Alcoholic Fermentation Monitoring and pH Prediction in Red and White Wine by Combining Spontaneous Raman Spectroscopy and Machine Learning Algorithms
Abstract
:1. Introduction
- Explore ways in which Raman spectroscopy can monitor the alcoholic fermentation of wine grapes;
- Address ways in which Raman signal obstruction due to fluorescence might be mitigated.
2. Materials and Methods
2.1. Chemicals
2.2. Sample Collection
2.3. Reference Analysis
2.4. Raman Analysis
2.5. Phenolic Reduction
2.6. Statistical Analysis
2.7. Software
3. Results and Discussion
3.1. Algorithm Comparison and Feature Selection
3.2. Ethanol and Total Sugar Model Performance
3.2.1. Post Fermentation Baseline Loss
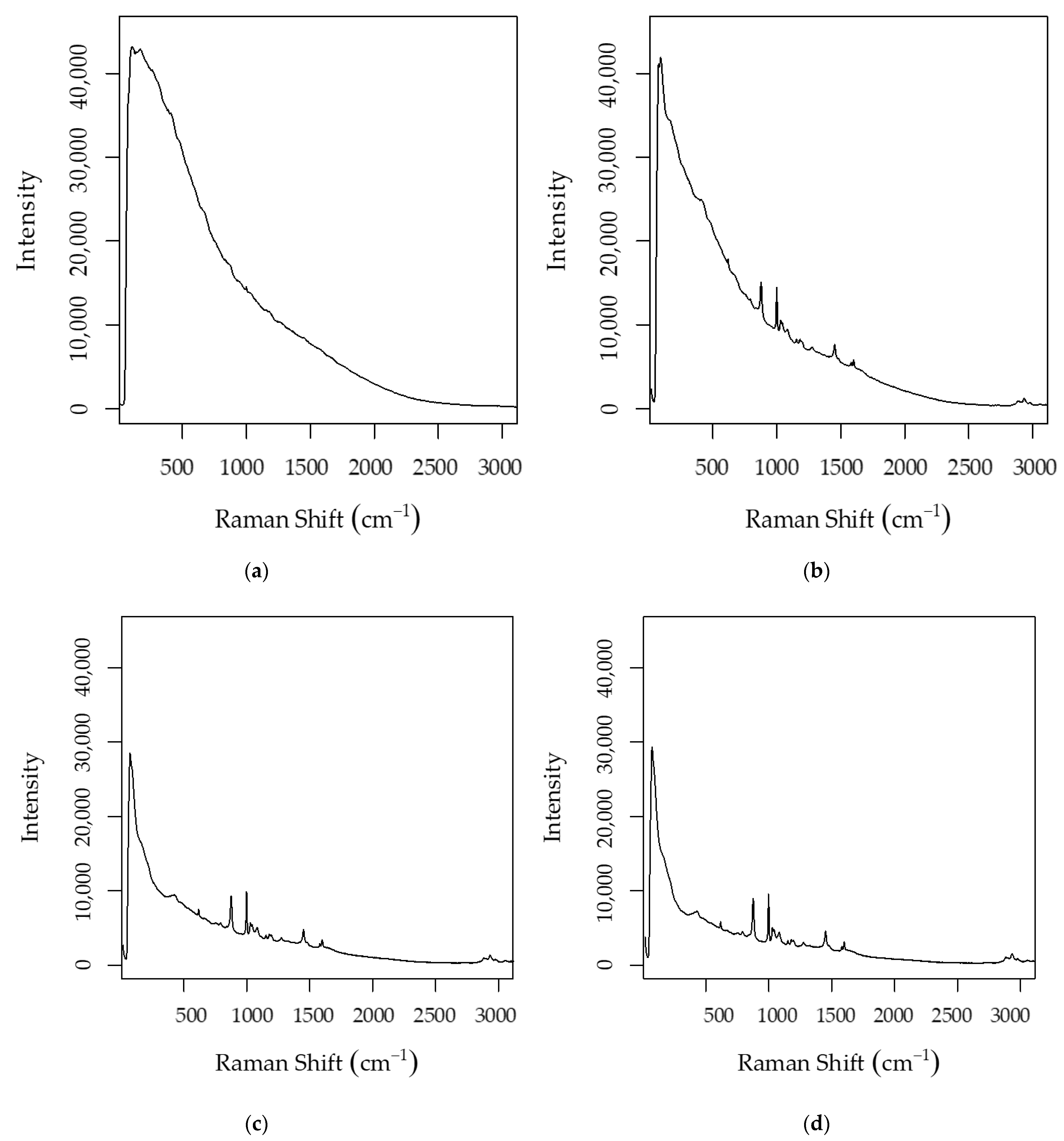
3.2.2. Fluorescence Reduction Using Polyvinylpolypyrrolidone (PVPP)
3.3. Limitations of Supervised Models
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Banerjee, S.C.V. Raman and colonial physics: Acoustics and the quantum. Phys. Perspect. 2014, 16, 146–178. [Google Scholar] [CrossRef]
- Raman, C.V.; Krishnan, K.S. A new type of secondary radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Radziemski, L.; Cremers, D. A Brief history of laser-Induced breakdown spectroscopy: From the concept of atoms to LIBS 2012. Spectrosc. Acta B 2013, 87, 3–10. [Google Scholar] [CrossRef]
- Huang, W.E.; Li, M.; Jarvis, B.M.; Goodacre, R.; Banwart, S.A. Shining light on the microbial world: The application of Raman microspectroscopy. Adv. Appl. Microbiol. 2010, 70, 153–186. [Google Scholar]
- Fini, G. Application of Raman spectroscopy in pharmacy. J. Raman Spectrosc. 2004, 35, 335–337. [Google Scholar] [CrossRef]
- Bunaciu, A.A.; Aboul-Enein, H.Y.; Hoang, V.D. Raman spectroscopy for protein analysis. Appl. Spectrosc. Rev. 2015, 50, 377–386. [Google Scholar] [CrossRef]
- Pyrak, E.; Kajczewski, J.; Kowalik, A.; Kubelski, A.; Jaworska, A. Surface enhanced Raman spectroscopy for DNA biosensors—How far are we? Molecules 2019, 24, 4423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strohm, E.M.; Moore, M.J.; Kolios, M.C. High resolution ultrasound and photoacoustic imaging of single cells. Photoacoustics 2016, 4, 36–42. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, A.; Larsen, R. Gemstone identification using Raman spectroscopy. Spectroscopy 2004, 19, 20–25. [Google Scholar]
- Nieuwoudt, M.K.; Shahlori, R.; Naot, D.; Patel, R.; Holtkamp, H.; Aguergaray, C.; Watson, M.; Musson, D.; Brown, C.; Dalbeth, N.; et al. Raman spectroscopy reveals age- and sex-related differences in cortical bone from people with osteoarthritis. Sci. Rep. 2020, 10, 19443. [Google Scholar] [CrossRef]
- Gomez-Alonso, S.; Garcia-Romero, E.; Hermosin-Gutierrez, I. HPLC analysis of diverse grape and wine phenolics using direct injection and multidetecion by DAD and fluorescence. J. Food Comp. Anal. 2007, 20, 618–626. [Google Scholar] [CrossRef]
- Cordella, C.; Moussa, I.; Martel, A.C.; Sbirrazzuoli, N.; Lizzani-Cuvelier, L. Recent developments in food characterization and adulteration detection: Technique-oriented perspectives. J. Agric. Food Chem. 2002, 50, 1751–1764. [Google Scholar] [CrossRef]
- Aleixandre-Tudo, J.L.; Buica, A.; Nieuwoudt, H.; Aleixandre, J.L.; Toit, W. Spectrophotometric analysis of phenolic compounds in grapes and wines. J. Agric. Food Chem. 2017, 65, 4009–4026. [Google Scholar] [CrossRef] [PubMed]
- Philippidis, A.; Poulakis, E.; Kontzedaki, R.; Orfanakis, E.; Symianaki, A.; Zoumi, A.; Velegrakis, M. Application of ultraviolet-visible absorption spectroscopy with machine learning techniques for the classification of cretan wines. Foods 2021, 10, 9. [Google Scholar] [CrossRef]
- Skogerson, K.; Downey, M.; Mazza, M.; Boulton, R. Rapid determination of phenolic components in red wines from UV-visible spectra and the method of partial least squares. Am. J. Enol. Vit. 2007, 58, 318–325. [Google Scholar]
- Chase, D.B. Fourier transform Raman spectroscopy. J. Am. Chem. Soc. 1986, 108, 7485–7488. [Google Scholar] [CrossRef]
- Guo, S.; Chernavskaia, O.; Popp, J.; Bockiltz, T. Spectral reconstruction of shift-excitation Raman difference spectroscopy. Talanta 2018, 186, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Ranatunge, I.; Adikary, S.; Dasanayake, P.; Fernando, C.D.; Soysa, P. Development of a rapid and simple method to remove polyphenols from plant extracts. Int. J. Anal. Chem. 2017, 2017, 7230145. [Google Scholar] [CrossRef] [Green Version]
- Mattick, L.R.; Rice, A.C. The use of PVPP of decolorizing wine in the determination of tartrate by metavanadate method. Am. J. Enol. Vit. 1981, 32, 297–298. [Google Scholar]
- Rodriguez, S.B.; Thornton, M.A.; Thornton, R.J. Raman Spectroscopy and chemometrics for identification and strain discrimination of the wine spoilage yeasts Saccharomyces cerevisiae, Zygosaccharomyces bailii, and Brettanomyces bruxellensis. App. Environ. Micro. 2013, 79, 6264–6270. [Google Scholar] [CrossRef] [Green Version]
- Frausto-Reyes, C.; Medina-Gutierrez, C.; Sato-Berru, R.; Sahagun, L.R. Qualitative study of ethanol content in tequila by Raman spectroscopy and principal components analysis. Spectrosc. Acta A 2005, 61, 2657–2662. [Google Scholar] [CrossRef] [PubMed]
- Boyaci, I.H.; Genis, H.E.; Guven, B.; Tamer, U.; Alper, N. A novel method for quantification of ethanol and methanol in distilled alcoholic beverages using Raman spectroscopy: Simultaneous detection of ethanol and methanol. J. Raman Spectrosc. 2012, 43, 1171–1176. [Google Scholar] [CrossRef]
- Delfino, I.; Camerlingo, C.; Portaccio, M.; Ventura, B.D.; Mita, L.; Mita, D.G.; Lepore, M. Visible micro-Raman spectroscopy for determining glucose content in beverage industry. Food Chem. 2011, 127, 735–742. [Google Scholar] [CrossRef]
- Richardson, P.I.C.; Muhamadali, H.; Ellis, D.I.; Goodacre, R. Rapid Quantification of the adulteration of fresh coconut water by dilution and sugars using Raman spectroscopy and chemometrics. Food Chem. 2019, 272, 157–164. [Google Scholar] [CrossRef]
- Pompeu, D.R.; Larondelle, Y.; Rogez, H.; Abbas, O.; Pierna, J.A.F.; Baeten, V. Characterization and discrimination of phenolic compounds using fourier transform Raman spectroscopy and chemometric tools. Biotechnol. Agron. Soc. Environ. 2018, 22, 13–28. [Google Scholar]
- Wu, Z.; Xu, E.; Long, J.; Pan, X.; Xu, X.; Jin, Z.; Jiao, A. Comparison between ATR-IR, Raman, concatenated ATR-IR and Raman spectroscopy for the determination of total antioxidant capacity and total phenolic content of chinese rice wine. Food Chem. 2016, 194, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Z.; Ma, Z.; Liang, L. Real time monitoring of multiple components in wine fermentation using an on-line suto-calibration Raman spectroscopy. Sens. Actuators B 2014, 202, 426–432. [Google Scholar] [CrossRef]
- Ramírez-Elías, M.G.; Guevara, E.; Zamora-Pedraza, C.; Rogelio Aguirre, J.R.; Juárez, B.I.F.; Bárcenas-Pezos, G.M.; Ruiz, F.; González, F.J. Assessment of mezcal aging combining Raman spectroscopy and multivariate analysis. Biomed. Spectrosc. Imaging 2017, 6, 75–81. [Google Scholar] [CrossRef]
- Magdas, D.A.; Guyon, F.; Feher, I.; Pinzaru, S.C. Wine Discrimination based on chemometric analysis of untargeted markers using FT-Raman spectroscopy. Food Control 2018, 85, 385–391. [Google Scholar] [CrossRef]
- Teixera dos Santos, C.A.; Pascoa, R.N.M.J.; Porto, P.A.L.S.; Cerdeira, A.L.; Gonzalez-Saiz, J.M.; Pizarro, C.; Lopes, J.A. Raman spectroscopy for wine analysis: A comparison with near and mid infrared spectroscopy. Talanta 2018, 186, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Pierna, J.A.F.; Abbas, O.; Dardenne, P.; Baeten, V. Discrimination of Corsican honey by FT-Raman spectroscopy and chemometrics. Biotechnol. Agron. Soc. Environ. 2011, 15, 75–84. [Google Scholar]
- Agati, G.; Matteini, P.; Oliveira, J.; de Freitas, V.; Mateus, N. Fluorescence approach for measuring anthocyanins and derived pigments in red wine. J. Agric. Food Chem. 2013, 61, 10156–10162. [Google Scholar] [CrossRef]
- Poustka, F.; Irani, N.G.; Feller, A.; Lu, Y.; Pourcel, L.; Frame, K.; Grotewold, E. Trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacule protein-sorting route in Arabidopsis and continues to the formation of vacuolar inclusions. Plant Phys. 2007, 145, 1323–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somers, T.C. The polymeric nature of wine pigments. Phytochemistry 1971, 10, 2175–2186. [Google Scholar] [CrossRef]
- Fe, H.; Na-na, L.; Duan, C.Q. Anthocyanins and their variation in red wines II: Anthocyanin derived pigments and their color evolution. Molecules 2012, 17, 1483–1519. [Google Scholar]
- Silva, G.T.M.; da Silva, K.M.; Silva, C.P.; Rodigues, A.C.B.; Oake, J.; Gehlen, M.H.; Bohne, C.; Quina, F.H. Highly fluorescent hybrid prigments from anthocyanin- and red wine pyranoanthocyanin- analogs absorbed on sepiolite clay. Photochem. Photobiol. Sci. 2019, 18, 1750–1760. [Google Scholar] [CrossRef] [PubMed]
- Lackey, H.E.; Nelson, G.L.; Lines, A.M.; Bryan, S.A. Reimagining pH measurements; utilizing Raman spectroscopy for enhanced accuracy in phosphoric acid systems. Anal. Chem. 2020, 92, 5882–5889. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Wang, J.; Lin, G.; Suo, H.; Zhao, C. Short-wave near-infrared spectrometer for alcohol determination and temperature correction. J. Anal. Methods Chem. 2012, 2012, 728128. [Google Scholar] [CrossRef]
- Fernandes, H.L.; Raimundo, I.M., Jr.; Pasquino, C.; Rohwedder, J.J.R. Simultaneous determination of methanol and ethanol in gasoline using NIR spectroscopy: Effect of gasoline composition. Talanta 2008, 75, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Golic, M.; Walsh, K.; Lawson, P. Short-wavelength near-infrared spectra of sucrose, glucose, and fructose with respect to sugar concentration and temperature. Appl. Spectrosc. 2003, 57, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.L.; Da-Ming, Z. On calibration of pH meters. Sensors 2005, 5, 209–219. [Google Scholar] [CrossRef]
- Magdas, D.A.; Cozar, B.I.; Feher, I.; Guyon, F.; Dehelean, A.; Pinzaru, S.C. Testing limits of FT-Raman spectroscopy for wine authentication: Cultivar, geographical origin, vintage and terroir effect influence. Sci. Rep. 2019, 9, 19954. [Google Scholar] [CrossRef] [PubMed]
- Zanuttin, F.; Gurian, E.; Ignat, I.; Fornasaro, S.; Calabretti, A.; Bigot, G.; Bonifacio, A. Characterization of white wines from north-eastern Italy with surface-enhanced Raman spectroscopy. Talanta 2019, 203, 99–105. [Google Scholar] [CrossRef]



| Algorithm | Ethanol–Raw Spectra | pH–Raw Spectra | Ethanol–Post PVPP Spectra | pH–Post PVPP Spectra | Total Sugars | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| RMEP | R2 | RMEP | R2 | RMEP | R2 | RMEP | R2 | RMEP | R2 | |
| SVR | 1.35 | 0.51 | 1.16 | 0.62 | 0.23 | 0.98 | 0.12 | 0.79 | 1.59 | 0.96 |
| PLSR | 1.22 | 0.50 | 1.17 | 0.61 | 0.21 | 0.99 | 0.12 | 0.84 | 1.57 | 0.95 |
| RR | 1.19 | 0.50 | 1.16 | 0.67 | 0.23 | 0.99 | 0.12 | 0.82 | 1.57 | 0.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuller, H.; Beaver, C.; Harbertson, J. Alcoholic Fermentation Monitoring and pH Prediction in Red and White Wine by Combining Spontaneous Raman Spectroscopy and Machine Learning Algorithms. Beverages 2021, 7, 78. https://doi.org/10.3390/beverages7040078
Fuller H, Beaver C, Harbertson J. Alcoholic Fermentation Monitoring and pH Prediction in Red and White Wine by Combining Spontaneous Raman Spectroscopy and Machine Learning Algorithms. Beverages. 2021; 7(4):78. https://doi.org/10.3390/beverages7040078
Chicago/Turabian StyleFuller, Harrison, Chris Beaver, and James Harbertson. 2021. "Alcoholic Fermentation Monitoring and pH Prediction in Red and White Wine by Combining Spontaneous Raman Spectroscopy and Machine Learning Algorithms" Beverages 7, no. 4: 78. https://doi.org/10.3390/beverages7040078






