Engineering CRISPR/Cas13 System against RNA Viruses: From Diagnostics to Therapeutics
Abstract
:1. Introduction
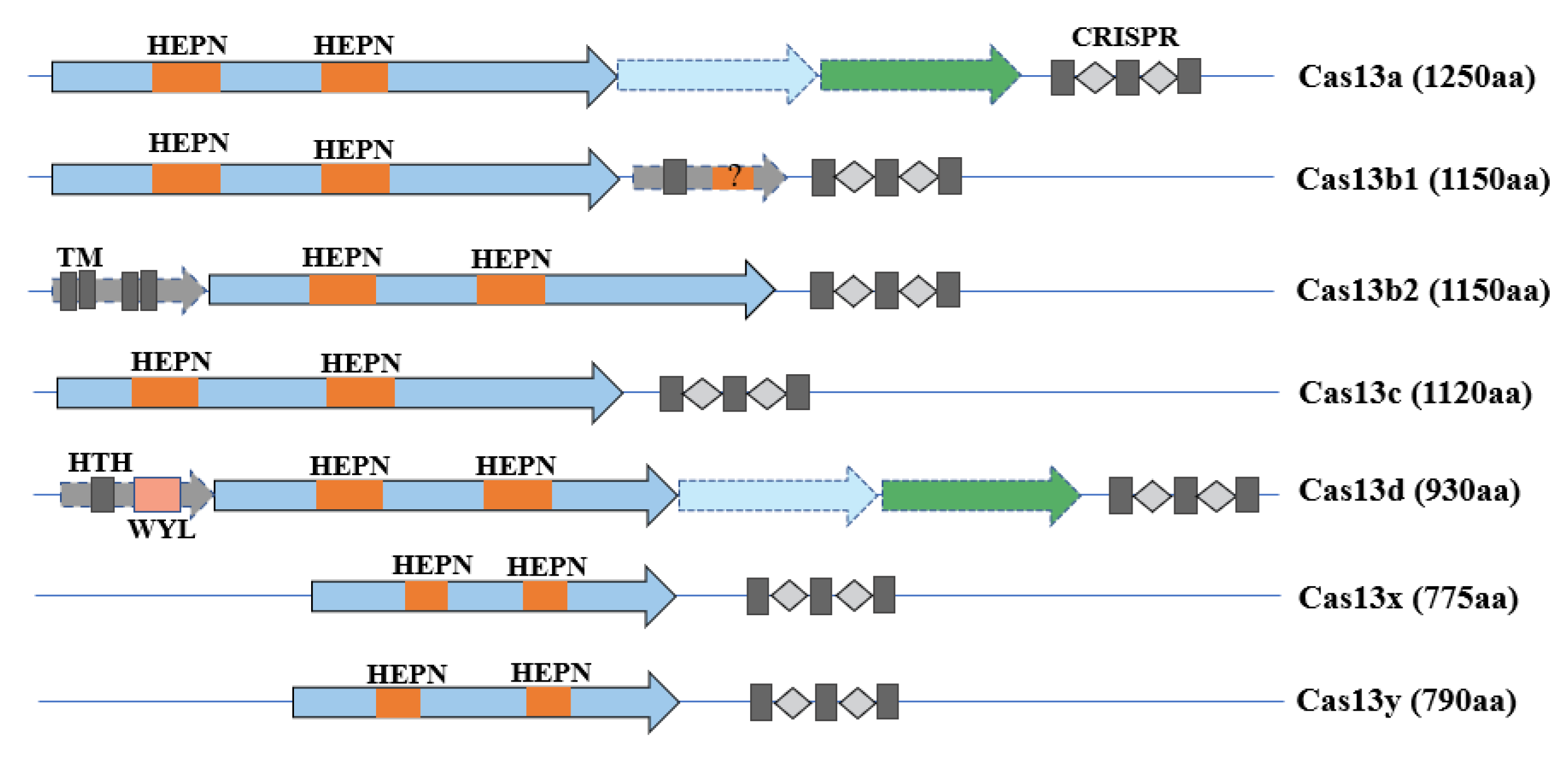
2. The CRISPR/Cas13 System
2.1. Cas13a
2.2. Cas13b
2.3. Cas13c
2.4. Cas13d
2.5. Cas13X and Cas13Y
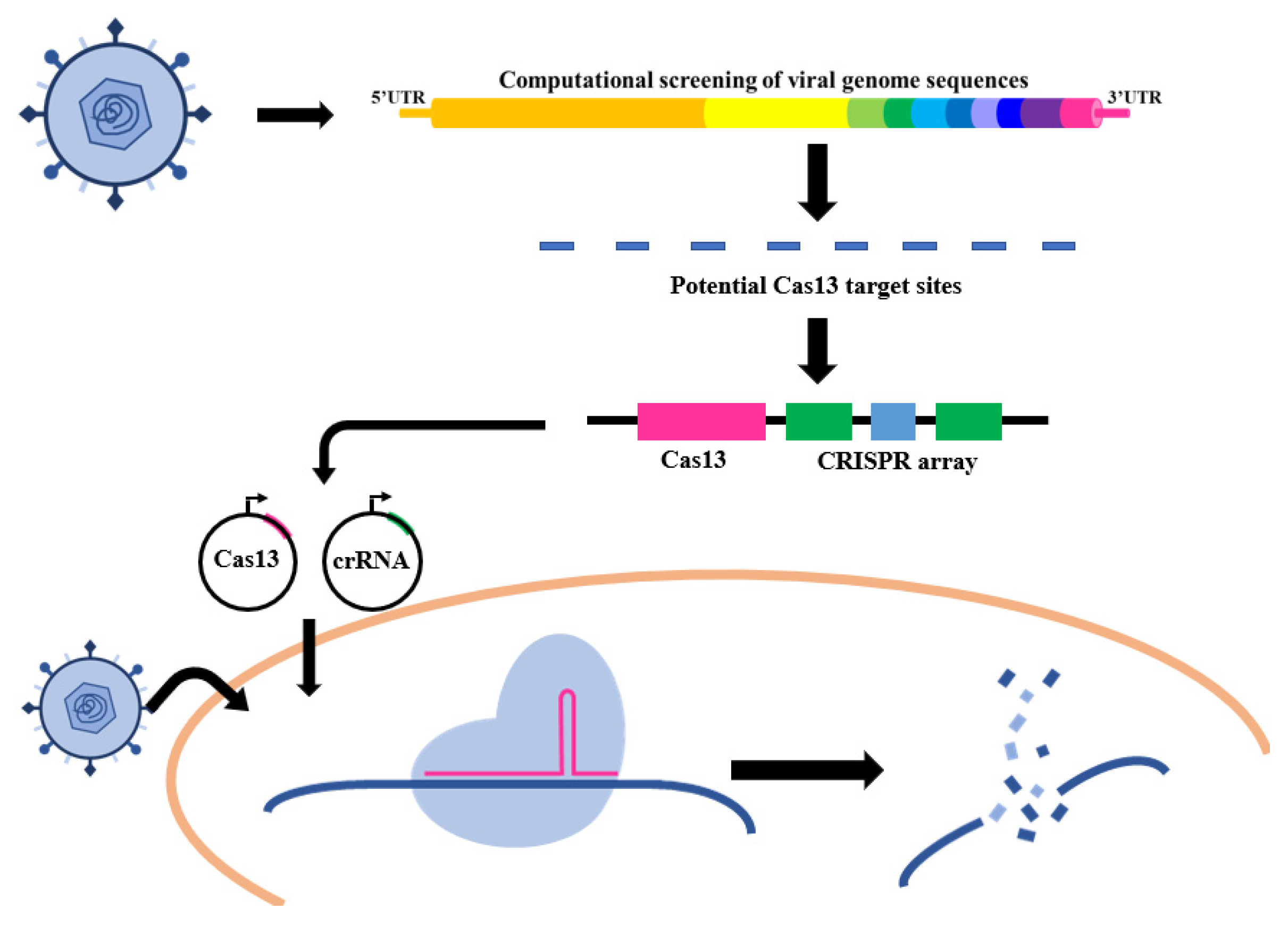
3. Applications of CRISPR/Cas13 in RNA Virus Diagnostics
3.1. Conventional RNA Virus Diagnostics
3.2. CRISPR-Cas13-Based RNA Virus Diagnostics
3.2.1. Applications of CRISPR-Cas13 in SARS-CoV-2 Detection
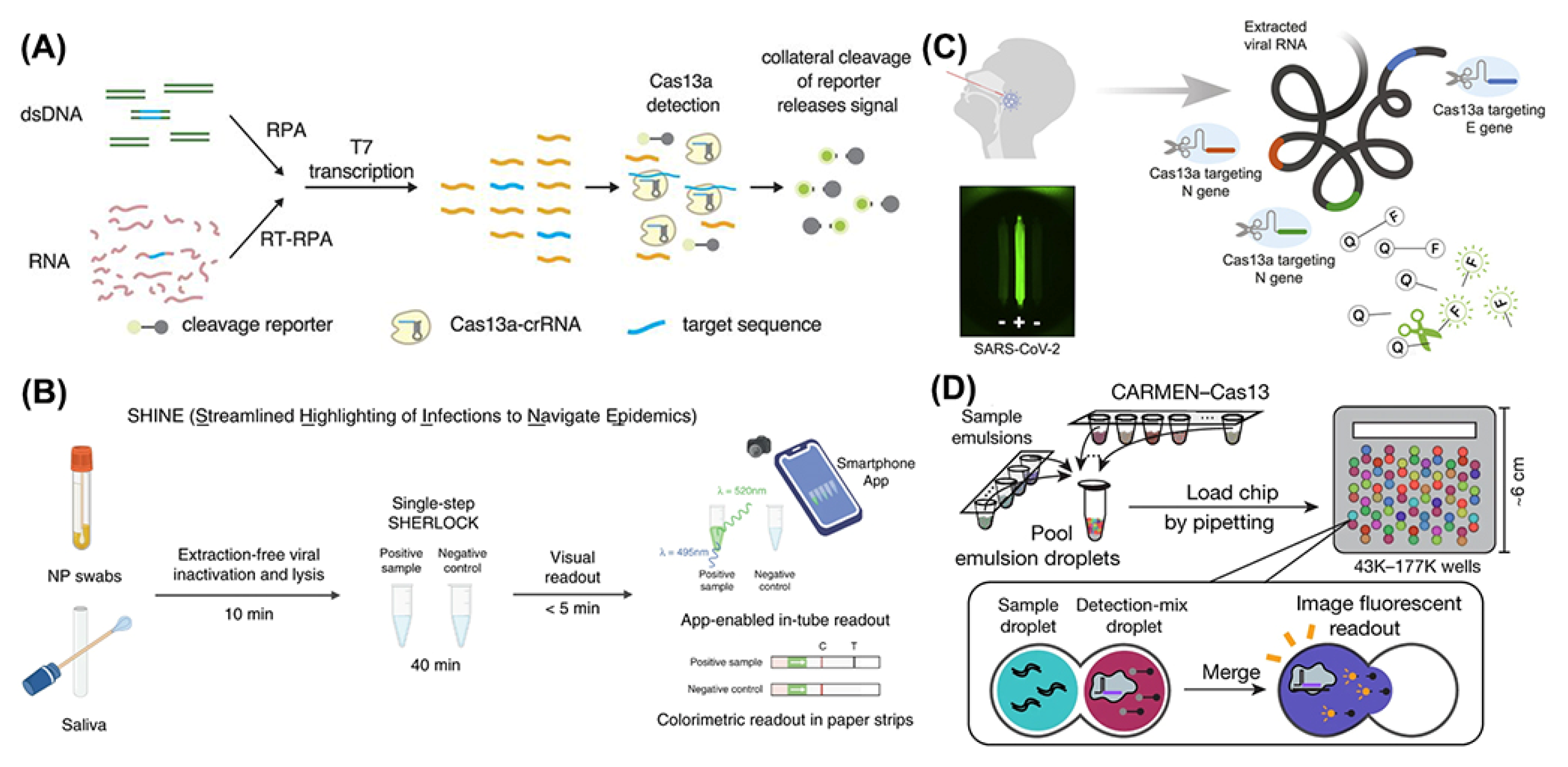
3.2.2. Applications of CRISPR-Cas13 in Dengue Virus Detection
3.2.3. Applications of CRISPR-Cas13 in HIV-1 Detection
3.2.4. Applications of CRISPR-Cas13 in Detection of Other RNA Viruses
4. Utilization of CRISPR/Cas13 in RNA Virus Therapeutics
4.1. Conventional RNA Virus Therapeutics
4.2. CRISPR/Cas13-Based Anti-RNA Virus Therapeutics
4.2.1. CRISPR-Cas13 in Anti-SARS-CoV-2 Therapy
4.2.2. CRISPR-Cas13 in Anti-DENV Therapy
4.2.3. CRISPR-Cas13 in Anti-HIV-1 Therapy
4.2.4. CRISPR-Cas13 in the Treatment of Diseases Caused by Other RNA Viruses
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Telenti, A.; Arvin, A.; Corey, L.; Corti, D.; Diamond, M.S.; Garcia-Sastre, A.; Garry, R.F.; Holmes, E.C.; Pang, P.S.; Virgin, H.W. After the pandemic: Perspectives on the future trajectory of COVID-19. Nature 2021, 596, 495–504. [Google Scholar] [CrossRef] [PubMed]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.X.; Liu, N.; Zhang, J.J. Functional Nucleic Acids as Modular Components against SARS-CoV-2: From Diagnosis to Therapeutics. Biosens. Bioelectron. 2022, 201, 13944. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Hlongwa, P. Current ethical issues in HIV/AIDS research and HIV/AIDS care. Oral Dis. 2016, 22, 61–65. [Google Scholar] [CrossRef]
- Liu, N.; Liu, R.; Zhang, J.J. CRISPR-Cas12a-mediated label-free electrochemical aptamer-based sensor for SARS-CoV-2 antigen detection. Bioelectrochemistry 2022, 146, 108105. [Google Scholar] [CrossRef]
- Zhang, W.X.; He, Y.; Feng, Z.; Zhang, J.J. Recent Advances of Functional Nucleic Acid-Based Sensors for Point-of-Care Detection of SARS-CoV-2. Microchim. Acta 2022, 189, 128. [Google Scholar] [CrossRef]
- Murugan, K.; Babu, K.; Sundaresan, R.; Rajan, R.; Sashital, D.G. The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit. Mol. Cell 2017, 68, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676. [Google Scholar] [CrossRef] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.M.; Zhang, Y.; Pandolfi, P.P. Virus against virus: A potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020, 30, 189–190. [Google Scholar] [CrossRef] [Green Version]
- Baddeley, H.J.E.; Isalan, M. The Application of CRISPR/Cas Systems for Antiviral Therapy. Front. Genome Ed. 2021, 3, 745559. [Google Scholar] [CrossRef] [PubMed]
- Jolany Vangah, S.; Katalani, C.; Booneh, H.A.; Hajizade, A.; Sijercic, A.; Ahmadian, G. CRISPR-Based Diagnosis of Infectious and Noninfectious Diseases. Biol. Proced. Online 2020, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. CRISPR-Cas13 as an Antiviral Strategy for Coronavirus Disease 2019. CRISPR J. 2020, 3, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef] [Green Version]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Li, X.; Wang, J.; Wang, M.; Chen, P.; Yin, M.; Li, J.; Sheng, G.; Wang, Y. Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell 2017, 168, 121–134.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, T.; Han, Y.; Wang, Y.; Huang, H.; Qian, P. Programmable System of Cas13-Mediated RNA Modification and Its Biological and Biomedical Applications. Front. Cell Dev. Biol. 2021, 9, 677587. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [Green Version]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630.e7. [Google Scholar] [CrossRef] [Green Version]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar]
- Xu, C.; Zhou, Y.; Xiao, Q.; He, B.; Geng, G.; Wang, Z.; Cao, B.; Dong, X.; Bai, W.; Wang, Y.; et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat. Methods 2021, 18, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Cassedy, A.; Parle-McDermott, A.; O’Kennedy, R. Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods. Front. Mol. Biosci. 2021, 8, 637559. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, S.; Ratnasri, K.; Valloly, P.; Sharma, R.; Maurya, S.; Gaigore, A.; Ardhya, C.; Biligi, D.S.; Desiraju, B.K.; Natchu, U.C.M.; et al. Evaluation of spike protein antigens for SARS-CoV-2 serology. J. Virol. Methods 2021, 296, 114222. [Google Scholar] [CrossRef] [PubMed]
- Lebani, K.; Jones, M.L.; Watterson, D.; Ranzoni, A.; Traves, R.J.; Young, P.R.; Mahler, S.M. Isolation of serotype-specific antibodies against dengue virus non-structural protein 1 using phage display and application in a multiplexed serotyping assay. PLoS ONE 2017, 12, e0180669. [Google Scholar] [CrossRef] [PubMed]
- Bystryak, S.; Acharya, C. Detection of HIV-1 p24 antigen in patients with varying degrees of viremia using an ELISA with a photochemical signal amplification system. Clin. Chim. Acta 2016, 456, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Teymouri, M.; Mollazadeh, S.; Mortazavi, H.; Naderi Ghale-Noie, Z.; Keyvani, V.; Aghababaei, F.; Hamblin, M.R.; Abbaszadeh-Goudarzi, G.; Pourghadamyari, H.; Hashemian, S.M.R.; et al. Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19. Pathol. Res. Pract. 2021, 221, 153443. [Google Scholar] [CrossRef]
- Ou, T.P.; Yun, C.; Auerswald, H.; In, S.; Leang, R.; Huy, R.; Choeung, R.; Dussart, P.; Duong, V. Improved detection of dengue and Zika viruses using multiplex RT-qPCR assays. J. Virol. Methods 2020, 282, 113862. [Google Scholar] [CrossRef]
- Falak, S.; Macdonald, R.; Busby, E.J.; O’Sullivan, D.M.; Milavec, M.; Plauth, A.; Kammel, M.; Zeichhardt, H.; Grunert, H.P.; Kummrow, A.; et al. An assessment of the reproducibility of reverse transcription digital PCR quantification of HIV-1. Methods 2022, 201, 34–40. [Google Scholar] [CrossRef]
- Jonathan, S.G.; Omar, O.A.; Jeong, W.L.; Patrick, E.; Aaron, J.D.; Julia, J.; Vanessa, V.; Nina, D.; Nichole, M.D.; Catherine, A.F.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef]
- Arizti-Sanz, J.; Freije, A.C.; Stanton, C.A.; Petros, A.B.; Boehm, K.C.; Siddiqui, C.; Shaw, M.B.; Adams, G.; Kosoko-Thoroddsen, F.T.; Kemball, E.M.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef] [PubMed]
- Fozouni, P.; Son, S.; Derby, M.D.d.L.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.T.; Knott, J.G.; Smock, C.J.D.; Desmarais, J.J.; Son, S.; Bhuiya, A.; Jakhanwal, S.; Prywes, N.; Agrawal, S.; Derby, M.; et al. Accelerated RNA detection using tandem CRISPR nucleases. Nat. Chem. Biol. 2021, 17, 982–988. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef]
- Welch, N.L.; Zhu, M.; Hua, C.; Weller, J.; Mirhashemi, M.E.; Nguyen, T.G.; Mantena, S.; Bauer, M.R.; Shaw, B.M.; Ackerman, C.M.; et al. Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants. Nat. Med. 2022, 28, 1083–1094. [Google Scholar] [CrossRef]
- Manning, B.J.; Khan, W.A.; Pena, J.M.; Fiore, E.S.; Boisvert, H.; Tudino, M.C.; Barney, R.E.; Wilson, M.K.; Singh, S.; Mowatt, J.A.; et al. High-Throughput CRISPR-Cas13 SARS-CoV-2 Test. Clin. Chem. 2021, 68, 172–180. [Google Scholar] [CrossRef]
- Brogan, J.D.; Chaverra-Rodriguez, D.; Lin, P.C.; Smidler, L.A.; Yang, T.; Alcantara, M.L.; Antoshechkin, I.; Liu, J.R.; Raban, R.R.; Belda-Ferre, P.; et al. Development of a Rapid and Sensitive CasRx-Based Diagnostic Assay for SARS-CoV-2. ACS Sens. 2021, 6, 3957–3966. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [Green Version]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Xia, Q.; Wu, J.; Lin, Y.; Ju, H. A sensitive electrochemical method for rapid detection of dengue virus by CRISPR/Cas13a-assisted catalytic hairpin assembly. Anal. Chim. Acta 2021, 1187, 339131. [Google Scholar] [CrossRef]
- McCutchan, J.A. Virology, immunology, and clinical course of HIV infection. J. Consult. Clin. Psychol. 1990, 58, 5–12. [Google Scholar] [CrossRef]
- Fozouni, P.; Melanie, O.; Jennifer, A.D. Hiv or Hcv Detection with Crispr-Cas13a. US Patent 17/273, 752, 11 November 2021. [Google Scholar]
- Liu, Y.; Xu, H.; Liu, C.; Peng, L.; Khan, H.; Cui, L.; Huang, R.; Wu, C.; Shen, S.; Wang, S.; et al. CRISPR-Cas13a Nanomachine Based Simple Technology for Avian Influenza A (H7N9) Virus On-Site Detection. J. Biomed. Nanotechnol. 2019, 15, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Deng, Y.; Li, T.; Wang, J.; Wang, T.; Tan, F.; Li, X.; Tian, K. Visual detection of porcine reproductive and respiratory syndrome virus using CRISPR-Cas13a. Transbound Emerg. Dis. 2020, 67, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Park, M.; Alfson, K.J.; Tamhankar, M.; Carrion, R.; Patterson, J.L.; Griffiths, A.; He, Q.; Yildiz, A.; Mathies, R.; et al. Rapid and Fully Microfluidic Ebola Virus Detection with CRISPR-Cas13a. ACS Sens. 2019, 4, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Barnes, K.G.; Lachenauer, A.E.; Nitido, A.; Siddiqui, S.; Gross, R.; Beitzel, B.; Siddle, K.J.; Freije, C.A.; Dighero-Kemp, B.; Mehta, S.B.; et al. Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time. Nat. Commun. 2020, 11, 4131. [Google Scholar] [CrossRef]
- Abbott, R.T.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R.; et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 2020, 181, 865–876. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, J.; Wang, Q.; Wang, Y.; Kang, C. Rapid design and development of CRISPR-Cas13a targeting SARS-CoV-2 spike protein. Theranostics 2021, 11, 649–664. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Dong, X.; Li, Q.; Li, M.; Li, J.; Guo, Y.; Jin, X.; Zhou, Y.; Song, H.; et al. CRISPR-Cas13a Cleavage of Dengue Virus NS3 Gene Efficiently Inhibits Viral Replication. Mol. Ther. Nucleic Acids 2020, 19, 1460–1469. [Google Scholar] [CrossRef]
- Singsuksawat, E.; Onnome, S.; Posiri, P.; Suphatrakul, A.; Srisuk, N.; Nantachokchawapan, R.; Praneechit, H.; Sae-kow, C.; Chidpratum, P.; Sa-ngiamsuntorn, K.; et al. Potent programmable antiviral against dengue virus in primary human cells by Cas13b RNP with short spacer and delivery by VLP. Mol. Ther. Methods Clin. Dev. 2021, 21, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhao, F.; Sun, H.; Wang, Z.; Huang, Y.; Zhu, W.; Xu, F.; Mei, S.; Liu, X.; Zhang, D.; et al. CRISPR-Cas13a Inhibits HIV-1 Infection. Mol. Ther. Nucleic Acids 2020, 21, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Hoang, N.; Wilson, H.; Jayakumar, S.; Kulkarni, V.; Kulkarni, S. Efficient Inhibition of HIV Using CRISPR/Cas13d Nuclease System. Viruses-Basel 2021, 13, 1850. [Google Scholar]
- Tng, P.Y.L.; Carabajal Paladino, L.; Verkuijl, S.A.N.; Purcell, J.; Merits, A.; Leftwich, P.T.; Fragkoudis, R.; Noad, R.; Alphey, L. Cas13b-dependent and Cas13b-independent RNA knockdown of viral sequences in mosquito cells following guide RNA expression. Commun. Biol. 2020, 3, 413. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Techakriengkrai, N.; Nedumpun, T.; Suradhat, S. Abrogation of PRRSV infectivity by CRISPR-Cas13b-mediated viral RNA cleavage in mammalian cells. Sci. Rep. 2020, 10, 9617. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.U.; Salman, H.M.; Khalid, M.F.; Khan, M.H.F.; Anwar, S.; Afzal, S.; Idrees, M.; Chaudhary, S.U. CRISPR-Cas13a mediated targeting of hepatitis C virus internal-ribosomal entry site (IRES) as an effective antiviral strategy. Biomed. Pharmacother. 2021, 136, 111239. [Google Scholar] [CrossRef] [PubMed]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Programmable Inhibition and Detection of RNA Viruses Using Cas13. Mol. Cell 2019, 76, 826–837.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; He, Y.; Hu, Y.S.; Luo, Z.F.; Zhang, J.J. A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem. Sci. 2020, 11, 12157–12164. [Google Scholar] [CrossRef]
- Liu, R.; Hu, Y.S.; He, Y.; Lan, T.; Zhang, J.J. Translating Daily COVID-19 Screening into a Simple Glucose Test: A Proof of Concept Study. Chem. Sci. 2021, 12, 9022–9030. [Google Scholar] [CrossRef]
- Zhang, J.J.; Lan, T.; Lu, Y. Overcoming Major Barriers to Developing Successful Sensors for Practical Applications Using Functional Nucleic Acids. Annu. Rev. Anal. Chem. 2022, 15, 3.1–3.21. [Google Scholar] [CrossRef]
- Nguyen, P.Q.; Soenksen, L.R.; Donghia, N.M.; Angenent-Mari, N.M.; de Puig, H.; Huang, A.; Lee, R.; Slomovic, S.; Galbersanini, T.; Lansberry, G.; et al. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat. Biotechnol. 2021, 39, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Daniels, J.; Wadekar, S.; DeCubellis, K.; Jackson, G.W.; Chiu, A.S.; Pagneux, Q.; Saada, H.; Engelmann, I.; Ogiez, J.; Loze-Warot, D.; et al. A mask-based diagnostic platform for point-of-care screening of Covid-19. Biosens. Bioelectron. 2021, 192, 113486. [Google Scholar] [CrossRef] [PubMed]

| Virus | Gene Targets | LoD | Reference |
|---|---|---|---|
| SARS-CoV-2 | S gene | 42 copies per reaction | SHERLOCK [32] |
| ORF1a | 10 cp/µL using a fluorescent readout; 100cp/μL using the lateral-flow-based colorimetric readout | SHINE [33] | |
| N gene | 30 copies/µL using a fluorescence plate reader; 100 cp/µL using mobile phone microscopy | [34] | |
| Total RNA | 31 copies/µL | FINF-IT [35] | |
| Total RNA | Not mentioned | CARMEN [36] | |
| Total RNA | 100 copies/µL | mCARMEN [37] | |
| N gene and ORF gene | 5 copies/µL | [38] | |
| S gene and N gene | 100 copies/µL | SENSR [39] | |
| Dengue virus | Total RNA | 2 aM | [40] |
| Total RNA Total RNA | 1 copy/µL 0.78 fM | [41] [42] | |
| HIV-1 | Not mentioned | Not mentioned | [44] |
| H7N9 | HA gene | 1 fM | [45] |
| PRRSV | M gene | 172 copies/µL | [46] |
| Ebola virus | Total RNA | 20 pfu/mL | [47] |
| Virus | Cas | Gene Targets | Knockdown Efficiency | Reference |
|---|---|---|---|---|
| SARS-CoV-2 | Cas13d | RdRP and N gene regions | 90% | [49] |
| Cas13a | S gene | 99% | [50] | |
| Cas13X | RdRP and E gene | 70% | [23] | |
| Dengue virus | Cas13a | NS3 gene | 95% | [51] |
| Cas13b | NS5 gene | 90% | [52] | |
| HIV-1 | Cas13a | LTR, gag, tat, and rev regions | 50~80% | [53] |
| Cas13d | Gag, pol, prot, int, cPPT, and CTS regions | 90% | [54] | |
| CHIKV | Cas13b | nsP2 gene | 35~50% | [55] |
| PRRSV | Cas13b | ORF5 and ORF7 genes | 55~70% | [56] |
| HCV | Cas13a | IRES | 70~84% | [57] |
| LCMV | Cas13a | L and S segments | 83.33% | [58] |
| IAV | Cas13b | mRNA and the complementary viral RNA | >85% | [58] |
| VSV | Cas13b | Single linear segment | >85% | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, Y.; Chen, Z.; Zhang, W.; Zhang, J. Engineering CRISPR/Cas13 System against RNA Viruses: From Diagnostics to Therapeutics. Bioengineering 2022, 9, 291. https://doi.org/10.3390/bioengineering9070291
Xue Y, Chen Z, Zhang W, Zhang J. Engineering CRISPR/Cas13 System against RNA Viruses: From Diagnostics to Therapeutics. Bioengineering. 2022; 9(7):291. https://doi.org/10.3390/bioengineering9070291
Chicago/Turabian StyleXue, Yi, Zhenzhen Chen, Wenxian Zhang, and Jingjing Zhang. 2022. "Engineering CRISPR/Cas13 System against RNA Viruses: From Diagnostics to Therapeutics" Bioengineering 9, no. 7: 291. https://doi.org/10.3390/bioengineering9070291
APA StyleXue, Y., Chen, Z., Zhang, W., & Zhang, J. (2022). Engineering CRISPR/Cas13 System against RNA Viruses: From Diagnostics to Therapeutics. Bioengineering, 9(7), 291. https://doi.org/10.3390/bioengineering9070291







