Information System for Selection of Conditions and Equipment for Mammalian Cell Cultivation
Abstract
:1. Introduction
2. Data Science: Theoretical Framework
2.1. Overview of Mammalian Cell Culture Technology
- The nutrient medium is easily and quickly replaced;
- Any type of cells can be introduced into the monolayer, indicating the flexibility of the system;
- Perfusion equipment can artificially increase cell density; and
- The possibility of changing the “cell–medium” attitude during the experiment, with the help of an instrumental solution.
- Difficulty of scaling;
- Lack of informative visual analysis;
- Difficulties in determining and maintaining parameters, such as acidity, O content, and cell homogeneity; and
- A lot of space is required.
2.2. Equipment for the Cultivation of Mammalian Cells
2.3. Matrix for the Cultivation of Mammalian Cells
3. Methodology Development
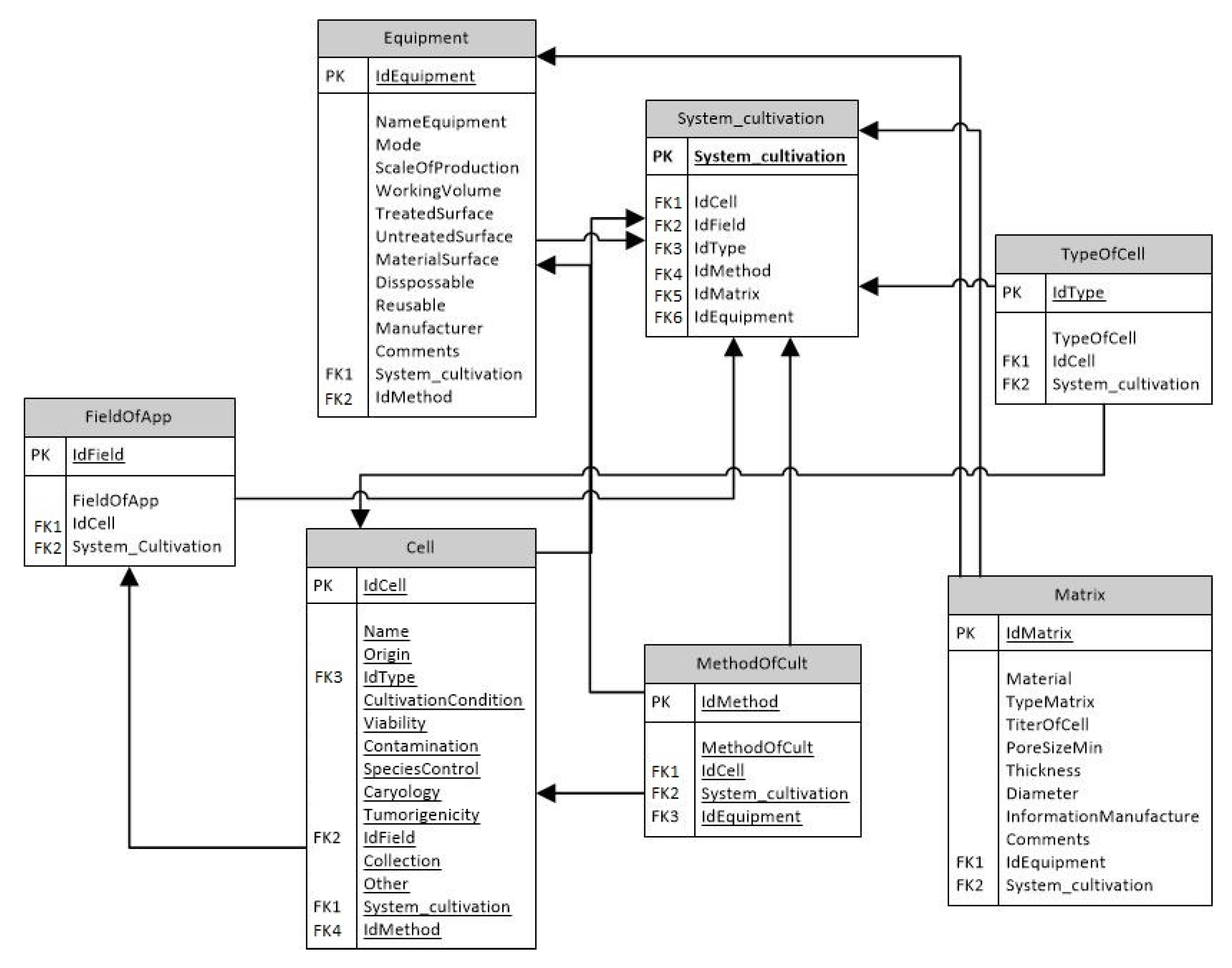
3.1. Design of the Database
3.2. Design of the Information System
- To organize all the data on conditions and equipment for mammalian cell cultivation;
- To provide the most comprehensive information on standard cultivation and choice of matrix. This information system includes a reference part, in the form of a database;
- To select the necessary system for carrying out the cultivation process.
4. Analysis of Results
4.1. Results of the Database Implementation
- One cell can have various fields of application;
- One cell may have different cultivation systems;
- Different types of cells can be cultivated within one equipment;
- One matrix can be applied to different cells; and
- One method of cultivation can be suitable for different types of cells.
4.2. Results of the Information System Implementation
- Database module, which handles data manipulation procedures (entering, modifying, deleting);
- Search module, which is used to find the information using various parameters and to sort the results, according to different criteria;
- Ontology module, which contains the ontology of bioreactors (i.e., the structure of selection of the cultivation system); and
- Help module, where the main objectives of the application are described.
4.2.1. Information Search Module
4.2.2. Module “Database”: Input–Output (I/O) of Information Data
4.2.3. Module “Ontology of Cultivation”
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DB | Database |
| IS | Information system |
| STN | Scientific technical information network |
| PC | Personal computer |
| PK | Primary key |
| I/O | Input/output |
References
- Azam, A.G. A Review on Artificial Intelligence (AI), Big Data and Block Chain: Future Impact and Business Opportunities. Available online: https://www.journalofbusiness.org/index.php/GJMBR/article/view/3246 (accessed on 17 December 2020).
- Galetsi, P.; Katsaliaki, K. A review of the literature on big data analytics in healthcare. J. Oper. Res. Soc. 2020, 71, 1511–1529. [Google Scholar] [CrossRef]
- Saura, J.R.; Herráez, B.R.; Reyes-Menendez, A. Comparing a traditional approach for financial Brand Communication Analysis with a Big Data Analytics technique. IEEE Access 2019, 7, 37100–37108. [Google Scholar] [CrossRef]
- Saura, J.R. Using Data Sciences in Digital Marketing: Framework, methods, and performance metrics. J. Innov. Knowl. 2020. [Google Scholar] [CrossRef]
- Srinivasan, R.; Chia, K.; Heikkila, A.M.; Schabel, J. A decision support database for inherently safer design. In Computer Aided Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2003; Volume 14, pp. 287–292. [Google Scholar] [CrossRef]
- de Freitas, L.H.; Roux, G.A.C.L. Exploiting R&D Databases for Efficient Product Design: Application to Brake Fluid Formulations. In Computer Aided Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2009; Volume 27, pp. 1161–1166. [Google Scholar] [CrossRef]
- Thai, Q.K.; Bös, F.; Pleiss, J. The Lactamase Engineering Database: a critical survey of TEM sequences in public databases. BMC Genom. 2009, 10, 390. [Google Scholar] [CrossRef] [Green Version]
- Mishra, R.K.; Mohamed, A.K.; Geissbühler, D.; Manzano, H.; Jamil, T.; Shahsavari, R.; Kalinichev, A.G.; Galmarini, S.; Tao, L.; Heinz, H.; et al. A force field database for cementitious materials including validations, applications and opportunities. Cem. Concr. Res. 2017, 102, 68–89. [Google Scholar] [CrossRef] [Green Version]
- Conte, C.; Vaysse, C.; Bosco, P.; Noize, P.; Fourrier-Reglat, A.; Despas, F.; Lapeyre-Mestre, M. The value of a health insurance database to conduct pharmacoepidemiological studies in oncology. Therapies 2019, 74, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Mante, J.; Gangadharan, N.; Sewell, D.J.; Turner, R.; Field, R.; Oliver, S.G.; Slater, N.; Dikicioglu, D. A heuristic approach to handling missing data in biologics manufacturing databases. Bioprocess Biosyst. Eng. 2019, 42, 657–663. [Google Scholar] [CrossRef] [Green Version]
- Gangadharan, N.; Turner, R.; Field, R.; Oliver, S.G.; Slater, N.; Dikicioglu, D. Metaheuristic approaches in biopharmaceutical process development data analysis. Bioprocess Biosyst. Eng. 2019, 42, 1399–1408. [Google Scholar] [CrossRef] [Green Version]
- Kawabata, T.; Ota, M.; Nishikawa, K. The Protein Mutant Database. Nucleic Acids Res. 1999, 27, 355–357. [Google Scholar] [CrossRef] [Green Version]
- Gromiha, M.M.; Uedaira, H.; An, J.; Selvaraj, S.; Prabakaran, P.; Sarai, A. ProTherm, Thermodynamic Database for Proteins and Mutants: developments in version 3.0. Nucleic Acids Res. 2002, 30, 301–302. [Google Scholar] [CrossRef] [Green Version]
- Braun, A.; Halwachs, B.; Geier, M.; Weinhandl, K.; Guggemos, M.; Marienhagen, J.; Ruff, A.J.; Schwaneberg, U.; Rabin, V.; Torres Pazmino, D.E.; et al. MuteinDB: The mutein database linking substrates, products and enzymatic reactions directly with genetic variants of enzymes. Database 2012, 2012, bas028. [Google Scholar] [CrossRef] [PubMed]
- Sirim, D.; Wagner, F.; Lisitsa, A.; Pleiss, J. The cytochrome P450 engineering database: Integration of biochemical properties. BMC Biochem. 2009, 10, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kourist, R.; Jochens, H.; Bartsch, S.; Kuipers, R.; Padhi, S.K.; Gall, M.; Böttcher, D.; Joosten, H.J.; Bornscheuer, U.T. The alpha/beta-hydrolase fold 3DM database (ABHDB) as a tool for protein engineering. ChemBioChem Eur. J. Chem. Biol. 2010, 11, 1635–1643. [Google Scholar] [CrossRef]
- Amirkia, V.; Qiubao, P. Cell-culture Database: Literature-based reference tool for human and mammalian experimentally-based cell culture applications. Bioinformation 2012, 8, 237–238. [Google Scholar] [CrossRef] [Green Version]
- Brunner, D. Serum-free cell culture: The serum-free media interactive online database. ALTEX 2010, 53–62. [Google Scholar] [CrossRef]
- Berks, A.H. Patent information in biotechnology. Trends Biotechnol. 1994, 12, 352–364. [Google Scholar] [CrossRef]
- Jossen, V.; Eibl, R.; Pörtner, R.; Kraume, M.; Eibl, D. Stirred Bioreactors. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2017; pp. 179–215. [Google Scholar] [CrossRef]
- Amoabediny, G.; Salehi-Nik, N.; Heli, B. The Role of Biodegradable Engineered Scaffold in Tissue Engineering. In Biomaterials Science and Engineering; Pignatello, R., Ed.; InTech: London, UK, 2011. [Google Scholar] [CrossRef] [Green Version]
- Mano, J.; Silva, G.; Azevedo, H.; Malafaya, P.; Sousa, R.; Silva, S.; Boesel, L.; Oliveira, J.; Santos, T.; Marques, A.; et al. Natural origin biodegradable systems in tissue engineering and regenerative medicine: Present status and some moving trends. J. R. Soc. Interface 2007, 4, 999–1030. [Google Scholar] [CrossRef] [Green Version]
- Cherry, R.S.; Papoutsakis, E.T. Growth and death rates of bovine embryonic kidney cells in turbulent microcarrier bioreactors. Bioprocess Eng. 1989, 4, 81–89. [Google Scholar] [CrossRef]
- Galvanauskas, V.; Simutis, R.; Volk, N.; Lübbert, A. Model based design of a biochemical cultivation process. Bioprocess Eng. 1998, 18, 227. [Google Scholar] [CrossRef]
- Ellis, M.; Jarman-Smith, M.; Chaudhuri, J. Bioreactor Systems for Tissue Engineering: A Four-Dimensional Challenge. In Bioreactors for Tissue Engineering; Chaudhuri, J., Al-Rubeai, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–18. [Google Scholar] [CrossRef]
- Awad, H.A.; Quinn Wickham, M.; Leddy, H.A.; Gimble, J.M.; Guilak, F. Chondrogenic differentiation of adipose-derived adult stem cells in agarose, alginate, and gelatin scaffolds. Biomaterials 2004, 25, 3211–3222. [Google Scholar] [CrossRef]
- Bellucci, D.; Sola, A.; Cannillo, V. A Revised Replication Method for Bioceramic Scaffolds. Bioceram. Dev. Appl. 2011, 1, 1–8. [Google Scholar] [CrossRef]
- Boccaccini, A.R.; Blaker, J.J. Bioactive composite materials for tissue engineering scaffolds. Exp. Rev. Med. Devices 2005, 2, 303–317. [Google Scholar] [CrossRef]
- Solovieva, A.; Kopylov, A.; Savko, M.; Zarkhina, T.; Lovskaya, D.; Lebedev, A.; Menshutina, N.; Krivandin, A.; Shershnev, I.; Kotova, S.; et al. Photocatalytic Properties of Tetraphenylporphyrins Immobilized on Calcium Alginate Aerogels. Sci. Rep. 2017, 7, 12640. [Google Scholar] [CrossRef] [Green Version]
- Chan, B.P.; Leong, K.W. Scaffolding in tissue engineering: general approaches and tissue-specific considerations. Eur. Spine J. 2008, 17, 467–479. [Google Scholar] [CrossRef] [Green Version]
- Hollister, S.J. Porous scaffold design for tissue engineering. Nat. Mater. 2005, 4, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Garcia-Fuentes, M. Bioactive Scaffolds for the Controlled Formation of Complex Skeletal Tissues. In Regenerative Medicine and Tissue Engineering—Cells and Biomaterials; Eberli, D., Ed.; InTech: London, UK, 2011. [Google Scholar] [CrossRef] [Green Version]
- Avramenko, Y.; Kraslawski, A. Similarity concept for case-based design in process engineering. Comput. Chem. Eng. 2006, 30, 548–557. [Google Scholar] [CrossRef]
- Leesley, M.; Buchmann, A. Databases for computer-aided process plant design. Comput. Chem. Eng. 1980, 4, 79–83. [Google Scholar] [CrossRef]
- Biesenbender, S.; Petersohn, S.; Thiedig, C. Using Current Research Information Systems (CRIS) to showcase national and institutional research (potential): Research information systems in the context of Open Science. Procedia Comput. Sci. 2019, 146, 142–155. [Google Scholar] [CrossRef]
- Verma, R.; Schwaneberg, U.; Roccatano, D. Computer-aided protein directed evolution: A review of web servers, databases and other computational tools for protein engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209008. [Google Scholar] [CrossRef] [Green Version]
- Griffith, L.G. Tissue Engineering–Current Challenges and Expanding Opportunities. Science 2002, 295, 1009–1014. [Google Scholar] [CrossRef]
- Liu, S.; Tao, D.; Zhang, L. Cellulose scaffold: A green template for the controlling synthesis of magnetic inorganic nanoparticles. Powder Technol. 2012, 217, 502–509. [Google Scholar] [CrossRef]
- Rosso, F.; Marino, G.; Giordano, A.; Barbarisi, M.; Parmeggiani, D.; Barbarisi, A. Smart materials as scaffolds for tissue engineering. J. Cell. Physiol. 2005, 203, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Stella, J.A.; D’Amore, A.; Wagner, W.R.; Sacks, M.S. On the biomechanical function of scaffolds for engineering load-bearing soft tissues. Acta Biomater. 2010, 6, 2365–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willerth, S.M.; Sakiyama-Elbert, S.E. Approaches to neural tissue engineering using scaffolds for drug delivery. Adv. Drug Deliv. Rev. 2007, 59, 325–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]


















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menshutina, N.; Guseva, E.; Batyrgazieva, D.; Mitrofanov, I. Information System for Selection of Conditions and Equipment for Mammalian Cell Cultivation. Data 2021, 6, 23. https://doi.org/10.3390/data6030023
Menshutina N, Guseva E, Batyrgazieva D, Mitrofanov I. Information System for Selection of Conditions and Equipment for Mammalian Cell Cultivation. Data. 2021; 6(3):23. https://doi.org/10.3390/data6030023
Chicago/Turabian StyleMenshutina, Natalia, Elena Guseva, Diana Batyrgazieva, and Igor Mitrofanov. 2021. "Information System for Selection of Conditions and Equipment for Mammalian Cell Cultivation" Data 6, no. 3: 23. https://doi.org/10.3390/data6030023
APA StyleMenshutina, N., Guseva, E., Batyrgazieva, D., & Mitrofanov, I. (2021). Information System for Selection of Conditions and Equipment for Mammalian Cell Cultivation. Data, 6(3), 23. https://doi.org/10.3390/data6030023





