From the Field to the Bottle—An Integrated Strategy for Wine Authenticity
Abstract
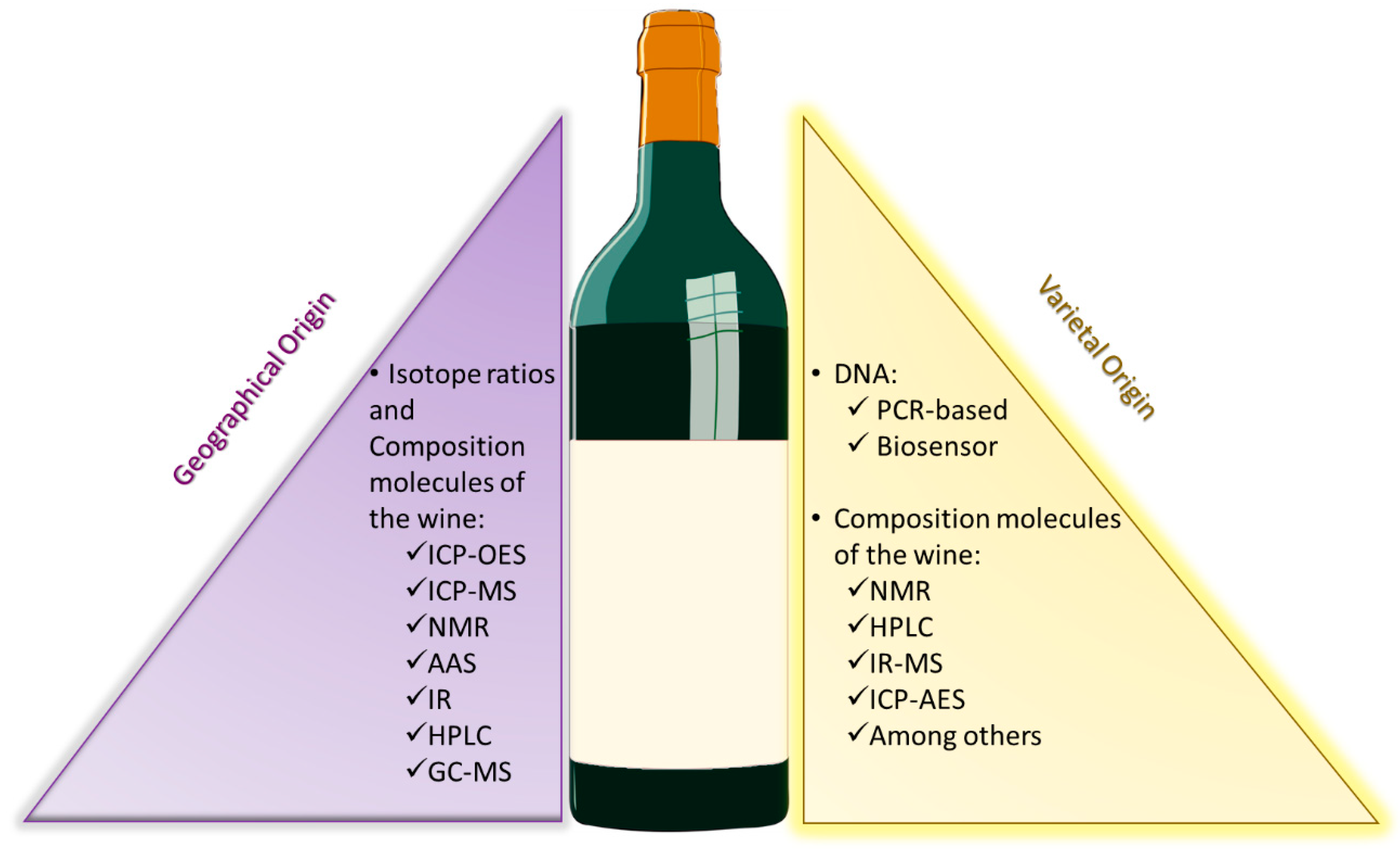
:1. Wine Authenticity
2. The Importance of an Integrated Strategy
3. Determination of the Region of Provenience
4. DNA Fingerprinting for Varietal Identification
5. Conclusions and Future Trends
Author Contributions
Funding
Conflicts of Interest
References
- Arvanitoyannis, I.S. Wine authenticity, traceability and safety monitoring. In Managing Wine Quality; Reynolds, A., Ed.; Woodhead: New York, NY, USA, 2010; pp. 218–270. ISBN 978-1-84569-484-5. [Google Scholar]
- Versari, A.; Laurie, V.F.; Ricci, A.; Laghi, L.; Parpinello, G.P. Progress in authentication, typification and traceability of grapes and wines by chemometric approaches. Food Res. Int. 2014, 60, 2–18. [Google Scholar] [CrossRef]
- Palade, M.; Popa, M.-E. Wine traceability and authenticity—A literature review. Sci. Bull. Ser. F Biotechnol. 2014, 18, 226–233. [Google Scholar]
- Pereira, L.; Martins-Lopes, P.; Batista, C.; Zanol, G.C.; Clímaco, P.; Brazão, J.; Eiras-Dias, J.E.; Guedes-Pinto, H. Molecular markers for assessing must varietal origin. Food Anal. Methods 2012, 5, 1252–1259. [Google Scholar] [CrossRef]
- Moreno-Arribas, M.V.; Polo, M.C. Winemaking biochemistry and microbiology: Current knowledge and future trends. Crit. Rev. Food Sci. Nutr. 2005, 45, 265–286. [Google Scholar] [CrossRef] [PubMed]
- Vukatana, K.; Sevrani, K.; Hoxha, E. Wine traceability: A data model and prototype in albanian context. Foods 2016, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Villano, C.; Lisanti, M.T.; Gambuti, A.; Vecchio, R.; Moio, L.; Frusciante, L.; Aversano, R.; Carputo, D. Wine varietal authentication based on phenolics, volatiles and DNA markers: State of the art, perspectives and drawbacks. Food Control 2017, 80, 1–10. [Google Scholar] [CrossRef]
- Pereira, L.; Gomes, S.; Barrias, S.; Fernandes, J.R.; Martins-Lopes, P. Applying high-resolution melting (HRM) technology to olive oil and wine authenticity. Food Res. Int. 2018, 103, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Sotirchos, D.G.; Danezis, G.P.; Georgiou, C.A. Introduction, Definitions and Legislation. In Food Authentication: Management, Analysis and Regulation; Georgiou, C.A., Danezis, G.P., Eds.; Wiley-Blackwell: West Sussex, UK, 2017; pp. 3–18. ISBN 978-1-118-81026-2. [Google Scholar]
- OIV. International Standard for the Labelling of Wines; Organisation Internationale de la Vigne et du Vin: Paris, France, 2015. [Google Scholar]
- Vineyards. Available online: https://vineyards.com (accessed on 31 July 2018).
- OIV. Distribution of the World’s Grapevine Varieties; Organisation Internationale de la Vigne et du Vin: Paris, France, 2017. [Google Scholar]
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food authentication: Techniques, trends & emerging approaches. Trends Anal. Chem. 2016, 85, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Basalekou, M.; Stratidaki, A.; Pappas, C.; Tarantilis, P.; Kotseridis, Y.; Kallithraka, S. Authenticity determination of greek-cretan mono-varietal white and red wines based on their phenolic content using Attenuated Total Reflectance Fourier Transform Infrared spectroscopy and chemometrics. Curr. Res. Nutr. Food Sci. J. 2016, 4, 54–62. [Google Scholar] [CrossRef]
- Lohumi, S.; Lee, S.; Lee, H.; Cho, B.-K. A review of vibrational spectroscopic techniques for the detection of food authenticity and adulteration. Trends Food Sci. Technol. 2015, 46, 85–98. [Google Scholar] [CrossRef]
- Perestrelo, R.; Silva, C.; Câmara, J.S. A useful approach for the differentiation of wines according to geographical origin based on global volatile patterns. J. Sep. Sci. 2014, 37, 1974–1981. [Google Scholar] [CrossRef] [PubMed]
- Duchowicz, P.R.; Giraudo, M.A.; Castro, E.A.; Pomilio, A.B. Amino acid profiles and quantitative structure-property relationship models as markers for Merlot and Torrontés wines. Food Chem. 2013, 140, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Rešetar, D.; Marchetti-Deschmann, M.; Allmaier, G.; Katalinić, J.P.; Kraljević Pavelić, S. Matrix assisted laser desorption ionization mass spectrometry linear time-of-flight method for white wine fingerprinting and classification. Food Control 2016, 64, 157–164. [Google Scholar] [CrossRef]
- González-Neves, G.; Favre, G.; Piccardo, D.; Gil, G. Anthocyanin profile of young red wines of Tannat, Syrah and Merlot made using maceration enzymes and cold soak. Int. J. Food Sci. Technol. 2016, 51, 260–267. [Google Scholar] [CrossRef]
- Kokkinofta, R.; Fotakis, C.; Zervou, M.; Zoumpoulakis, P.; Savvidou, C.; Poulli, K.; Louka, C.; Economidou, N.; Tzioni, E.; Damianou, K.; et al. Isotopic and elemental authenticity markers: A case study on cypriot wines. Food Anal. Methods 2017, 10, 3902–3913. [Google Scholar] [CrossRef]
- Pereira, L.; Guedes-Pinto, H.; Martins-Lopes, P. An enhanced method for Vitis vinifera L. DNA extraction from wines. Am. J. Enol. Vitic. 2011, 62, 547–552. [Google Scholar] [CrossRef]
- Catalano, V.; Moreno-Sanz, P.; Lorenzi, S.; Grando, M.S. Experimental review of DNA-based methods for wine traceability and development of a single-nucleotide polymorphism (SNP) genotyping assay for quantitative varietal authentication. J. Agric. Food Chem. 2016, 64, 6969–6984. [Google Scholar] [CrossRef] [PubMed]
- OIV. 2nd Edition of the OIV Descriptor List for Grape Varieties and Vitis Species; OIV: Paris, France, 2007. [Google Scholar]
- Cabezas, J.A.; Ibáñez, J.; Lijavetzky, D.; Vélez, D.; Bravo, G.; Rodríguez, V.; Carreño, I.; Jermakow, A.M.; Carreño, J.; Ruiz-García, L.; et al. A 48 SNP set for grapevine cultivar identification. BMC Plant Biol. 2011, 11, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- REGULATION (EC) No 178/2002 of the European Parliament and of the Council of 28 January 2002 laying down the general principles and requirements of food law, establishing the European Food Safety Authority and laying down procedures in matters of food safety. Off. J. Eur. Commun. 2002, 31, 1–24.
- Gomes, S.; Castro, C.; Barrias, S.; Pereira, L.; Jorge, P.; Fernandes, J.R.; Martins-Lopes, P. Alternative SNP detection platforms, HRM and biosensors, for varietal identification in Vitis vinifera L. using F3H and LDOX genes. Sci. Rep. 2018, 8, 5850. [Google Scholar] [CrossRef] [PubMed]
- Barrias, S.; Fernandes, J.R.; Eiras-Dias, J.E.; Brazão, J.; Martins-Lopes, P. Label free DNA-based optical biosensor as a potential system for wine authenticity. Food Chem. 2019, 270, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Martins, P.; Madeira, M.; Monteiro, F.; Bruno de Sousa, R.; Curvelo-Garcia, A.S.; Catarino, S. 87Sr/86Sr ratio in vineyard soils from Portuguese denominations of origin and its potential for origin authentication. J. Int. Sci. Vigne Vin 2014, 48, 21–29. [Google Scholar] [CrossRef]
- Esslinger, S.; Riedl, J.; Fauhl-Hassek, C. Potential and limitations of non-targeted fingerprinting for authentication of food in official control. Food Res. Int. 2014, 60, 189–204. [Google Scholar] [CrossRef]
- Fang, F.; Li, J.-M.; Zhang, P.; Tang, K.; Wang, W.; Pan, Q.-H.; Huang, W.-D. Effects of grape variety, harvest date, fermentation vessel and wine ageing on flavonoid concentration in red wines. Food Res. Int. 2008, 41, 53–60. [Google Scholar] [CrossRef]
- Fernandes, J.R.; Pereira, L.; Jorge, P.; Moreira, L.; Gonçalves, H.; Coelho, L.; Alexandre, D.; Eiras-Dias, J.; Brazão, J.; Clímaco, P.; et al. Wine fingerprinting using a bio-geochemical approach. BIO Web Conf. 2015, 5, 02021. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. MBio 2016, 7, e00631-16. [Google Scholar] [CrossRef] [PubMed]
- Vinciguerra, V.; Stevenson, R.; Pedneault, K.; Poirier, A.; Hélie, J.-F.; Widory, D. Strontium isotope characterization of wines from Quebec, Canada. Food Chem. 2016, 210, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Zarraonaindia, I.; Perisin, M.; Palacios, A.; Acedo, A. From vineyard soil to wine fermentation: microbiome approximations to explain the “terroir” concept. Front. Microbiol. 2017, 8, 821. [Google Scholar] [CrossRef] [PubMed]
- Luykx, D.M.A.M.; van Ruth, S.M. An overview of analytical methods for determining the geographical origin of food products. Food Chem. 2008, 107, 897–911. [Google Scholar] [CrossRef]
- Petrini, R.; Sansone, L.; Slejko, F.F.; Buccianti, A.; Marcuzzo, P.; Tomasi, D. The 87Sr/86Sr strontium isotopic systematics applied to Glera vineyards: A tracer for the geographical origin of the Prosecco. Food Chem. 2015, 170, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.; Heaton, K.; Hoogewerff, J. Tracing the geographical origin of food: The application of multi-element and multi-isotope analysis. Trends Food Sci. Technol. 2005, 16, 555–567. [Google Scholar] [CrossRef]
- Rodríguez-Delgado, M.-Á.; González-Hernández, G.; Conde-González, J.-E.; Pérez-Trujillo, J.-P. Principal component analysis of the polyphenol content in young red wines. Food Chem. 2002, 78, 523–532. [Google Scholar] [CrossRef]
- Nakano, T. Potential uses of stable isotope ratios of Sr, Nd, and Pb in geological materials for environmental studies. Proc. Jpn. Acad. Ser. B 2016, 92, 167–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, M.P.; Zhang, B.; Martin, G.J. Determination of the geographical origin of wine using joint analysis of elemental and isotopic composition. II—Differentiation of the principal production zones in france for the 1990 vintage. J. Sci. Food Agric. 1995, 67, 113–123. [Google Scholar] [CrossRef]
- Braschi, E.; Marchionni, S.; Priori, S.; Casalini, M.; Tommasini, S.; Natarelli, L.; Buccianti, A.; Bucelli, P.; Costantini, E.A.C.; Conticelli, S. Tracing the 87Sr/86Sr from rocks and soils to vine and wine: An experimental study on geologic and pedologic characterisation of vineyards using radiogenic isotope of heavy elements. Sci. Total Environ. 2018, 628–629, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Marchionni, S.; Buccianti, A.; Bollati, A.; Braschi, E.; Cifelli, F.; Molin, P.; Parotto, M.; Mattei, M.; Tommasini, S.; Conticelli, S. Conservation of 87Sr/86Sr isotopic ratios during the winemaking processes of “Red” wines to validate their use as geographic tracer. Food Chem. 2016, 190, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, L.A.; Moreira, M.Z.; Ometto, J.P.H.B.; Alcarde, A.R.; Rizzon, L.A.; Stange, E.; Ehleringer, J.R. Stable carbon isotopic composition of the wine and CO2 bubbles of sparkling wines: Detecting C4 sugar additions. J. Agric. Food Chem. 2003, 51, 2625–2631. [Google Scholar] [CrossRef] [PubMed]
- Marchionni, S.; Braschi, E.; Tommasini, S.; Bollati, A.; Cifelli, F.; Mulinacci, N.; Mattei, M.; Conticelli, S. High-precision 87Sr/86Sr analyses in wines and their use as a geological fingerprint for tracing geographic provenance. J. Agric. Food Chem. 2013, 61, 6822–6831. [Google Scholar] [CrossRef] [PubMed]
- Durante, C.; Baschieri, C.; Bertacchini, L.; Bertelli, D.; Cocchi, M.; Marchetti, A.; Manzini, D.; Papotti, G.; Sighinolfi, S. An analytical approach to Sr isotope ratio determination in Lambrusco wines for geographical traceability purposes. Food Chem. 2015, 173, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, P.P.; Steffens, F.E.; Eiselen, R.J.; Augustyn, O.P.; Balcaen, L.; Vanhaecke, F. Multi-element analysis of South African wines by ICP−MS and their classification according to geographical origin. J. Agric. Food Chem. 2005, 53, 5060–5066. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, M.J.; Martins, N.; Barrulas, P.; Garcia, R.; Dias, C.B.; Pérez-Álvarez, E.P.; Costa Freitas, A.M.; Garde-Cerdán, T. Multi-element composition of red, white and palhete amphora wines from Alentejo by ICPMS. Food Control 2018, 92, 80–85. [Google Scholar] [CrossRef]
- Fan, S.; Zhong, Q.; Gao, H.; Wang, D.; Li, G.; Huang, Z. Elemental profile and oxygen isotope ratio (δ18O) for verifying the geographical origin of Chinese wines. J. Food Drug Anal. 2018, 26, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.-F.; Peng, Z.-X.; Du, H.-J.; Duan, C.-Q.; Reeves, M.J.; Pan, Q.-H. Elemental patterns of wines, grapes, and vineyard soils from Chinese wine-producing regions and their association. Am. J. Enol. Vitic. 2012, 63, 232–240. [Google Scholar] [CrossRef]
- Azcarate, S.M.; Martinez, L.D.; Savio, M.; Camiña, J.M.; Gil, R.A. Classification of monovarietal Argentinean white wines by their elemental profile. Food Control 2015, 57, 268–274. [Google Scholar] [CrossRef]
- Martin, G.J.; Mazure, M.; Jouitteau, C.; Martin, Y.-L.; Aguile, L.; Allain, P. Characterization of the geographic origin of Bordeaux wines by a combined use of isotopic and trace element Measurements. Am. J. Enol. Vitic. 1999, 50, 409–417. [Google Scholar]
- Ogrinc, N.; Košir, I.J.; Kocjančič, M.; Kidrič, J. Determination of authenticy, regional origin, and vintage of Slovenian wines using a combination of IRMS and SNIF-NMR analyses. J. Agric. Food Chem. 2001, 49, 1432–1440. [Google Scholar] [CrossRef] [PubMed]
- Ogrinc, N.; Košir, I.J.; Spangenberg, J.E.; Kidrič, J. The application of NMR and MS methods for detection of adulteration of wine, fruit juices, and olive oil. A review. Anal. Bioanal. Chem. 2003, 376, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Košir, I.J.; Kocjančič, M.; Ogrinc, N.; Kidrič, J. Use of SNIF-NMR and IRMS in combination with chemometric methods for the determination of chaptalisation and geographical origin of wines (the example of Slovenian wines). Anal. Chim. Acta 2001, 429, 195–206. [Google Scholar] [CrossRef]
- Brescia, M.A.; Košir, I.J.; Caldarola, V.; Kidrič, J.; Sacco, A. Chemometric classification of apulian and slovenian wines using 1H NMR and ICP-OES together with HPICE data. J. Agric. Food Chem. 2003, 51, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Picque, D.; Cattenoz, T.; Corrieu, G.; Berger, J. Discrimination of red wines according to their geographical origin and vintage year by the use of mid-infrared spectroscopy. Sci. Aliments 2005, 25, 207–220. [Google Scholar] [CrossRef]
- Liu, L.; Cozzolino, D.; Cynkar, W.U.; Gishen, M.; Colby, C.B. Geographic classification of spanish and australian Tempranillo red wines by visible and near-infrared spectroscopy combined with multivariate analysis. J. Agric. Food Chem. 2006, 54, 6754–6759. [Google Scholar] [CrossRef] [PubMed]
- Frías, S.; Trujillo, J.P.; Peña, E.; Conde, J.E. Classification and differentiation of bottled sweet wines of Canary Islands (Spain) by their metallic content. Eur. Food Res. Technol. 2001, 213, 145–149. [Google Scholar] [CrossRef]
- Makris, D.P.; Kallithraka, S.; Mamalos, A. Differentiation of young red wines based on cultivar and geographical origin with application of chemometrics of principal polyphenolic constituents. Talanta 2006, 70, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Pavloušek, P.; Kumšta, M. Authentication of Riesling wines from the Czech Republic on the basis of the non-flavonoid phenolic compounds. Czech J. Food Sci. 2013, 31, 474–482. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Lourido, D.; Saurina, J.; Hernández-Cassou, S.; Checa, A. Classification and characterisation of Spanish red wines according to their appellation of origin based on chromatographic profiles and chemometric data analysis. Food Chem. 2012, 135, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
- Ziółkowska, A.; Wąsowicz, E.; Jeleń, H.H. Differentiation of wines according to grape variety and geographical origin based on volatiles profiling using SPME-MS and SPME-GC/MS methods. Food Chem. 2016, 213, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Etièvant, P.; Schlich, P.; Cantagrel, R.; Bertrand, M.; Bouvier, J.-C. Varietal and geographic classification of french red wines in terms of major acids. J. Sci. Food Agric. 1989, 46, 421–438. [Google Scholar] [CrossRef]
- Robinson, A.L.; Adams, D.O.; Boss, P.K.; Heymann, H.; Solomon, P.S.; Trengove, R.D. Influence of geographic origin on the sensory characteristics and wine composition of Vitis vinifera cv. Cabernet Sauvignon wines from Australia. Am. J. Enol. Vitic. 2012, 63, 467–476. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, F.; Ye, J. Quantitative and qualitative analysis of flavonoid markers in Frucus aurantii of different geographical origin by capillary electrophoresis with electrochemical detection. J. Chromatogr. B 2006, 830, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Ohta, M.; Richardson, P.M.; Mills, D.A. Monitoring seasonal changes in winery-resident microbiota. PLoS ONE 2013, 8, e66437. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.W.; Tsai, P.; Anfang, N.; Ross, H.A.; Goddard, M.R. Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 2014, 16, 2848–2858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuller, D.; Cardoso, F.; Sousa, S.; Gomes, P.; Gomes, A.C.; Santos, M.A.S.; Casal, M. Genetic diversity and population structure of Saccharomyces cerevisiae strains isolated from different grape varieties and winemaking regions. PLoS ONE 2012, 7, e32507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drumonde-Neves, J.; Franco-Duarte, R.; Lima, T.; Schuller, D.; Pais, C. Association between grape yeast communities and the vineyard ecosystems. PLoS ONE 2017, 12, e0169883. [Google Scholar] [CrossRef] [PubMed]
- Buratti, S.; Benedetti, S.; Scampicchio, M.; Pangerod, E.C. Characterization and classification of Italian Barbera wines by using an electronic nose and an amperometric electronic tongue. Anal. Chim. Acta 2004, 525, 133–139. [Google Scholar] [CrossRef]
- Beltrán, N.H.; Duarte-Mermoud, M.A.; Muñoz, R.E. Geographical classification of Chilean wines by an electronic nose. Int. J. Wine Res. 2009, 2009, 209–219. [Google Scholar] [CrossRef]
- Knight, S.; Klaere, S.; Fedrizzi, B.; Goddard, M.R. Regional microbial signatures positively correlate with differential wine phenotypes: Evidence for a microbial aspect to terroir. Sci. Rep. 2015, 5, 14233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, C.; Pinho, D.; Cardoso, R.; Custódio, V.; Fernandes, J.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Wine fermentation microbiome: A landscape from different Portuguese wine appellations. Front. Microbiol. 2015, 6, 905. [Google Scholar] [CrossRef] [PubMed]
- Mezzasalma, V.; Ganopoulos, I.; Galimberti, A.; Cornara, L.; Ferri, E.; Labra, M. Poisonous or non-poisonous plants? DNA-based tools and applications for accurate identification. Int. J. Leg. Med. 2017, 131, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bigliazzi, J.; Scali, M.; Paolucci, E.; Cresti, M.; Vignani, R. DNA extracted with optimized protocols can be genotyped to reconstruct the varietal composition of monovarietal wines. Am. J. Enol. Vitic. 2012, 63, 568–573. [Google Scholar] [CrossRef]
- Fatima, F.; Pathak, N.; Rastogi Verma, S. An improved method for soil DNA extraction to study the microbial assortment within rhizospheric region. Mol. Biol. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, M.A.; Ye, G.-N.; Weeden, N.F.; Reisch, B.I. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 1994, 12, 6–13. [Google Scholar] [CrossRef]
- Baleiras-Couto, M.M.; Eiras-Dias, J.E. Detection and identification of grape varieties in must and wine using nuclear and chloroplast microsatellite markers. Anal. Chim. Acta 2006, 563, 283–291. [Google Scholar] [CrossRef]
- Işçi, B.; Kalkan Yildirim, H.; Altindisli, A. Evaluation of methods for DNA extraction from must and wine. J. Inst. Brew. 2014, 120, 238–243. [Google Scholar] [CrossRef] [Green Version]
- Pereira, L.; Gomes, S.; Castro, C.; Eiras-Dias, J.E.; Brazão, J.; Graça, A.; Fernandes, J.R.; Martins-Lopes, P. High Resolution Melting (HRM) applied to wine authenticity. Food Chem. 2017, 216, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Barcaccia, G.; Lucchin, M.; Cassandro, M. DNA barcoding as a molecular tool to track down mislabeling and food piracy. Diversity 2015, 8, 2. [Google Scholar] [CrossRef]
- Valentini, P.; Galimberti, A.; Mezzasalma, V.; De Mattia, F.; Casiraghi, M.; Labra, M.; Pompa, P.P. DNA barcoding meets nanotechnology: Development of a universal colorimetric test for food authentication. Angew. Chem. Int. Ed. 2017, 56, 8094–8098. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Martins-Lopes, P. Vitis vinifera L. Single-nucleotide polymorphism detection with high-resolution melting analysis based on the UDP-glucose:Flavonoid 3-O-Glucosyltransferase gene. J. Agric. Food Chem. 2015, 63, 9165–9174. [Google Scholar] [CrossRef] [PubMed]
- Giusti, A.; Armani, A.; Sotelo, C.G. Advances in the analysis of complex food matrices: Species identification in surimi-based products using next generation sequencing technologies. PLoS ONE 2017, 12, e0185586. [Google Scholar] [CrossRef] [PubMed]
- Tillmar, A.O.; Dell’Amico, B.; Welander, J.; Holmlund, G. A universal method for species identification of mammals utilizing next generation sequencing for the analysis of DNA mixtures. PLoS ONE 2013, 8, e83761. [Google Scholar] [CrossRef] [PubMed]
- Navarro, E.; Serrano-Heras, G.; Castaño, M.J.; Solera, J. Real-time PCR detection chemistry. Clin. Chim. Acta 2015, 439, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Salihah, N.T.; Hossain, M.M.; Lubis, H.; Ahmed, M.U. Trends and advances in food analysis by real-time polymerase chain reaction. J. Food Sci. Technol. 2016, 53, 2196–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; García-Fernández, D.; Mas, A.; Esteve-Zarzoso, B. Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front. Microbiol. 2015, 6, 1156. [Google Scholar] [CrossRef] [PubMed]
- Scarano, D.; Rao, R. DNA markers for food products authentication. Diversity 2014, 6, 579–596. [Google Scholar] [CrossRef]
- Mendes-Ferreira, A.; lí del Olmo, M.; García-Martínez, J.; Pérez-Ortín, J.E. Functional genomics in wine yeast: DNA arrays and next generation sequencing. In Biology of Microorganisms on Grapes, in Must and in Wine; König, H., Unden, G., Fröhlich, J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 573–604. ISBN 978-3-319-60021-5. [Google Scholar] [CrossRef]
- Consolandi, C.; Palmieri, L.; Severgnini, M.; Maestri, E.; Marmiroli, N.; Agrimonti, C.; Baldoni, L.; Donini, P.; De Bellis, G.; Castiglioni, B. A procedure for olive oil traceability and authenticity: DNA extraction, multiplex PCR and LDR–universal array analysis. Eur. Food Res. Technol. 2008, 227, 1429–1438. [Google Scholar] [CrossRef]
- Nummelin, S.; Kommeri, J.; Kostiainen, M.A.; Linko, V. Evolution of structural DNA nanotechnology. Adv. Mater. 2018, 30, 1703721. [Google Scholar] [CrossRef] [PubMed]
- Stephen Inbaraj, B.; Chen, B.H. Nanomaterial-based sensors for detection of foodborne bacterial pathogens and toxins as well as pork adulteration in meat products. J. Food Drug Anal. 2016, 24, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, P. Biosensors and their applications—A review. J. Oral Biol. Craniofacial Res. 2016, 6, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, H.M.R.; Moreira, L.; Pereira, L.; Jorge, P.; Gouveia, C.; Martins-Lopes, P.; Fernandes, J.R.A. Biosensor for label-free DNA quantification based on functionalized LPGs. Biosens. Bioelectron. 2016, 84, 30–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, L.; Gonçalves, H.M.R.; Pereira, L.; Castro, C.; Jorge, P.; Gouveia, C.; Fernandes, J.R.; Martins-Lopes, P. Label-free optical biosensor for direct complex DNA detection using Vitis vinifera L. Sens. Actuators B Chem. 2016, 234, 92–97. [Google Scholar] [CrossRef]


| Samples | Analytical Technique | Data Analyzed/Analyte | Purpose of Analysis | References |
|---|---|---|---|---|
| Mass Spectrometry | ||||
| Grape, wine, and soil | IR-MS | 87SR/86SR | Geographic origin of wine from Canada | [33] |
| Rocks, soils, and wine | IR-MS | 87SR/86SR | Geologic and pedologic traceability of Italian wines | [41] |
| Red wines, musts grape juices, soils, and rocks | IR-MS | 87SR/86SR | Fingerprinting wine geographic provenance. | [42] |
| Musts, soils, and grape components (skin, seeds, must, and stem) | TIMS and XRD spectra | 87SR/86SR | Geographic traceability study of Italian white wine labelled with the Controlled Designation of Origin (DOC) | [36] |
| Sparkling wines | IR-MS | δ13C | The δ13C evaluation in the sparkling wines to detect adulteration—wines chaptalization | [43] |
| Wines and rocks | TIMS | 87SR/86SR | Radiogenic isotopic evaluation for tracing geographic provenance of wines | [44] |
| Soils, grapes, and wines | AAS, IR-MS, MC-ICP-MS | δ18O, (D/H)I, (D/H)II, δ13C, δ15N, and 87Sr/86Sr | Development of a geographical traceability model | [45] |
| Vineyard soils | ICP-MS | 87SR/86SR | Evaluation of 87Sr/86Sr ratio in vineyard soils from Portuguese Denominations of Origin and its potential for origin authentication | [28] |
| Wines | ICP−MS and multi-element analysis | Li, B, Mg, Al, Si, Cl, Sc, Mn, Ni, Ga, Se, Rb, Sr, Nb, Cs, Ba, La, W, Tl, and U | South African wines classification according to geographical origin | [46] |
| Red, white, and palhete amphora wines | ICP-MS | Mineral content | Elemental composition characterization of Alentejo wines to establish the geographic origin | [47] |
| Wine | ICP-MS, ICP-OES and IRMS | Elemental profile (Ca, Al, Mg, B, Fe, K, Rb, Mn, Na, P, Co, Ga, As, Sr), and Isotope ratio (δ18O) | Geographical origin of Chinese wines | [48] |
| Soils, grapes, and wines | ICP-MS | Cr, Co, Ni, Ga, Se, Y, Zr, Nb, Mo, Pd, In, La, Pr, Sm, Eu, Gd, Tm, Yb, Au, Tl, Th, U | Elemental patterns of wines, grapes, and vineyard soils from Chinese wine-producing regions and their origin association | [49] |
| Monovarietal wines | ICP-MS | Ba, As, Pb, Mo, and Co | Geographical origin differentiation of Argentinean white wines by their elemental profile | [50] |
| Spectroscopy | ||||
| Wines | SNIF-NMR | Isotopic and trace elements | Characterization of the geographic origin of Bordeaux wines | [51] |
| Wines | IRMS and SNIF-NMR | Isotopic ratios hydrogen (2H/1H), carbon (13C/12C), nitrogen (15N/14N), oxygen (18O/16O) | Regional origin discrimination of Slovenian Wines | [52] |
| Wines | NMR and MS | Cd, Cr, Cs, Er, Ga, Mn, and Sr | Wine adulterations | [53] |
| Wines | SNIF-NMR and IRMS in combination with chemometric | Multielement analysis | Geographical origin | [54] |
| Wines authentication | 1H NMR, ICP-AES, HPIC | 1H and 13C | Classification of wines from Slovenia and from Apulia | [55] |
| Red wines | MIR | Multielement analysis | Discrimination of wines based on their geographical origin and vintage year | [56] |
| Red wines | NIR combined with multivariate analysis (PCA, PLS-DA, LDA) | Chemometrics | Geographic classification of Spanish and Australian tempranillo wines | [57] |
| Sweet wines | F-AAS | Metallic content (Na, K, Ca, Mg, Fe, and Cu) | Classification and geographical differentiation of wines from Canary Islands (Spain) | [58] |
| Separation | ||||
| Red wines | HPLC, UV, and fluorescence detection | Polyphenol content | Polyphenolic compounds quantification to typify wines according to their geographical origin | [38] |
| Red wines | HPLC-DAD | Polyphenolic components | Red wines differentiation based on cultivar and geographical origin with application of chemometrics of principal polyphenolic constituents | [59] |
| Monovarietal wines | HPLC | Non-flavonoid phenolic compounds: hydroxybenzoic acids, hydroxycinnamates, and Stilbenes | Czech Republic wines authentication: Wine discrimination according to the geographical origin | [60] |
| Red wines | RP-HPLC-DAD-F | Chromatographic profiles and chemometric data analysis | Classification and characterization of Spanish wines according to their appellation of origin. | [61] |
| Monovarietal red and white wines | SPME-MS and SPME-GC/MS | Volatiles compounds | Differentiation of wines according to grape variety and geographical origin | [62] |
| Red wines | HPLC | Organic acids (Shikimic and galacturonic acids); plenolic compounds (e.g., alkanes, aldehydes, alcohols, acids). | Varietal and geographic classification of wines according to their geographical origin | [63] |
| HS-SPME GC×GC-TOFMS | Volatile compounds | [64] | ||
| Red wines | CE | Metals content (Na, K, Ca, Mg, Mn, and Li) | Wines classification according to their geographical origin. | [65] |
| Others | ||||
| Must and grapes microbiota | DNA | High-throughput sequencing, molecular markers (SSR) | Biogeographical wines characteristics | [66] |
| Grapevines’ fungal communities | DNA | Pyrosequencing of the 26S rRNA gene region | Vine fungi biogeography | [67] |
| Grape varieties | DNA | Ribosomal ITS region | Geographical region and grape varieties are drivers of population structures of fermentative vineyard-associated S. cerevisiae strains | [68] |
| Grape yeast biota | DNA | RFLP and DNA sequencing | Azorean geographical indications wines: Grape-associated microbial biogeography from five islands of Azores Archipelago | [69] |
| Sensory | ||||
| Wine | Electronic nose and amperometric electronic tongue | Aroma | Characterization and classification of Italian Barbera wines | [70] |
| Wine | Electronic nose (fast gas chromatograph) | Aroma profile | Geographical classification of Chilean wines | [71] |
| Method | Pros | Cons |
|---|---|---|
| HRM |
|
|
| qPCR |
|
|
| dPCR |
|
|
| NGS |
|
|
| Biosensors Nanotechnology DNA chips |
| |
| DNA markers |
| |
| SSR |
|
|
| SNP |
|
|
| DNA barcoding |
|
|
| Data analysis |
|
|
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, L.; Gomes, S.; Barrias, S.; Gomes, E.P.; Baleiras-Couto, M.; Fernandes, J.R.; Martins-Lopes, P. From the Field to the Bottle—An Integrated Strategy for Wine Authenticity. Beverages 2018, 4, 71. https://doi.org/10.3390/beverages4040071
Pereira L, Gomes S, Barrias S, Gomes EP, Baleiras-Couto M, Fernandes JR, Martins-Lopes P. From the Field to the Bottle—An Integrated Strategy for Wine Authenticity. Beverages. 2018; 4(4):71. https://doi.org/10.3390/beverages4040071
Chicago/Turabian StylePereira, Leonor, Sónia Gomes, Sara Barrias, Elisa Preto Gomes, Margarida Baleiras-Couto, José Ramiro Fernandes, and Paula Martins-Lopes. 2018. "From the Field to the Bottle—An Integrated Strategy for Wine Authenticity" Beverages 4, no. 4: 71. https://doi.org/10.3390/beverages4040071





