraborty, N.; Chakraborty, S.; Datta, A. Enhancement of Fruit Shelf Life by Suppressing N-Glycan Processing Enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 2413–2418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Meli, V.S.; Kumar, A.; Thakur, A.; Chakraborty, N.; Chakraborty, S.; Datta, A. The N-Glycan Processing Enzymes α-Mannosidaseand β-D-N-Acetylhexosaminidase Are Involvedin Ripening-Associated Softeninginthe Non-Climacteric Fruits of Capsicum. J. Exp. Bot. 2011, 62, 571–582. [Google Scholar] [CrossRef]
- Irfan, M.; Datta, A. Improving Food Nutritional Quality and Productivity through Genetic Engineering. IJCSMB 2017, 2, 555576. [Google Scholar] [CrossRef]
- Ghosh, S.; Irfan, M.; Datta, A. Application of RNA Silencing in Improving Plant Traits for Industrial Use. In Plant Gene Silencing: Mechanisms and Applications; Dalmay, T., Ed.; CABI: Wallingford, UK, 2017; pp. 128–146. [Google Scholar] [CrossRef]
- Kramer, M.G.; Redenbaugh, K. Commercialization of a Tomato with an Antisense Polygalacturonase Gene: The FLAVRSAVRTM Tomato Story. Euphytica 1994, 79, 293–297. [Google Scholar] [CrossRef]
- Rommens, C.M.; Ye, J.; Richael, C.; Swords, K. Improving Potato Storage and Processing Characteristics through All-Native DNA Transformation. J. Agric. Food Chem. 2006, 54, 9882–9887. [Google Scholar] [CrossRef]
- Rommens, C.M.; Yan, H.; Swords, K.; Richael, C.; Ye, J. Low-Acrylamide French Fries and Potato Chips. Plant Biotechnol. J. 2008, 6, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Savin, K.W.; Baudinette, S.C.; Graham, M.W.; Michael, M.Z.; Nugent, G.D.; Lu, C.-Y.; Chandler, S.F.; Cornish, E.C. Antisense ACC Oxidase RNA Delays Carnation Petal Senescence. HortScience 1995, 30, 970–972. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.-C.; Lai, U.-L.; Yang, S.-F.; Chu, M.-J.; Kuo, C.-I.; Tsai, M.-F.; Sun, C.-W. Delayed Flower Senescence of Petunia Hybrida Plants Transformed with Antisense Broccoli ACC Synthaseand ACC Oxidase Genes. Postharvest Biol. Technol. 2007, 46, 47–53. [Google Scholar] [CrossRef]
- Smith, C.J.S.; Watson, C.F.; Morris, P.C.; Bird, C.R.; Seymour, G.B.; Gray, J.E.; Arnold, C.; Tucker, G.A.; Schuch, W.; Harding, S.; et al. Inheritance and Effecton Ripening of Antisense Polygalacturonase Genesin Transgenic Tomatoes. Plant Mol. Biol. 1990, 14, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; Abbott, J.A.; Gross, K.C. Down-Regulation of Tomato β-Galactosidase 4 Results in Decreased Fruit Softening. Plant Physiol. 2002, 129, 1755–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, B.; Ström, A.; Tasker, A.; West, G.; Tucker, G.A. Effect of Silencing the Two Major Tomato Fruit Pectin Methylesterase Isoformson Cell Wall Pectin Metabolism. Plant Biol. J. 2013, 15, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Bermúdez, S.; Redondo-Nevado, J.; Muñoz-Blanco, J.; Caballero, J.L.; López-Aranda, J.M.; Valpuesta, V.; Pliego-Alfaro, F.; Quesada, M.A.; Mercado, J.A. Manipulation of Strawberry Fruit Softening by Antisense Expression of a Pectate Lyase Gene. Plant Physiol. 2002, 128, 751–759. [Google Scholar] [CrossRef]
- Santiago-Domenech, N.; Jimenez-Bemudez, S.; Matas, A.J.; Rose, J.K.C.; Munoz-Blanco, J.; Mercado, J.A.; Quesada, M.A. Antisense Inhibitionofa Pectate Lyase Gene Supportsa Role for Pectin Depolymerizationin Strawberry Fruit Softening. J. Exp. Bot. 2008, 59, 2769–2779. [Google Scholar] [CrossRef] [Green Version]
- Posé, S.; Paniagua, C.; Cifuentes, M.; Blanco-Portales, R.; Quesada, M.A.; Mercado, J.A. Insights into the Effects of Polygalacturonase FaPG1 Gene Silencingon Pectin Matrix Disassembly, Enhanced Tissue Integrity, and Firmnessin Ripe Strawberry Fruits. J. Exp. Bot. 2013, 64, 3803–3815. [Google Scholar] [CrossRef]
- Paniagua, C.; Blanco-Portales, R.; Barceló-Muñoz, M.; García-Gago, J.A.; Waldron, K.W.; Quesada, M.A.; Muñoz-Blanco, J.; Mercado, J.A. Antisense Down-Regulation of the Strawberry β-Galactosidase Gene FaβGal4 Increases Cell Wall Galactose Levels and Reduces Fruit Softening. EXBOTJ 2016, 67, 619–631. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Si, X.; Ji, X.; Fan, R.; Liu, J.; Chen, K.; Wang, D.; Gao, C. Genome Editing of Upstream Open Reading Frames Enables Translational Controlin Plants. Nat. Biotechnol. 2018, 36, 894–898. [Google Scholar] [CrossRef]
- Behboodian, B.; Mohd Ali, Z.; Ismail, I.; Zainal, Z. Postharvest Analysis of Lowland Transgenic Tomato Fruits Harboring HpRNAi-ACO1 Construct. Sci. World J. 2012, 2012, 439870. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of SlPL, Which Encodesa Pectate Lyasein Tomato, Confers Enhanced Fruit Firmness, Prolonged Shelf-Lifeand Reduced Susceptibility to Grey Mould. Plant Biotechnol. J. 2017, 15, 1544–1555. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Tavano, E.C.D.; Lammers, M.; Martinelli, A.P.; Angenent, G.C.; deMaagd, R.A. Re-Evaluation of Transcription Factor Function in Tomato Fruit Development and Ripening with CRISPR/Cas9-Mutagenesis. Sci. Rep. 2019, 9, 1696. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Ghosh, S.; Kumar, V.; Chakraborty, N.; Chakraborty, S.; Datta, A. Insights into Transcriptional Regulation of β-D-N-Acetylhexosaminidase, anN-Glycan-Processing Enzyme Involved in Ripening-Associated Fruit Softening. J. Exp. Bot 2014, 65, 5835–5848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irfan, M.; Ghosh, S.; Meli, V.S.; Kumar, A.; Kumar, V.; Chakraborty, N.; Chakraborty, S.; Datta, A. Fruit Ripening Regulation of α-Mannosidase Expression by the MADS Box Transcription Factor RIPENINGINHIBITOR and Ethylene. Front. Plant Sci. 2016, 7, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quesada, M.A.; Blanco-Portales, R.; Posé, S.; García-Gago, J.A.; Jiménez-Bermúdez, S.; Muñoz-Serrano, A.; Caballero, J.L.; Pliego-Alfaro, F.; Mercado, J.A.; Muñoz-Blanco, J. Antisense Down-Regulation of the FaPG1 Gene Reveal san Unexpected Central Role for Polygalacturonasein Strawberry Fruit Softening. Plant Physiol. 2009, 150, 1022–1032. [Google Scholar] [CrossRef] [Green Version]
- Oliveira Filho, J.G.D.; daCruz Silva, G.; deAguiar, A.C.; Cipriano, L.; deAzeredo, H.M.C.; Bogusz Junior, S.; Ferreira, M.D. Chemical Composition and Antifungal Activity of Essential Oilsand Their Combinations against Botrytis Cinereain Strawberries. Food Meas. 2021, 15, 1815–1825. [Google Scholar] [CrossRef]
- Elitzur, T.; Yakir, E.; Quansah, L.; Zhangjun, F.; Vrebalov, J.; Khayat, E.; Giovannoni, J.J.; Friedman, H. Banana MaMADS Transcription Factors Are Necessary for Fruit Ripening and Molecular Tools to Promote Shelf-Life and Food Security. Plant Physiol. 2016, 171, 380–391. [Google Scholar] [CrossRef] [Green Version]
- Bhaskar, P.B.; Wu, L.; Busse, J.S.; Whitty, B.R.; Hamernik, A.J.; Jansky, S.H.; Buell, C.R.; Bethke, P.C.; Jiang, J. Suppression of the Vacuolar Invertase Gene Prevents Cold-Induced Sweetening in Potato. Plant Physiol. 2010, 154, 939–948. [Google Scholar] [CrossRef] [Green Version]
- Waltz, E. USDA Approves Next-Generation GM Potato. Nat. Biotechnol. 2015, 33, 12–13. [Google Scholar] [CrossRef]
- Waltz, E. Nonbrowning GM Apple Cleared for Market. Nat. Biotechnol. 2015, 33, 326–327. [Google Scholar] [CrossRef]
- Prado, J.R.; Segers, G.; Voelker, T.; Carson, D.; Dobert, R.; Phillips, J.; Cook, K.; Cornejo, C.; Monken, J.; Grapes, L.; et al. Genetically Engineered Crops: From Idea to Product. Annu. Rev. Plant Biol. 2014, 65, 769–790. [Google Scholar] [CrossRef] [Green Version]
- Carzoli, A.K.; Aboobucker, S.I.; Sandall, L.L.; Lübberstedt, T.T.; Suza, W.P. Risks and Opportunities of GM Crops: Bt Maize Example. Glob. Food Secur. 2018, 19, 84–91. [Google Scholar] [CrossRef]
- Mackelprang, R.; Lemaux, P.G. Genetic Engineering and Editing of Plants: An Analysis of New and Persisting Questions. Annu. Rev. Plant Biol. 2020, 71, 659–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef] [PubMed]
- Ku, H.-K.; Ha, S.-H. Improving Nutritional and Functional Quality by Genome Editing of Crops: Statusand Perspectives. Front. Plant Sci. 2020, 11, 577313. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid Restriction Enzymes: Zinc Finger Fusions to FokI Cleavage Domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [Green Version]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Que, Q.; Chen, Z.; Kelliher, T.; Skibbe, D.; Dong, S.; Chilton, M.-D. Plant DNA Repair Pathway sand Their Application sin Genome Engineering. In Plant Genome Editing with CRISPR Systems; Qi, Y., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 1917, pp. 3–24. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, R. An Efficient Tool for Improving Biotic Stress in Plants. In Plant-Microbial Interactionsand Smart Agricultural Biotechnology; CRC Press: Boca Raton, FL, USA, 2021; pp. 185–198. [Google Scholar]
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR/Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 Structuresand Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [Green Version]
- Subburaj, S.; Chung, S.J.; Lee, C.; Ryu, S.M.; Kim, D.H.; Kim, J.S.; Bae, S.; Lee, G.J. Site-directedmutagenesisin Petunia × hybrida protoplastsystem using direct delivery of purifie drecombinant Cas9 ribonucleo proteins. Plant Cell Rep. 2016, 35, 1535–1544. [Google Scholar] [CrossRef]
- Liu, W.; Rudis, M.R.; Cheplick, M.H.; Millwood, R.J.; Yang, J.-P.; Ondzighi-Assoume, C.A.; Montgomery, G.A.; Burris, K.P.; Mazarei, M.; Chesnut, J.D.; et al. Lipofection-mediated genomee ditingusing DNA-free delivery of the Cas9/gRNA ribonucleo proteininto plant cells. Plant Cell Rep. 2019, 39, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shen, J.; Li, D.; Cheng, Y. Strategies in the delivery of Cas9 ribonucleo protein for CRISPR/Cas9 genomeediting. Theranostics 2021, 11, 614–648. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.M.; Zhang, Y.; Yuan, G.; De, K.; Chen, J.-G.; Muchero, W.; Tuskan, G.A.; Qi, Y.; Yang, X. Construct Design for CRISPR/Cas-Based Genome Editing in Plants. Trends Plant Sci. 2021, 26, 1133–1152. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lecourieux, F.; Zhang, R.; Dai, Z.; Lecourieux, D.; Li, S.; Liang, Z. Data ComparisonandSoftware DesignforEasy SelectionandApplicationofCRISPR-Based GenomeEditing SystemsinPlants. Genom. Proteom. Bioinform. 2021, S1672022921001509. [Google Scholar] [CrossRef]
- Metje-Sprink, J.; Menz, J.; Modrzejewski, D.; Sprink, T. DNA-Free Genome Editing: Past, Presentand Future. Front. Plant Sci. 2019, 9, 1957. [Google Scholar] [CrossRef]
- EFSA Panelon Genetically Modified Organisms (EFSAGMOPanel); Naegeli, H.; Bresson, J.; Dalmay, T.; Dewhurst, I.C.; Epstein, M.M.; Firbank, L.G.; Guerche, P.; Hejatko, J.; Moreno, F.J.; et al. Applicability of the EFSA Opinionon Site-directed Nucleases Type3 for the Safety Assessment of Plants Developed Using Site-directed Nucleases Type1 and 2 and Oligonucleotide-directed Mutagenesis. EFS2 2020, 18, e06299. [Google Scholar] [CrossRef]
- Podevin, N.; Davies, H.V.; Hartung, F.; Nogué, F.; Casacuberta, J.M. Site-Directed Nucleases: A Paradigm Shiftin Predictable, Knowledge-Based Plant Breeding. Trends Biotechnol. 2013, 31, 375–383. [Google Scholar] [CrossRef]
- Entine, J.; Felipe, M.S.S.; Groenewald, J.-H.; Kershen, D.L.; Lema, M.; Mc Hughen, A.; Nepomuceno, A.L.; Ohsawa, R.; Ordonio, R.L.; Parrott, W.A.; et al. Regulatory Approaches for Genome Edited Agricultural Plants in Select Countries and Jurisdictions around the World. Transgenic Res. 2021, 30, 551–584. [Google Scholar] [CrossRef]
- Matas, A.J.; Gapper, N.E.; Chung, M.-Y.; Giovannoni, J.J.; Rose, J.K. Biologyand Genetic Engineering of Fruit Maturation for Enhanced Quality and Shelf-Life. Curr. Opin. Biotechnol. 2009, 20, 197–203. [Google Scholar] [CrossRef]
- Xiong, J.-S.; Ding, J.; Li, Y. Genome-Editing Technologies and Their Potential Applicationin Horticultural Crop Breeding. Hortic. Res. 2015, 2, 15019. [Google Scholar] [CrossRef] [Green Version]
- Tayal, R.; Kumar, V.; Irfan, M. Harnessing the Power of Hydrogen Sulphide (H2S) for Improving Fruit Quality Traits. Plant Biol. J. 2021, plb.13372. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Kumar, P.; Ahmad, I.; Datta, A. Unraveling the Role of Tomato Bcl-2-Associated Athanogene(BAG) Proteinsduring Abiotic Stress Response and Fruit Ripening. Sci. Rep. 2021, 11, 21734. [Google Scholar] [CrossRef] [PubMed]
- Albornoz, K.; Cantwell, M.I.; Zhang, L.; Beckles, D.M. Integrative Analysis of Postharvest Chilling Injury in Cherry Tomato Fruit Reveals Contrapuntal Spatio-Temporal Responses to Ripening and ColdStress. Sci. Rep. 2019, 9, 2795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lü, P.; Yu, S.; Zhu, N.; Chen, Y.-R.; Zhou, B.; Pan, Y.; Tzeng, D.; Fabi, J.P.; Argyris, J.; Garcia-Mas, J.; et al. Genome Encode Analyses Reveal the Basis of Convergent Evolution of Fleshy Fruit Ripening. Nat. Plants 2018, 4, 784–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, Y.J.; Lee, G.-J.; Bae, S.; Kang, K.K. Reduced Ethylene Productionin Tomato Fruitsupon CRSPR/Cas9-Mediated LeMADS-RIN Mutagenesis. Korean J. Hortic. Sci. Technol. 2018, 36, 396–405. [Google Scholar] [CrossRef]
- Yu, Q.; Wang, B.; Li, N.; Tang, Y.; Yang, S.; Yang, T.; Xu, J.; Guo, C.; Yan, P.; Wang, Q.; et al. CRISPR/Cas9-Induced Targeted Mutagenesis and Gene Replace ment to Generate Long-Shelf Life Tomato Lines. Sci. Rep. 2017, 7, 11874. [Google Scholar] [CrossRef]
- Gao, Y.; Zhu, N.; Zhu, X.; Wu, M.; Jiang, C.-Z.; Grierson, D.; Luo, Y.; Shen, W.; Zhong, S.; Fu, D.-Q.; et al. Diversity and Redundancy of the Ripening Regulatory Networks Revealed by the Fruit ENCODE and the New CRISPR/Cas9CNR and NOR Mutants. Hortic. Res. 2019, 6, 39. [Google Scholar] [CrossRef] [Green Version]
- Clasen, B.M.; Stoddard, T.J.; Luo, S.; Demorest, Z.L.; Li, J.; Cedrone, F.; Tibebu, R.; Davison, S.; Ray, E.E.; Daulhac, A.; et al. Improving Cold Storage and Processing Traitsin Potato through Targeted Gene Knockout. Plant Biotechnol. J. 2016, 14, 169–176. [Google Scholar] [CrossRef]
- González, M.N.; Massa, G.A.; Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Storani, L.; Décima Oneto, C.A.; Hofvander, P.; Feingold, S.E. Reduced Enzymatic Browning in Potato Tubersby Specific Editing of a Polyphenol Oxidase Gene via Ribonucleo protein Complexes Delivery of the CRISPR/Cas9 System. Front. Plant Sci. 2020, 10, 1649. [Google Scholar] [CrossRef]
- Waltz, E. Gene-Edited CRISPR Mushroom Escapes US Regulation. Nature 2016, 532, 293. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Kang, B.; Naing, A.H.; Bae, S.; Kim, J.; Kim, H.; Kim, C.K. CRISPR/Cas9-mediated Editing of 1-amino cyclopropane-1-carboxylate Oxidase1 Enhances Petunia Flower Longevity. Plant Biotechnol J. 2020, 18, 287–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uluisik, S.; Chapman, N.H.; Smith, R.; Poole, M.; Adams, G.; Gillis, R.B.; Besong, T.M.D.; Sheldon, J.; Stiegelmeyer, S.; Perez, L.; et al. Genetic Improvement of Tomatoby Targeted Control of Fruit Softening. Nat. Biotechnol. 2016, 34, 950–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral Organ Boundaries1 Isa Disease Susceptibility Gene for Citrus Bacterial Canker Disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome Editing of the Disease Susceptibility Gene CsLOB1 in Citrus Confers Resistance to Citrus Canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wang, L.; Zhao, R.; Yu, W.; Li, R.; Li, Y.; Sheng, J.; Shen, L. Knock out of SlMAPK3 Reduced Disease Resistance to Botrytis Cinereain Tomato Plants. J. Agric. Food Chem. 2018, 66, 8949–8956. [Google Scholar] [CrossRef]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-Mediated Efficient Targeted Mutagenesis in Grape in the First Generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef] [Green Version]
- Mishra, R.; Mohanty, J.N.; Mahanty, B.; Joshi, R.K. A Single Transcript CRISPR/Cas9 Mediated Mutagenesis of CaERF28 Confers An thracnose Resistance in Chilli Pepper (Capsicum Annuum L.). Planta 2021, 254, 5. [Google Scholar] [CrossRef]
- Gago, C.; Drosou, V.; Paschalidis, K.; Guerreiro, A.; Miguel, G.; Antunes, D.; Hilioti, Z. Targeted Gene Disruption Coupled with Metabolic Screen Approach to Uncover the LEAFYCOTYLEDON1-LIKE4(L1L4) Functionin Tomato Fruit Metabolism. Plant Cell Rep. 2017, 36, 1065–1082. [Google Scholar] [CrossRef]
- Ma, J.; Xiang, H.; Donnelly, D.J.; Meng, F.-R.; Xu, H.; Durnford, D.; Li, X.-Q. Genome Editing in Potato Plants by Agrobacterium-Mediated Transient Expression of Transcription Activator-like Effector Nucleases. Plant Biotechnol. Rep. 2017, 11, 249–258. [Google Scholar] [CrossRef]
- Ren, C.; Liu, X.; Zhang, Z.; Wang, Y.; Duan, W.; Li, S.; Liang, Z. CRISPR/Cas9-Mediated Efficient Targeted Mutagenesis in Chardonnay(Vitis Vinifera L.). Sci. Rep. 2016, 6, 32289. [Google Scholar] [CrossRef]
- Osakabe, Y.; Liang, Z.; Ren, C.; Nishitani, C.; Osakabe, K.; Wada, M.; Komori, S.; Malnoy, M.; Velasco, R.; Poli, M.; et al. CRISPR–Cas9-Mediated Genome Editing in Apple and Grapevine. Nat. Protoc. 2018, 13, 2844–2863. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.-S.; Samuelsson, M.; Hofvander, P. Efficient Targeted Multiallelic Mutagenesis in Tetraploid Potato (Solanum Tuberosum) by Transient CRISPR/Cas9 Expressionin Protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusano, H.; Ohnuma, M.; Mutsuro-Aoki, H.; Asahi, T.; Ichinosawa, D.; Onodera, H.; Asano, K.; Noda, T.; Horie, T.; Fukumoto, K.; et al. Establishment of a Modified CRISPR/Cas9 System with Increased Mutagenesis Frequency Using the Translational Enhancer DMac3 and Multiple Guide RNAs in Potato. Sci. Rep. 2018, 8, 13753. [Google Scholar] [CrossRef] [PubMed]
- Tuncel, A.; Corbin, K.R.; Ahn-Jarvis, J.; Harris, S.; Hawkins, E.; Smedley, M.A.; Harwood, W.; Warren, F.J.; Patron, N.J.; Smith, A.M. Cas9-mediated Mutagenesis of Potato Starch-branching Enzymes Generates a Range of Tuber Starch Phenotypes. Plant Biotechnol. J. 2019, 17, 2259–2271. [Google Scholar] [CrossRef] [PubMed]
- Veillet, F.; Chauvin, L.; Kermarrec, M.-P.; Sevestre, F.; Merrer, M.; Terret, Z.; Szydlowski, N.; Devaux, P.; Gallois, J.-L.; Chauvin, J.-E. The Solanum tuberosum GBSSI Gene:A Target for Assessing Geneand Base Editing in Tetraploid Potato. Plant Cell Rep. 2019, 38, 1065–1080. [Google Scholar] [CrossRef]
- Waltz, E. GABA-enriched tomato is first CRISPR-edited food to enter market. Nat. Biotechnol. 2022, 40, 9–11. [Google Scholar] [CrossRef]
- Kumar, V.; Irfan, M.; Ghosh, S.; Chakraborty, N.; Chakraborty, S.; Datta, A. Fruit Ripening Mutants Reveal Cell Metabolism and Redox State during Ripening. Protoplasma 2016, 253, 581–594. [Google Scholar] [CrossRef]
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Toki, S. CRISPR/Cas9-Mediated Mutagenesis of the RINL ocus That Regulates Tomato Fruit Ripening. Biochem. Biophys. Res. Commun. 2015, 467, 76–82. [Google Scholar] [CrossRef]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of Wild Tomato Is Accelerated by Genome Editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef]
- Gao, Y.; Wei, W.; Fan, Z.; Zhao, X.; Zhang, Y.; Jing, Y.; Zhu, B.; Zhu, H.; Shan, W.; Chen, J.; et al. Re-Evaluation of then or Mutation and the Role of the NAC-NOR Transcription Factor in Tomato Fruit Ripening. J. Exp. Bot. 2020, 71, 3560–3574. [Google Scholar] [CrossRef]
- Dev, S.S.; Joseph, J.; Lambert D’Rosario, L. Prospects for Genome Editing of Potato. In Solanum Tuberosum—A Promising Cropfor Starvation Problem; Yildiz, M., Ozgen, Y., Eds.; Intechopen: London, UK, 2021. [Google Scholar] [CrossRef]
- Wenneker, M.; Thomma, B.P.H.J. Latent Post harvest Pathogens of Pome Fruit and Their Management: From Single Measures to a Systems Intervention Approach. Eur. J. Plant Pathol. 2020, 156, 663–681. [Google Scholar] [CrossRef] [Green Version]
- Hua, L.; Yong, C.; Zhanquan, Z.; Boqiang, L.; Guozheng, Q.; Shiping, T. Pathogenic Mechanisms and Control Strategies of Botrytis Cinerea Causing Post-Harvest Decay in Fruits and Vegetables. Food Qual. Saf. 2018, 2, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Sonker, N.; Pandey, A.K.; Singh, P. Strategies to Control Post-Harvest Diseases of Table Grape: A Review. J. Wine Res. 2016, 27, 105–122. [Google Scholar] [CrossRef]
- Wang, W.; Liu, S.; Deng, L.; Ming, J.; Yao, S.; Zeng, K. Control of Citrus Post-Harvest Green Molds, Blue Molds, and Sour Rot by the CecropinA-Melitt in Hybrid Peptide BP21. Front. Microbiol. 2018, 9, 2455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramírez-Gil, J.G.; Henao-Rojas, J.C.; Morales-Osorio, J.G. Post harvest Diseases and Disorders in AvocadoCv.Hassand Their Relationship to Preharvest Management Practices. Heliyon 2021, 7, e05905. [Google Scholar] [CrossRef]
- Nelson, S. Postharvest RotsofBanana. Plant Dis. 2008, 54, 1–4. [Google Scholar]
- Papoutsis, K.; Edelenbos, M. Post harvest Environmentally and Human-Friendly Pre-Treatments to Minimize Carrot Waste in the Supply Chain Caused by Physiological Disorders and Fungi. Trends FoodSci. Technol. 2021, 112, 88–98. [Google Scholar] [CrossRef]
- Kumar, V.; Neeraj, S.; Sagar, N. Post Harvest Management of Fungal Diseases in Onion—A Review. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 737–752. [Google Scholar]
- Benkeblia, N. Post harvest Diseases and Disorders of Potato Tuber Solanum tuberosum L. Potatoes Prod. Consum. Health Benefits 2012, 7, 99–114. [Google Scholar]
- Brummell, D.A.; Harpster, M.H.; Civello, P.M.; Palys, J.M.; Bennett, A.B.; Dunsmuir, P. Modification of Expans in Protein Abundance in Tomato Fruit Alters Softening and Cell Wall Polymer Metabolism during Ripening. Plant Cell 1999, 11, 2203–2216. [Google Scholar] [CrossRef] [Green Version]
- Yahaya, S.; Mardiyya, A. Review of Post-Harvest Losses of Fruits and Vegetables. BJSTR 2019, 13, 10192–10200. [Google Scholar] [CrossRef]
- Coates, L.; Johnson, G.; Dale, M. Post harvest Pathology of Fruit and Vegetables. In Plant Disease; Rockvale Publications Editors: Armidale, Australia, 1997; pp. 533–547. [Google Scholar]
- Nayak, S.; Mukherjee, A.; Sengupta, C.; Samanta, S. Association of Microbial Diversity with Post Harvest Crops and Bioprospecting of Endophytic Microorganisms for Management. In Trends & Prospectsin Post Harvest Management of Horticultural Crops; ICAR-NRRI: Cuttack, India, 2019; p. 263. [Google Scholar]
- Park, M.-H.; Kim, J.-G. Low-DoseUV-CIrradiation Reduces the Microbial Population and Preserves Antioxidant Levels in Peeled Garlic (Allium Sativum L.) during Storage. Postharvest Biol. Technol. 2015, 100, 109–112. [Google Scholar] [CrossRef]
- Jin, P.; Wang, H.; Zhang, Y.; Huang, Y.; Wang, L.; Zheng, Y. UV-C Enhances Resistance against Gray Mold Decay Caused by Botryt is Cinerea in Strawberry Fruit. Sci. Hortic. 2017, 225, 106–111. [Google Scholar] [CrossRef]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic Engineering Strategies for Biotic and Abiotic Stress Tolerance and Quality Enhancement in Horti cultural Crops: A Comprehensive Review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, M.; Sakina, A.; Wani, S.H.; Shikari, A.B.; Tripathi, P.; Zaid, A.; Galla, A.; Abdelrahman, M.; Sharma, M.; Singh, A.K.; et al. Harnessing Genome Editing Techniques to Engineer Disease Resistancein Plants. Front. Plant Sci. 2019, 10, 550. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, S.; Kumar, R.; Kumar, V.; Won, S.Y.; Shukla, P. Engineering Disease Resistant Plants through CRISPR/Cas9 Technology. GM Crops Food 2021, 12, 125–144. [Google Scholar] [CrossRef]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering Canker-Resistant Plants through CRISPR/Cas9-Targeted Editing of the Susceptibility Gene CsLOB1 Promoterin Citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef] [Green Version]
- Hao, G.; Stover, E.; Gupta, G. Overexpression of a Modified Plant Thion in Enhances Disease Resistance to Citrus Canker and Huang long bing (HLB). Front. Plant Sci. 2016, 7, 1078. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Ke, F.; Nie, Z.; Wang, P.; Xu, J. Citrus Genetic Engineering for Disease Resistance: Past, Present and Future. Int. J. Mol. Sci. 2019, 20, 5256. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Li, D.; Wang, G.; Wang, F.; Kunjal, M.; Joldersma, D.; Liu, Z. Application and future perspective of CRISPR/Cas9 genome editing in fruit crops. J. Integrat Plant Biol. 2019, 62, 269–286. [Google Scholar] [CrossRef] [Green Version]
- Eckerstorfer, M.F.; Engelhard, M.; Heissenberger, A.; Simon, S.; Teichmann, H. Plants Developed by New Genetic Modification Techniques-Comparison of Existing Regulatory Frameworks in the EU and Non-EU Countries. Front. Bioeng. Biotechnol. 2019, 7, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-Free Genetically Edited Grapevine and Apple Proto plast Using CRISPR/Cas9 Ribonucleo proteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P. Genome Editing in Potato via CRISPR/Cas9 Ribonucleo protein Delivery. Physiol. Plant. 2018, 164, 378–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| S.N. | Crop Species | Gene Editing Tool | Transformation Method | Target Gene | Function of Target Gene | Outcome | Reference |
|---|---|---|---|---|---|---|---|
| Shelf life | |||||||
| 1. | Tomato | CRISPR/Cas9 | Agrobacterium tumefaciens-mediated transformation | ALC | Inhibit ethylene synthesis (SN1 is an insertion of an actual inhibitor gene ALC) | Mutants with longer shelf life as compared to wild type | [57] |
| 2. | Tomato | CRISPR/Cas9 (SDN1) | Agrobacterium tumefaciens-mediated transformation | RIN | Inhibit ethylene synthesis and specific biochemical processes related to fruit ripening | Mutant lines exhibited lower ethylene contents and delayed fruit ripening | [58] |
| 3. | Tomato | CRISPR/Cas9 (SDN2) | Agrobacterium tumefaciens-mediated transformation | ALC | Inhibit ethylene synthesis (SN2 seems to be a knockout mutant of the RIN gene) | Mutants with longer shelf life as compared to wild type | [59] |
| 4. | Tomato | CRISPR/Cas9 (SDN1) | Not mentioned | SBP-CNR&NAC-NOR | Transcription factor of ripening genes | Mutants displayed partial non-ripening phenotypes | [60] |
| 5. | Potato | TALEN (SDN1) | Protoplast transfection using PEG mediated transformation system | Vinv | Hydrolyzes the sucrose produced from starch breakdown into one molecule of glucose and one of fructose | Mutant lines with improved cold storage and processing traits | [61] |
| 6. | Potato | CRISPR/Cas9 (SDN1) | Protoplast transfection with RNPs using PEG mediated transformation system | StPPO2 | Catalyzes the oxidation of phenolic compounds into compounds into quinones (highly reactive form) | Mutant lines exhibited reduction in enzymatic browning and PPO gene. | [62] |
| 7. | White button mushroom | CRISPR/Cas9 (SDN1) | Protoplast transfection using PEG mediated transformation system | StPPO2 | Catalyzes the oxidation of phenolic compounds into quinones (highly reactive form) | Mutants lines showed 30% reduction in enzymatic browning with improved appearance and shelf life | [63] |
| 8. | Petunia | CRISPR/Cas9 (SDN1) | Agrobacterium tumefaciens-mediated transformation | PhACO | Catalyzes aminocyclopropane-1-carboxylic acid to ethylene in ethylene biosynthesis pathway | Mutant lines exhibited significant reduction in ethylene production and enhanced flower longevity as compared to wild-type | [64] |
| Texture | |||||||
| 9. | Tomato | CRISPR/Cas9 (SDN1) | Not mentioned | PL | Involved in plant cell wall degradation | Higher fruit firmness efficiency were found in mutants plants | [65] |
| Post-harvest pathogen resistance | |||||||
| 10. | Citrus | CRISPR/Cas9 (SDN1) | Agrobacterium tumefaciens-mediated transformation | CsLOB1 | Disease susceptibility gene for citrus bacterial canker | Mutant lines showed lower host pustule development with improved fungal resistance against Xanthomonas citri subsp.citri. | [66] |
| 11. | Citrus | CRISPR/Cas9 (SDN1) | Agrobacterium tumefaciens-mediated transformation | CsLOB1 | Disease susceptibility gene for citrus bacterial canker | Improved fungal resistance against citrus bacterial canker in mutant plants | [67] |
| 12. | Tomato | CRISPR/Cas9 (SDN1) | Not mentioned | SlMAPK3 | MAPKs genes play an important role in defense responses to biotic and abiotic stresses | Mutants lines were prepared by knocking out SIMAPK3 gene that showed resistance to Botrytis cinerea | [68] |
| 13. | Grape | CRISPR/Cas9 (SDN1) | Agrobacterium tumefaciens-mediated transformation | VvWRKY52 | Important in plant biotic stresses responses | Mutants lines with knocked out VvWRKY52 gene showed higher resistance to Botrytis cinerea | [69] |
| 14. | Chili pepper | CRISPR/Cas9 (SDN1) | Agrobacterium tumefaciens-mediated transformation | CaERF28 | Susceptibility gene for anthracnose disease | Mutant lines showed higher resistance toward anthracnose | [70] |
| Nutritional and flavor quality | |||||||
| 15. | Tomato | ZFNs (SDN1) | Not mentioned | NF-Y, L1L4, NF-YB6 | Responsible for biosynthesis for seed storage proteins and fatty acids | Mutants showed varied metabolite profiles and high amounts of OA as compared to wild type | [71] |
| 16. | Potato | TALEN (SDN1) | Agrobacterium tumefaciens-mediated transformation | SBE1 and INV2 | SBE1 enzymes are responsible forformation of amylopectin. INV2 catalyze the irreversible hydrolysis of sucrose into glucose and fructose | Improved amylopectin content and cold sweetening | [72] |
| 17. | Grape | CRISPR/Cas9 (SDN1) | Agrobacterium tumefaciens-mediated transformation | IdnDH | Important enzyme in tartaric acid (TA) biosynthetic pathway | Significant accumulation of tartaric acid (TA) in mutants lines | [73] |
| 18. | Apple | CRISPR/Cas9 | Agrobacterium tumefaciens-mediated transformation and PEG transformation system | IdnDH | Important enzyme in TA biosynthetic pathway | Stable accumulation of TA in mutant plants | [74] |
| 19. | Potato | CRISPR/Cas9 (SDN1) | Protoplast transfection using PEG mediated transformation system | StGBSS | Responsible for amylase synthesis | Mutant lines showed higher amylopectin content than wild type | [75] |
| 20. | Potato | CRISPR/Cas9 (SDN1) | Agrobacterium tumefaciens-mediated transformation | StGBSS | Responsible for the synthesis of amylase in starch biosynthetic pathway | Improved amylopectin content in potato plants | [76] |
| 21. | Potato | CRISPR/Cas9 (SDN) | Agrobacteriumtumefaciens-mediated transformation and PEG transformation system | SBE1, SBE2 | Starch branching enzymes which introduce α -1,6 -linkages into starch | Mutant lines showed reduced amylopectin content during granule growth | [77] |
| 22. | Potato | CRISPR/Cas9 | Agrobacterium tumefaciens-mediated transformation | StGBSS | Responsible for the synthesis of amylase in starch biosynthetic pathway | Mutant plants showed higher amylopectin content by using a CBE | [78] |
| 23. | Tomato | CRISPR/Cas9 (SDN1) | Not mentioned | CaMBD | Improved GABA content (4–5 times) | [79] | |
| Crop | Disease | Causal Pathogen | Reference |
|---|---|---|---|
| Fruit crops | |||
| Pome Fruit | Blue mold Gray mold Bitter rot Alternaria rot Mucor rot | Pencillium spp. Botrytis cinerea Colletotrtchum gloeosporioides Alternaria spp. Mucor piriformis | [87] |
| Stone Fruit | Brown rot Rhizopus rot Graymold Blue mold Alternaria rot | Monilia spp. Rhizopus spp. (mostly R. stolonde) Botrytis cinerea Penicillium spp. Alternaria alternate | [96] |
| Berries | Graymold Rhizopus rot Cladosporium rot Blue mold | Botrytis cinerea Rhizopus spp. Cladosporium spp. Pencillium spp. | [96] |
| Mango | Anthracnose Stem end rot Rhizopus rot Black mold Alternaria rot Graymold Blue mold Mucor rot | Colletotrichum gloeosporioides, C. Acutatum Dothiorella spp. Phomopsis mangiferae Rhizopus stolonifer Aspergillus niger Alternaria alternate Botrytis cinerea Penicillium expansum Mucor circinelloides | [96] |
| Papaya | Anthracnose Black rot Phomopsis rot Rhizopus rot Phytophthora fruit rot | Colletotrichum spp. Phomacaricae-papayae Phomopsis caricae-papayae Rhizopus stolonifer Phytophthora palmivora | [96] |
| Grapes | Blue mold Graymold Rhizopus rot | Pencillium spp. Botrytis cinerea Rhizopus spp. | [97] |
| Citrus Fruit | Blue mold Green mold Black center rot Stem end rot Brown rot | Penicillium italicum Penicillium digitatum Alternartacitri Phomopsis citri Phytophthora citrophthora and/or P. Parasitica | [98] |
| Avocado | Anthracnose Stem end rot | Colletotrichum gloeosporoides, C. Acutatum Dothiorellaspp., Lasiodiplodiatheobromae | [99] |
| Banana | Anthracnose Crown rot Black end Ceratocystis fruit rot | Colletotrichummusae Various fungi including Fusarium spp., Vertcillium spp., Acremonium sp. and Colletotrichum musae Various fungi including Colletotrichum musae, Fusarium spp., Nigrospora sphaerica Ceratocystis paradoxa | [100] |
| Vegetable crops | |||
| Carrot | Bacterial soft rot Rhizopus rot Watery soft rot Graymold Sclerotium rot | Various Erwinia spp. and Pseudomonas spp. Rhizopus spp. Sclerotinia spp. Botrytis cinerea Sclerotium rolfsii | [88] |
| Cucurbits | Bacterial soft rots Graymold Fusarium rot Alternaria rot Charcoal rot Cottony leak Rhizopus rot | Various Erwinia spp., Bacillus polymgyxa, Pseudomonas syringae, Xanthomonas campestris Botrytis cinerea Fusarium spp. Alternaria spp. Macrophomina phaseolina Pythium spp. Rhizopus spp. | [96] |
| Tomato, Eggplant, and Capsicum | Bacterial soft rots Graymold Fusarium rot Alternaria rot Cladosporium rot Rhizopus rot Watery soft rot Cottony leak Sclerotium rot | Various Erwinia spp., Bacillus polymyxa, Pseudomonas spp., and Xanthomonas campestris Botrytis cinerea Fusarium spp. Alternaria spp. Cladosporium spp. Rhizopus spp. Sclerotinia spp. Pythium spp. Sclerotium rolfsii | [96] |
| Brassicas, Leafy Vegetables | Bacterial soft rots Graymold Alternaria rot Watery soft rot Phytophthora rot | Various Erwinia spp., Bacillus polymyxa, Pseudomonas spp., and Xanthomonas campestris Botrytis cinerea Alternaria spp. Sclerotiniaspp. Phytophthora porri | [96] |
| Onion | Bacterial soft rots Black mold rot Fusarium basal rot Smudge | Various Erwinia spp., Lactobacillus spp., and Pseudomonas spp. Aspergillus niger Fusarium oxysporum f. sp. cepae Colletotrichum circinans | [101] |
| Potato | Bacterial soft rot Dry rot Silver scurf | Erwinia spp. Fusarium spp. Helminthosporium solani | [102] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumari, C.; Sharma, M.; Kumar, V.; Sharma, R.; Kumar, V.; Sharma, P.; Kumar, P.; Irfan, M. Genome Editing Technology for Genetic Amelioration of Fruits and Vegetables for Alleviating Post-Harvest Loss. Bioengineering 2022, 9, 176. https://doi.org/10.3390/bioengineering9040176
Kumari C, Sharma M, Kumar V, Sharma R, Kumar V, Sharma P, Kumar P, Irfan M. Genome Editing Technology for Genetic Amelioration of Fruits and Vegetables for Alleviating Post-Harvest Loss. Bioengineering. 2022; 9(4):176. https://doi.org/10.3390/bioengineering9040176
Chicago/Turabian StyleKumari, Chanchal, Megha Sharma, Vinay Kumar, Rajnish Sharma, Vinay Kumar, Parul Sharma, Pankaj Kumar, and Mohammad Irfan. 2022. "Genome Editing Technology for Genetic Amelioration of Fruits and Vegetables for Alleviating Post-Harvest Loss" Bioengineering 9, no. 4: 176. https://doi.org/10.3390/bioengineering9040176
APA StyleKumari, C., Sharma, M., Kumar, V., Sharma, R., Kumar, V., Sharma, P., Kumar, P., & Irfan, M. (2022). Genome Editing Technology for Genetic Amelioration of Fruits and Vegetables for Alleviating Post-Harvest Loss. Bioengineering, 9(4), 176. https://doi.org/10.3390/bioengineering9040176








