The Relationship between Changes in MYBPC3 Single-Nucleotide Polymorphism-Associated Metabolites and Elite Athletes’ Adaptive Cardiac Function
Abstract
:1. Introduction
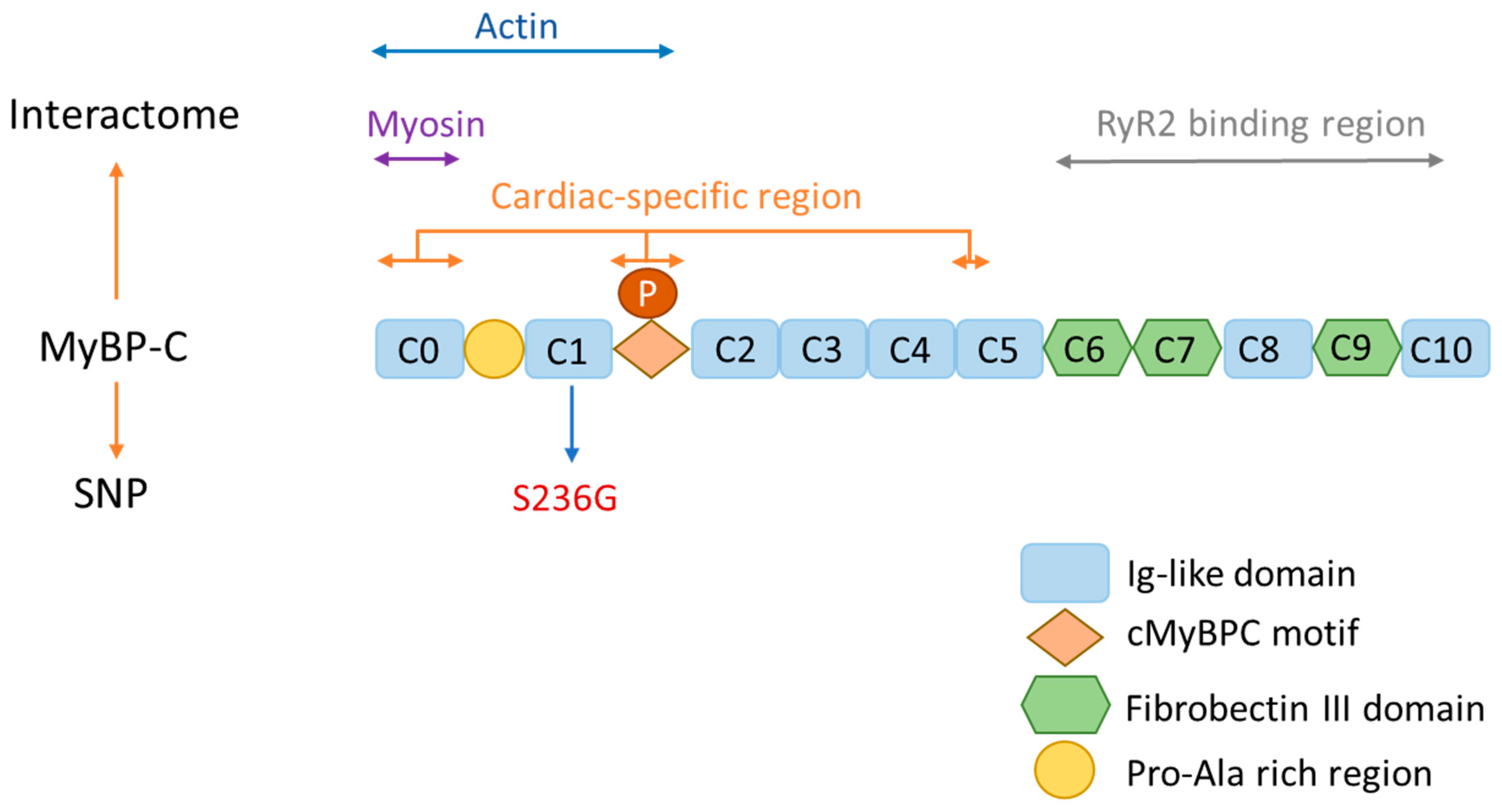
2. MyBP-C Structure and Function
3. MyBP-C and Its Association with Cardiac Disease
4. MyBP-C and Its Association with Elite Athlete Performance
| Elite Athlete’s Endurance | Hypertrophy Cardiomyopathy | |
|---|---|---|
| Left ventricular thickness | Mild wall thickness | Unusual hypertrophy pattern |
| Left ventricular cavity size | >55 mm | <45 mm |
| Left atrium size | ≥40 mm | <40 mm |
| Left ventricular filling | Normal | Abnormal |
| Family History of HCM | Negative | Positive |
5. MYBPC3 SNPs and Endurance
6. MYBPC3 SNP-Associated Metabolites
6.1. Quinate
6.2. Theophylline
6.3. Decanoyl-Carnitine
6.4. Ursodeoxycholic Acid
7. Limitations, Future Work, and Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Georgiades, E.; Klissouras, V.; Baulch, J.; Wang, G.; Pitsiladis, Y. Why Nature Prevails over Nurture in the Making of the Elite Athlete. BMC Genom. 2017, 18, 835. [Google Scholar] [CrossRef] [PubMed]
- Al-Khelaifi, F.; Yousri, N.A.; Diboun, I.; Semenova, E.A.; Kostryukova, E.S.; Kulemin, N.A.; Borisov, O.V.; Andryushchenko, L.B.; Larin, A.K.; Generozov, E.V.; et al. Genome-Wide Association Study Reveals a Novel Association between MYBPC3 Gene Polymorphism, Endurance Athlete Status, Aerobic Capacity and Steroid Metabolism. Front. Genet. 2020, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, D.G.; North, K.N. Genes and Human Elite Athletic Performance. Hum. Genet. 2005, 116, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Al-Khelaifi, F.; Donati, F.; Botre, F.; Latiff, A.; Abraham, D.; Hingorani, A.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. Metabolic Profiling of Elite Athletes with Different Cardiovascular Demand. Scand. J. Med. Sci. Sports 2019, 29, 933–943. [Google Scholar] [CrossRef]
- Znazen, H.; Mejri, A.; Touhami, I.; Chtara, M.; Siala, H.; LE Gallais, D.; Ahmetov, I.I.; Messaoud, T.; Chamari, K.; Soussi, N. Genetic Advantageous Predisposition of Angiotensin Converting Enzyme Id Polymorphism in Tunisian Athletes. J. Sports Med. Phys. Fit. 2016, 56, 724–730. [Google Scholar]
- Ahmetov, I.I.; Hall, E.C.R.; Semenova, E.A.; Pranckevičienė, E.; Ginevičienė, V. Advances in Sports Genomics. Adv. Clin. Chem. 2022, 107, 215–263. [Google Scholar] [CrossRef]
- Bers, D.M. Cardiac Excitation-Contraction Coupling. Nature 2002, 415, 198–205. [Google Scholar] [CrossRef]
- Clark, K.A.; McElhinny, A.S.; Beckerle, M.C.; Gregorio, C.C. Striated Muscle Cytoarchitecture: An Intricate Web of Form and Function. Annu. Rev. Cell Dev. Biol. 2002, 18, 637–706. [Google Scholar] [CrossRef]
- Moss, R.L.; Fitzsimons, D.P.; Ralphe, J.C. Cardiac MyBP-C Regulates the Rate and Force of Contraction in Mammalian Myocardium. Circ. Res. 2015, 116, 183–192. [Google Scholar] [CrossRef]
- Flashman, E.; Redwood, C.; Moolman-Smook, J.; Watkins, H. Cardiac Myosin Binding Protein C: Its Role in Physiology and Disease. Circ. Res. 2004, 94, 1279–1289. [Google Scholar] [CrossRef]
- Lin, B.; Govindan, S.; Lee, K.; Zhao, P.; Han, R.; Runte, K.E.; Craig, R.; Palmer, B.M.; Sadayappan, S. Cardiac Myosin Binding Protein-C Plays No Regulatory Role in Skeletal Muscle Structure and Function. PLoS ONE 2013, 8, e69671. [Google Scholar] [CrossRef] [PubMed]
- Sadayappan, S.; de Tombe, P.P. Cardiac Myosin Binding Protein-C: Redefining Its Structure and Function. Biophys. Rev. 2012, 4, 93–106. [Google Scholar] [CrossRef]
- Barefield, D.; Sadayappan, S. Phosphorylation and Function of Cardiac Myosin Binding Protein-C in Health and Disease. J. Mol. Cell Cardiol. 2010, 48, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.R.; McNamara, J.W.; Sadayappan, S. Mutations in Myosin S2 Alter Cardiac Myosin-Binding Protein-C Interaction in Hypertrophic Cardiomyopathy in a Phosphorylation-Dependent Manner. J. Biol. Chem. 2021, 297, 100836. [Google Scholar] [CrossRef] [PubMed]
- Zacharchenko, T.; von Castelmur, E.; Rigden, D.J.; Mayans, O. Structural Advances on Titin: Towards an Atomic Understanding of Multi-Domain Functions in Myofilament Mechanics and Scaffolding. Biochem. Soc. Trans. 2015, 43, 850–855. [Google Scholar] [CrossRef]
- Okagaki, T.; Weber, F.E.; Fischman, D.A.; Vaughan, K.T.; Mikawa, T.; Reinach, F.C. The Major Myosin-Binding Domain of Skeletal Muscle MyBP-C (C Protein) Resides in the COOH-Terminal, Immunoglobulin C2 Motif. J. Cell Biol. 1993, 123, 619–626. [Google Scholar] [CrossRef]
- Inchingolo, A.V.; Previs, S.B.; Previs, M.J.; Warshaw, D.M.; Kad, N.M. Revealing the Mechanism of How Cardiac Myosin-Binding Protein C N-Terminal Fragments Sensitize Thin Filaments for Myosin Binding. Proc. Natl. Acad. Sci. USA 2019, 116, 6828–6835. [Google Scholar] [CrossRef] [PubMed]
- Stanczyk, P.J.; Seidel, M.; White, J.; Viero, C.; George, C.H.; Zissimopoulos, S.; Lai, F.A. Association of Cardiac Myosin-Binding Protein-C with the Ryanodine Receptor Channel—Putative Retrograde Regulation? J. Cell Sci. 2018, 131, jcs210443. [Google Scholar] [CrossRef]
- Bayram, Y.; Karaca, E.; Coban Akdemir, Z.; Yilmaz, E.O.; Tayfun, G.A.; Aydin, H.; Torun, D.; Bozdogan, S.T.; Gezdirici, A.; Isikay, S.; et al. Molecular Etiology of Arthrogryposis in Multiple Families of Mostly Turkish Origin. J. Clin. Investig. 2016, 126, 762–778. [Google Scholar] [CrossRef]
- Desai, D.; Stiene, D.; Song, T.; Sadayappan, S. Distal Arthrogryposis and Lethal Congenital Contracture Syndrome—An Overview. Front. Physiol. 2020, 11, 689. [Google Scholar] [CrossRef]
- Kuster, D.W.; Bawazeer, A.C.; Zaremba, R.; Goebel, M.; Boontje, N.M.; van der Velden, J. Cardiac Myosin Binding Protein C Phosphorylation in Cardiac Disease. J. Muscle Res. Cell Motil. 2012, 33, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Heling LW, H.J.; Geeves, M.A.; Kad, N.M. MyBP-C: One Protein to Govern Them All. J. Muscle Res. Cell Motil. 2020, 41, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Rowin, E.J.; Casey, S.A.; Maron, M.S. How Hypertrophic Cardiomyopathy Became a Contemporary Treatable Genetic Disease with Low Mortality: Shaped by 50 Years of Clinical Research and Practice. JAMA Cardiol. 2016, 1, 98–105. [Google Scholar] [CrossRef]
- Seidman, C.E.; Seidman, J.G. Identifying Sarcomere Gene Mutations in Hypertrophic Cardiomyopathy: A Personal History. Circ. Res. 2011, 108, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsdottir, B.; Burke, M.; Maron, B.J.; Danielsen, R.; Lopez, B.; Diez, J.; Jarolim, P.; Seidman, J.; Seidman, C.E.; Ho, C.Y.; et al. Hypertrophic Cardiomyopathy in Myosin-Binding Protein C (MYBPC3) Icelandic Founder Mutation Carriers. Open Heart 2020, 7, e001220. [Google Scholar] [CrossRef] [PubMed]
- Hartmannova, H.; Kubanek, M.; Sramko, M.; Piherova, L.; Noskova, L.; Hodanova, K.; Stranecky, V.; Pristoupilova, A.; Sovova, J.; Marek, T.; et al. Isolated X-Linked Hypertrophic Cardiomyopathy Caused by a Novel Mutation of the Four-and-a-Half LIM Domain 1 Gene. Circ. Cardiovasc. Genet. 2013, 6, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Cirino, A.L.; Harris, S.; Lakdawala, N.K.; Michels, M.; Olivotto, I.; Day, S.M.; Abrams, D.J.; Charron, P.; Caleshu, C.; Semsarian, C.; et al. Role of Genetic Testing in Inherited Cardiovascular Disease: A Review. JAMA Cardiol. 2017, 2, 1153–1160. [Google Scholar] [CrossRef]
- Maron, B.J.; Maron, M.S.; Maron, B.A.; Loscalzo, J. Moving Beyond the Sarcomere to Explain Heterogeneity in Hypertrophic Cardiomyopathy: JACC Review Topic of the Week. J. Am. Coll. Cardiol. 2019, 73, 1978–1986. [Google Scholar] [CrossRef]
- Maron, B.J.; Maron, M.S.; Maurer, M.S.; Rowin, E.J.; Maron, B.A.; Galie, N. Cardiovascular Diseases That Have Emerged From the Darkness. J. Am. Heart Assoc. 2021, 10, e021095. [Google Scholar] [CrossRef]
- Ingles, J.; Doolan, A.; Chiu, C.; Seidman, J.; Seidman, C.; Semsarian, C. Compound and Double Mutations in Patients with Hypertrophic Cardiomyopathy: Implications for Genetic Testing and Counselling. J. Med. Genet. 2005, 42, e59. [Google Scholar] [CrossRef]
- Ingles, J.; Yeates, L.; Semsarian, C. The Emerging Role of the Cardiac Genetic Counselor. Heart Rhythm. 2011, 8, 1958–1962. [Google Scholar] [CrossRef] [PubMed]
- Tudurachi, B.-S.; Zăvoi, A.; Leonte, A.; Țăpoi, L.; Ureche, C.; Bîrgoan, S.G.; Chiuariu, T.; Anghel, L.; Radu, R.; Sascău, R.A.; et al. An Update on MYBPC3 Gene Mutation in Hypertrophic Cardiomyopathy. Int. J. Mol. Sci. 2023, 24, 10510. [Google Scholar] [CrossRef] [PubMed]
- Carrier, L. Making Sense of Inhibiting Nonsense in Hypertrophic Cardiomyopathy. Circulation 2019, 139, 812–814. [Google Scholar] [CrossRef]
- Helms, A.S.; Tang, V.T.; O’Leary, T.S.; Friedline, S.; Wauchope, M.; Arora, A.; Wasserman, A.H.; Smith, E.D.; Lee, L.M.; Wen, X.W.; et al. Effects of MYBPC3 Loss-of-Function Mutations Preceding Hypertrophic Cardiomyopathy. JCI Insight 2020, 5, e133782. [Google Scholar] [CrossRef]
- Kelly, M.; Semsarian, C. Multiple Mutations in Genetic Cardiovascular Disease: A Marker of Disease Severity? Circ. Cardiovasc. Genet. 2009, 2, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, M.; Ho, C.Y. Cardiovascular Genetics: The Role of Genetic Testing in Diagnosis and Management of Patients with Hypertrophic Cardiomyopathy. Heart 2021, 107, 183–189. [Google Scholar] [CrossRef]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botrè, F.; Alsayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A Pilot Study Comparing the Metabolic Profiles of Elite-Level Athletes from Different Sporting Disciplines. Sports Med.-Open 2018, 4, 2. [Google Scholar] [CrossRef]
- Gomez-Cabrera, M.C.; Domenech, E.; Vina, J. Moderate Exercise Is an Antioxidant: Upregulation of Antioxidant Genes by Training. Free Radic. Biol. Med. 2008, 44, 126–131. [Google Scholar] [CrossRef]
- Baggish, A.L.; Wood, M.J. Athlete’s Heart and Cardiovascular Care of the Athlete. Circulation 2011, 123, 2723–2735. [Google Scholar] [CrossRef]
- Maron, B.J. Distinguishing Hypertrophic Cardiomyopathy from Athlete’s Heart: A Clinical Problem of Increasing Magnitude and Significance. Heart 2005, 91, 1380–1382. [Google Scholar] [CrossRef]
- Prutkin, J.M.; Ackerman, M.J.; Drezner, J.A. Athletes with Implantable Cardioverter Defibrillators: Can They Return to Competitive Sports? Heart 2016, 102, 93–94. [Google Scholar] [CrossRef] [PubMed]
- Campbell, R.M.; Berger, S.; Drezner, J. Sudden Cardiac Arrest in Children and Young Athletes: The Importance of a Detailed Personal and Family History in the Pre-Participation Evaluation. Br. J. Sports Med. 2009, 43, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botrè, F.; Abraham, D.; Hingorani, A.; Albagha, O.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; et al. Metabolic GWAS of Elite Athletes Reveals Novel Genetically-Influenced Metabolites Associated with Athletic Performance. Sci. Rep. 2019, 9, 19889. [Google Scholar] [CrossRef]
- Verwijs, S.M.; Pinto, Y.M.; Kuster, D.W.; van der Velden, J.; Limpens, J.; van Hattum, J.C.; van der Crabben, S.N.; Lekanne Deprez, R.H.; Wilde, A.A.; Jørstad, H.T. Beneficial Effects of Cardiomyopathy-Associated Genetic Variants on Physical Performance: A Hypothesis-Generating Scoping Review. Cardiology 2022, 147, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Jaguri, A.; Al Thani, A.A.; Elrayess, M.A. Exercise Metabolome: Insights for Health and Performance. Metabolites 2023, 13, 694. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, T.; Sakuraba, K. Changes in Urine Components and Characteristics During a 415-Km Mountain Ultra-Marathon. Juntendo Med. J. 2019, 65, 368–377. [Google Scholar] [CrossRef]
- Morville, T.; Sahl, R.E.; Moritz, T.; Helge, J.W.; Clemmensen, C. Plasma Metabolome Profiling of Resistance Exercise and Endurance Exercise in Humans. Cell Rep. 2020, 33, 108554. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, Y.; Zhao, D.; Zhang, L.; Chen, P.; Xu, X. Integration of Metabolomics and Proteomics to Reveal the Metabolic Characteristics of High-Intensity Interval Training. Analyst 2020, 145, 6500–6510. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. Analysis of Metabonomic Characteristics after Exercise Fatigue Based on NMR. Contrast Media Mol. Imaging 2022, 2022, e9041293. [Google Scholar] [CrossRef]
- Nayor, M.; Shah, R.V.; Miller, P.E.; Blodgett, J.B.; Tanguay, M.; Pico, A.R.; Murthy, V.L.; Malhotra, R.; Houstis, N.E.; Deik, A.; et al. Metabolic Architecture of Acute Exercise Response in Middle-Aged Adults in the Community. Ciculation 2020, 142, 1905–1924. [Google Scholar] [CrossRef]
- Li, K.; Schön, M.; Naviaux, J.C.; Monk, J.M.; Alchus-Laiferová, N.; Wang, L.; Straka, I.; Matejička, P.; Valkovič, P.; Ukropec, J.; et al. Cerebrospinal Fluid and Plasma Metabolomics of Acute Endurance Exercise. FASEB J. 2022, 36, e22408. [Google Scholar] [CrossRef] [PubMed]
- Gallois, A.; Mefford, J.; Ko, A.; Vaysse, A.; Julienne, H.; Ala-Korpela, M.; Laakso, M.; Zaitlen, N.; Pajukanta, P.; Aschard, H. A Comprehensive Study of Metabolite Genetics Reveals Strong Pleiotropy and Heterogeneity across Time and Context. Nat. Commun. 2019, 10, 4788. [Google Scholar] [CrossRef] [PubMed]
- Lawless, C.E.; Olshansky, B.; Washington, R.L.; Baggish, A.L.; Daniels, C.J.; Lawrence, S.M.; Sullivan, R.M.; Kovacs, R.J.; Bove, A.A. Sports and exercise cardiology in the United States: Cardiovascular specialists as members of the athlete healthcare team. J. Am. Coll. Cardiol. 2014, 63, 1461–1472. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.V.; Burgess, S.; Fan, B.; Schooling, C.M. L-Carnitine, a Friend or Foe for Cardiovascular Disease? A Mendelian Randomization Study. BMC Med. 2022, 20, 272. [Google Scholar] [CrossRef]
- Matthay, R.A. Effects of Theophylline on Cardiovascular Performance in Chronic Obstructive Pulmonary Disease. Chest 1985, 88, 112S–117S. [Google Scholar] [CrossRef]
- Tousoulis, D.; Papageorgiou, N.; Stefanadis, C. Ursodeoxycholic acid in patients with chronic heart failure. J. Am. Coll. Cardiol. 2012, 60, 1579–1580. [Google Scholar] [CrossRef]
- Nickolas, T.L. Treating Osteoporosis with Denosumab in Patients on Hemodialysis: The Good, the Bad, and the Ugly. Clin. J. Am. Soc. Nephrol. 2023, 18, 1116–1118. [Google Scholar] [CrossRef]
- Li, Y.; Sekula, P.; Wuttke, M.; Wahrheit, J.; Hausknecht, B.; Schultheiss, U.T.; Gronwald, W.; Schlosser, P.; Tucci, S.; Ekici, A.B.; et al. Genome-Wide Association Studies of Metabolites in Patients with CKD Identify Multiple Loci and Illuminate Tubular Transport Mechanisms. J. Am. Soc. Nephrol. 2018, 29, 1513–1524. [Google Scholar] [CrossRef]
- Kennedy, M. Effects of Theophylline and Theobromine on Exercise Performance and Implications for Competition Sport: A Systematic Review. Drug Test. Anal. 2021, 13, 36–43. [Google Scholar] [CrossRef]
- Chorostowska-Wynimko, J.; Kus, J.; Skopińska-Rózewska, E. Theophylline Inhibits Free Oxygen Radicals Production by Human Monocytes via Phosphodiesterase Inhibition. J. Physiol. Pharmacol. 2007, 58 (Suppl. 5), 95–103. [Google Scholar]
- McLellan, T.M.; Caldwell, J.A.; Lieberman, H.R. A Review of Caffeine’s Effects on Cognitive, Physical and Occupational Performance. Neurosci. Biobehav. Rev. 2016, 71, 294–312. [Google Scholar] [CrossRef] [PubMed]
- Greer, F.; Friars, D.; Graham, T.E. Comparison of Caffeine and Theophylline Ingestion: Exercise Metabolism and Endurance. J. Appl. Physiol. (1985) 2000, 89, 1837–1844. [Google Scholar] [CrossRef] [PubMed]
- Seo, W.-K.; Jo, G.; Shin, M.-J.; Oh, K. Medium-Chain Acylcarnitines Are Associated with Cardioembolic Stroke and Stroke Recurrence. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2245–2253. [Google Scholar] [CrossRef]
- Saud Gany, S.L.; Tan, J.K.; Chin, K.Y.; Hakimi, N.H.; Ab Rani, N.; Ihsan, N.; Makpol, S. Untargeted Muscle Tissue Metabolites Profiling in Young, Adult, and Old Rats Supplemented with Tocotrienol-Rich Fraction. Front. Mol. Biosci. 2022, 9, 1008908. [Google Scholar] [CrossRef]
- Shulman, R.G.; Rothman, D.L. The “Glycogen Shunt” in Exercising Muscle: A Role for Glycogen in Muscle Energetics and Fatigue. Proc. Natl. Acad. Sci. USA 2001, 98, 457–461. [Google Scholar] [CrossRef]
- Pla, R.; Pujos-Guillot, E.; Durand, S.; Brandolini-Bunlon, M.; Centeno, D.; Pyne, D.B.; Toussaint, J.F.; Hellard, P. Non-Targeted Metabolomics Analyses by Mass Spectrometry to Explore Metabolic Stress after Six Training Weeks in High Level Swimmers. J. Sports Sci. 2021, 39, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Hanafi, N.I.; Mohamed, A.S.; Sheikh Abdul Kadir, S.H.; Othman, M.H.D. Overview of Bile Acids Signaling and Perspective on the Signal of Ursodeoxycholic Acid, the Most Hydrophilic Bile Acid, in the Heart. Biomolecules 2018, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, J.; Shevchuk, A.I.; Diakonov, I.; de Swiet, M.; Lab, M.; Korchev, Y.; Williamson, C. Dexamethasone and Ursodeoxycholic Acid Protect against the Arrhythmogenic Effect of Taurocholate in an in Vitro Study of Rat Cardiomyocytes. BJOG 2003, 110, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Abdul Kadir, S.H.S.; Ali, N.N.; Mioulane, M.; Brito-Martins, M.; Abu-Hayyeh, S.; Foldes, G.; Moshkov, A.V.; Williamson, C.; Harding, S.E.; Gorelik, J. Embryonic Stem Cell-Derived Cardiomyocytes as a Model to Study Fetal Arrhythmia Related to Maternal Disease. J. Cell Mol. Med. 2009, 13, 3730–3741. [Google Scholar] [CrossRef]
- Mohamed, A.S.; Hanafi, N.I.; Sheikh Abdul Kadir, S.H.; Md Noor, J.; Abdul Hamid Hasani, N.; Ab Rahim, S.; Siran, R. Ursodeoxycholic Acid Protects Cardiomyocytes against Cobalt Chloride Induced Hypoxia by Regulating Transcriptional Mediator of Cells Stress Hypoxia Inducible Factor 1α and P53 Protein. Cell Biochem. Funct. 2017, 35, 453–463. [Google Scholar] [CrossRef]
| rs 1052373 | rs 7120118 | |||
|---|---|---|---|---|
| Genotype | GG | TT | ||
| GWAS | p OR (95% CI) | 5.48 × 10−6 2.61 (1.72–3.94) | p OR (95% CI) | 1.26 × 10−5 2.49 (1.65–3.75) |
| Russian cohort | p OR (95% CI) | 1.2 × 10−2 1.67 (1.12–2.49) | p OR (95% CI) | 1.6 × 10−2 1.64 (1.10–2.45) |
| Japanese cohort | p OR (95% CI) | 2.7 × 10−3 2.92 (1.41–6.05) | p OR (95% CI) | 3.52 × 10−2 2.48 (1.10–5.56) |
| Combined | p OR (95% CI) | 1.43 × 10−8 2.17 (1.67–2.84) | p OR (95% CI) | 1.66 × 10−7 2.07 (1.59–2.7) |
| Exercise | Findings | Reference |
|---|---|---|
| Trans Japan Alps race | Increase in lipid metabolism and hemolysis. | [46] |
| Endurance exercise (bike) | Significant alterations in metabolites linked to cellular energy processes. Induction of metabolites associated with glycolytic pathways. | [47] |
| Shuttle runs | Significant upregulation of metabolites associated with amino acids after exercise. Downregulation of steroid hormone metabolism after exercise. | [48] |
| Endurance and speed endurance training | Exercise leads to significant changes in metabolism. Before and after exercise, there is a rise in lactic acid and glycine levels while the concentration of creatinine decreases. | [49] |
| Cardiopulmonary exercise | Reduction in produced metabolites associated with insulin resistance. Increase in metabolites linked to lipolysis. | [50] |
| Outdoor running | Activation of metabolic pathways of amino acid and fatty acid metabolism. Mitochondrial and metabolic changes induced by ATP signaling. | [51] |
| Metabolite | rsID | SNPpos | HapMap Allele | Beta GWAS Server | p-Value | Reference |
|---|---|---|---|---|---|---|
| Theophylline | 10769255 | chr11:47367371 | C/T | 0.0523 | 5.217 × 10−4 | [55] |
| Urso-deoxycholate | rs10838696 | chr11:47363285 | G/A | 0.04233 | 8.696 × 10−4 | [56] |
| Quinate | rs2856650 | chr11:47365199 | C/T | −0.082 | 2.257 × 10−4 | [57] |
| Decanoyl-carnitine | rs11570058 | chr11:47369443 | C/T | −0.0232 | 6.64 × 10−5 | [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riguene, E.; Theodoridou, M.; Barrak, L.; Elrayess, M.A.; Nomikos, M. The Relationship between Changes in MYBPC3 Single-Nucleotide Polymorphism-Associated Metabolites and Elite Athletes’ Adaptive Cardiac Function. J. Cardiovasc. Dev. Dis. 2023, 10, 400. https://doi.org/10.3390/jcdd10090400
Riguene E, Theodoridou M, Barrak L, Elrayess MA, Nomikos M. The Relationship between Changes in MYBPC3 Single-Nucleotide Polymorphism-Associated Metabolites and Elite Athletes’ Adaptive Cardiac Function. Journal of Cardiovascular Development and Disease. 2023; 10(9):400. https://doi.org/10.3390/jcdd10090400
Chicago/Turabian StyleRiguene, Emna, Maria Theodoridou, Laila Barrak, Mohamed A. Elrayess, and Michail Nomikos. 2023. "The Relationship between Changes in MYBPC3 Single-Nucleotide Polymorphism-Associated Metabolites and Elite Athletes’ Adaptive Cardiac Function" Journal of Cardiovascular Development and Disease 10, no. 9: 400. https://doi.org/10.3390/jcdd10090400
APA StyleRiguene, E., Theodoridou, M., Barrak, L., Elrayess, M. A., & Nomikos, M. (2023). The Relationship between Changes in MYBPC3 Single-Nucleotide Polymorphism-Associated Metabolites and Elite Athletes’ Adaptive Cardiac Function. Journal of Cardiovascular Development and Disease, 10(9), 400. https://doi.org/10.3390/jcdd10090400







