Embryonic Mouse Cardiodynamic OCT Imaging
Abstract
:1. Introduction
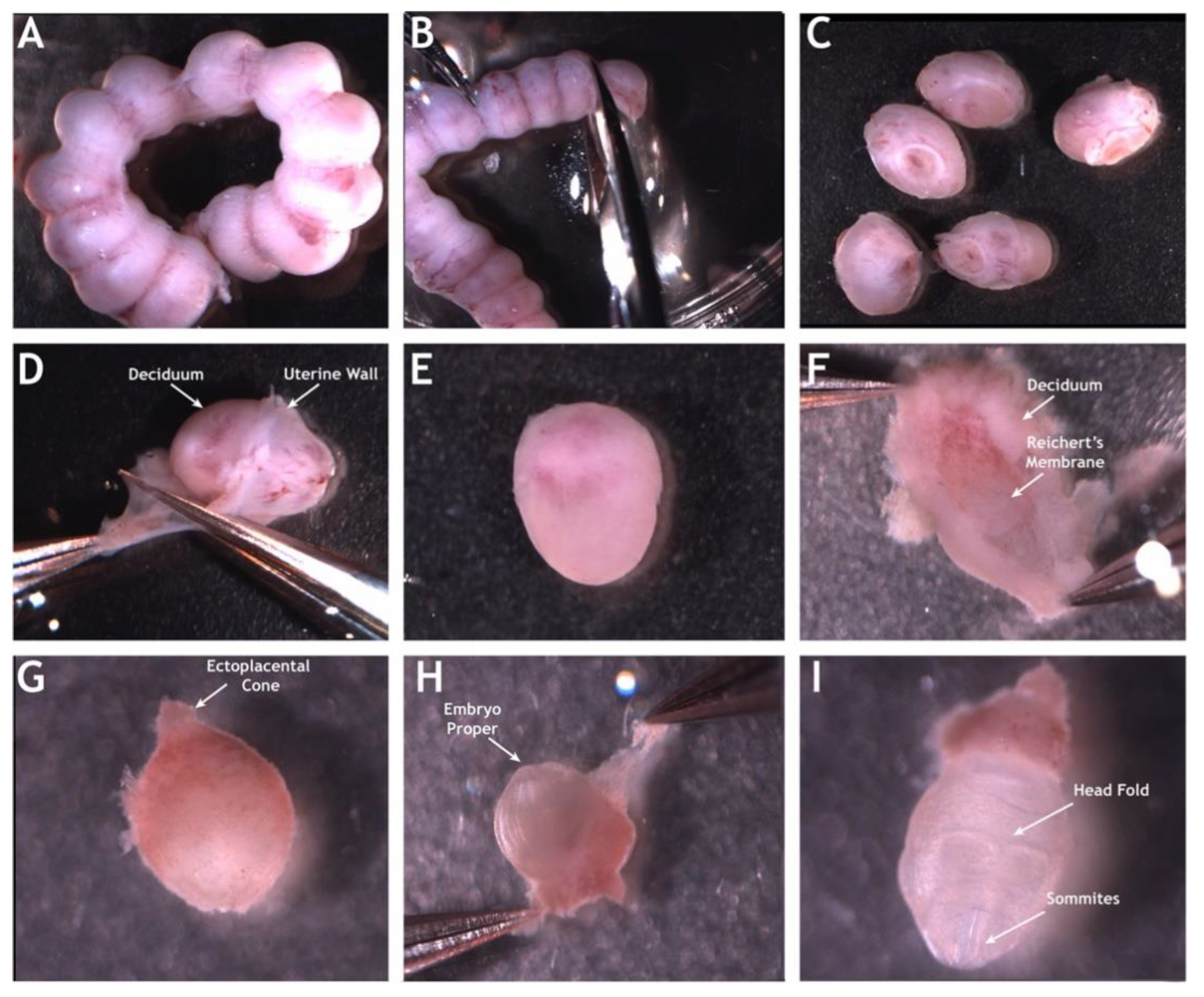
2. Mouse Embryonic Imaging in Static Culture
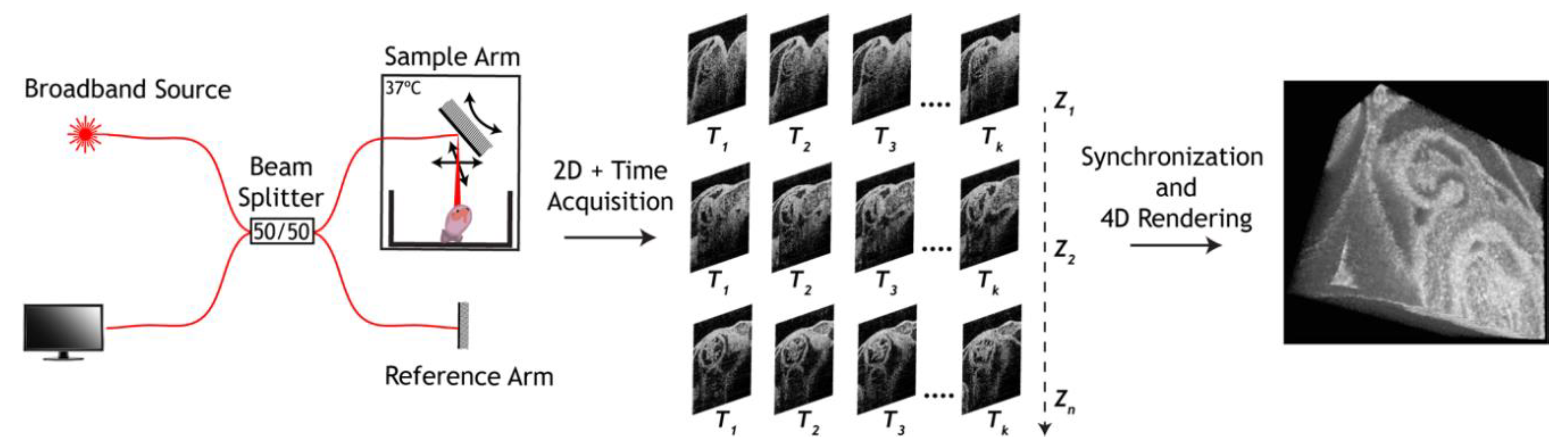
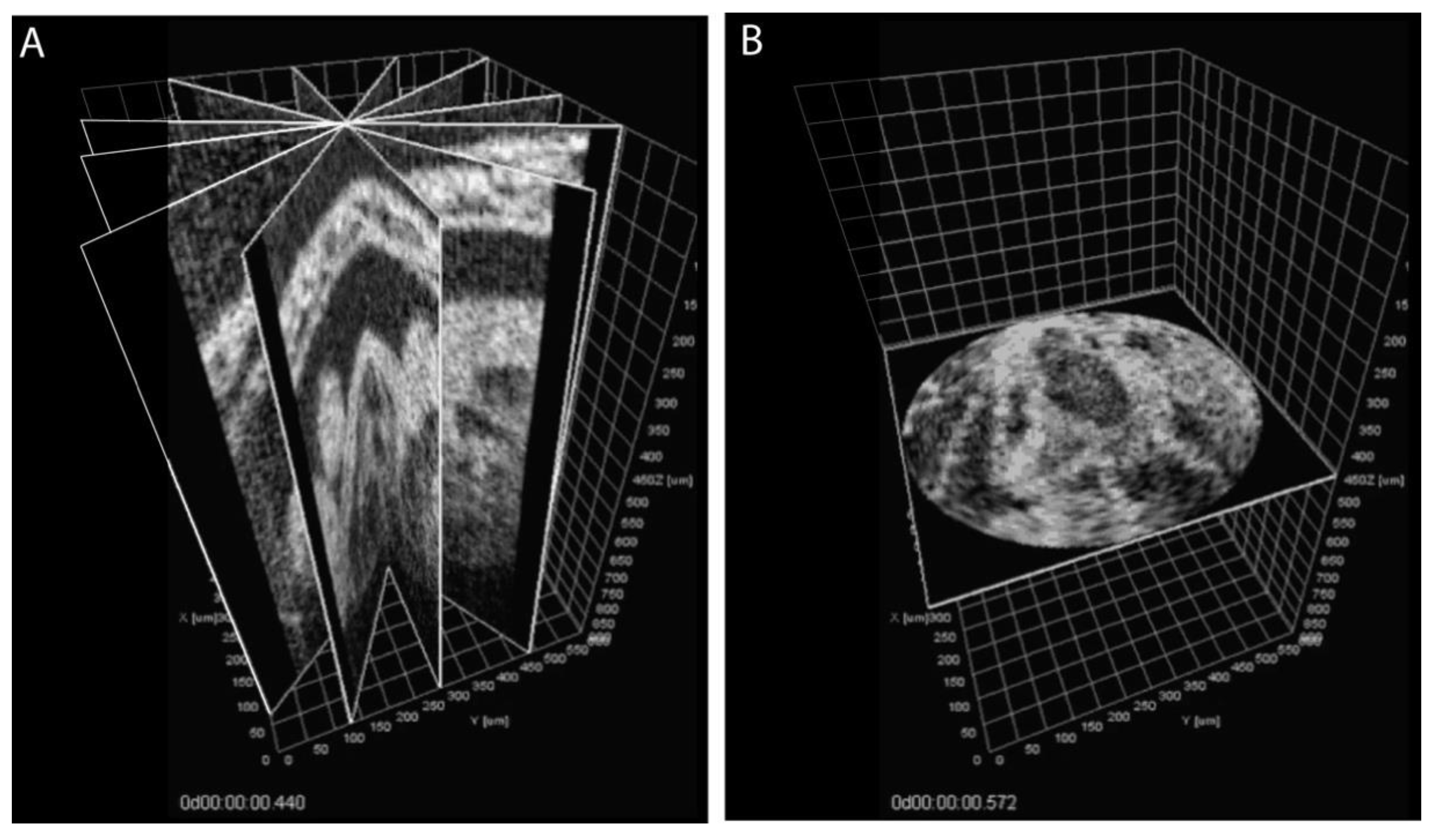
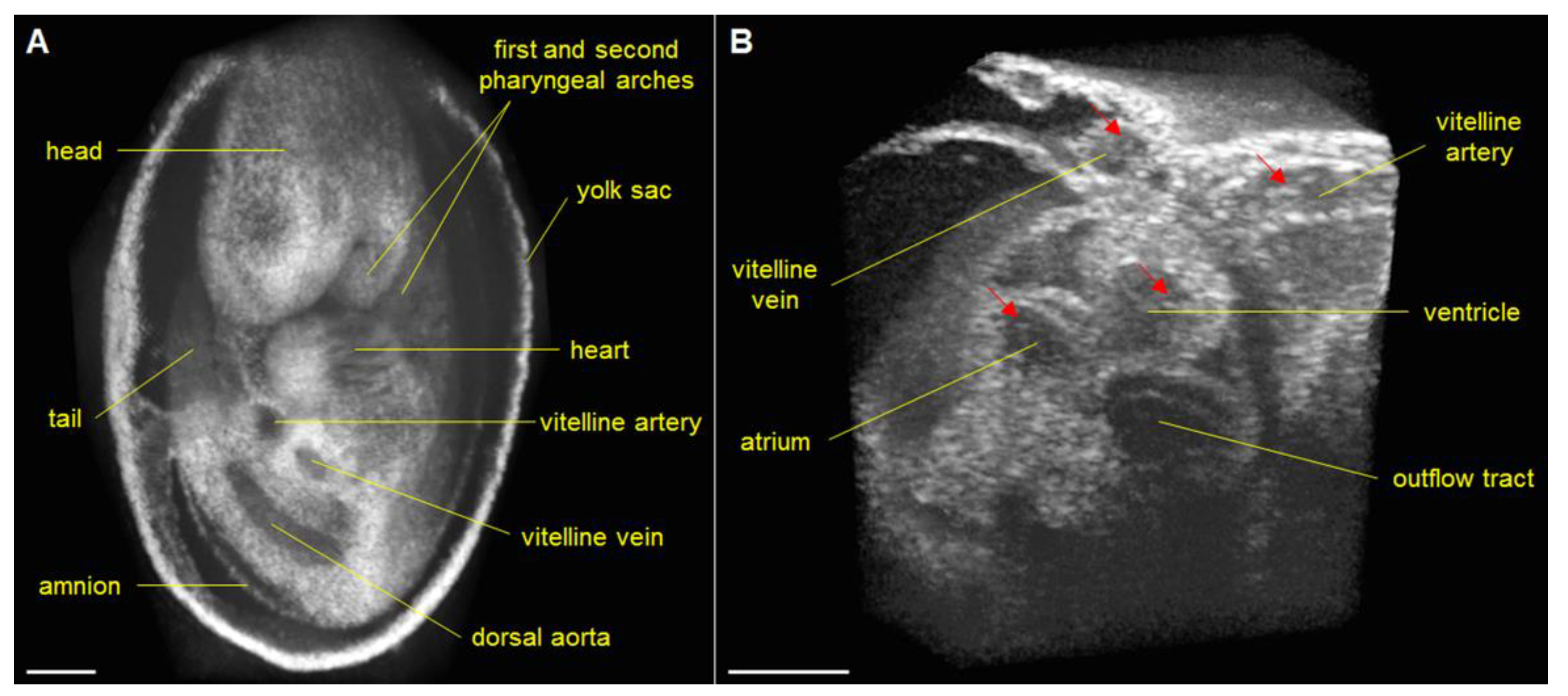
3. Approaches to Volumetric Cardiodynamic Imaging
4. Cardiac Phenotyping of Genetic Mouse Models
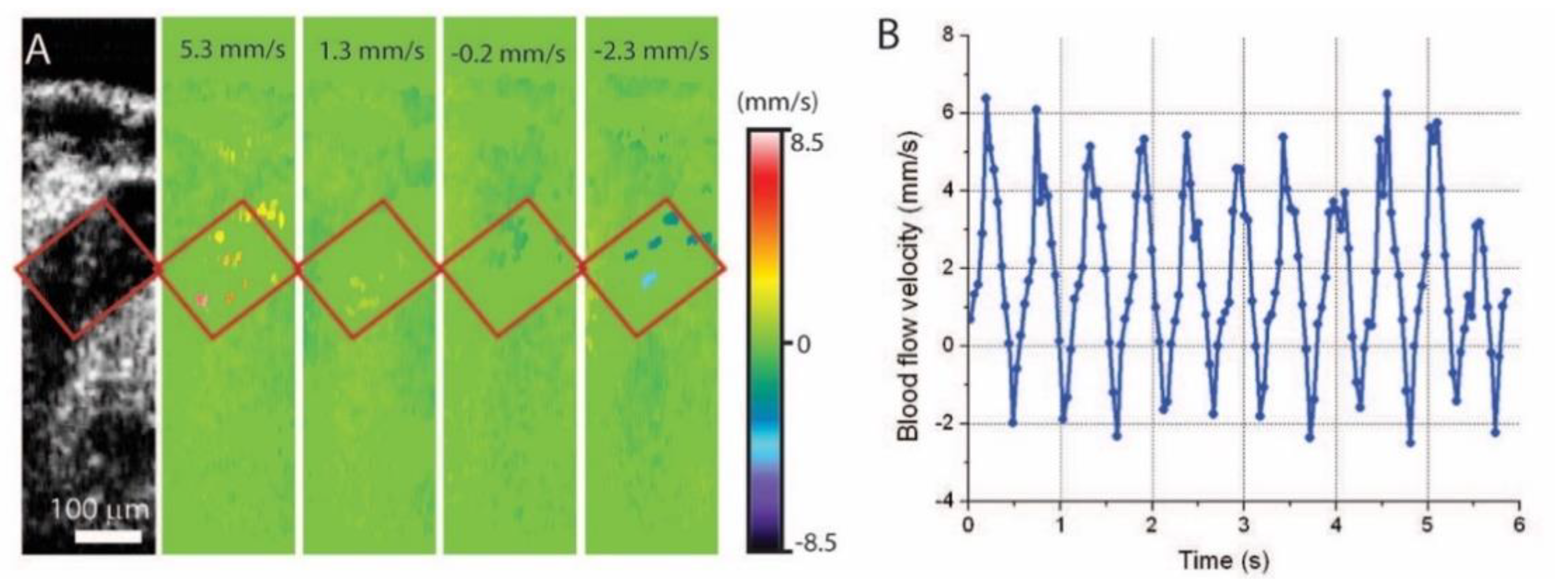
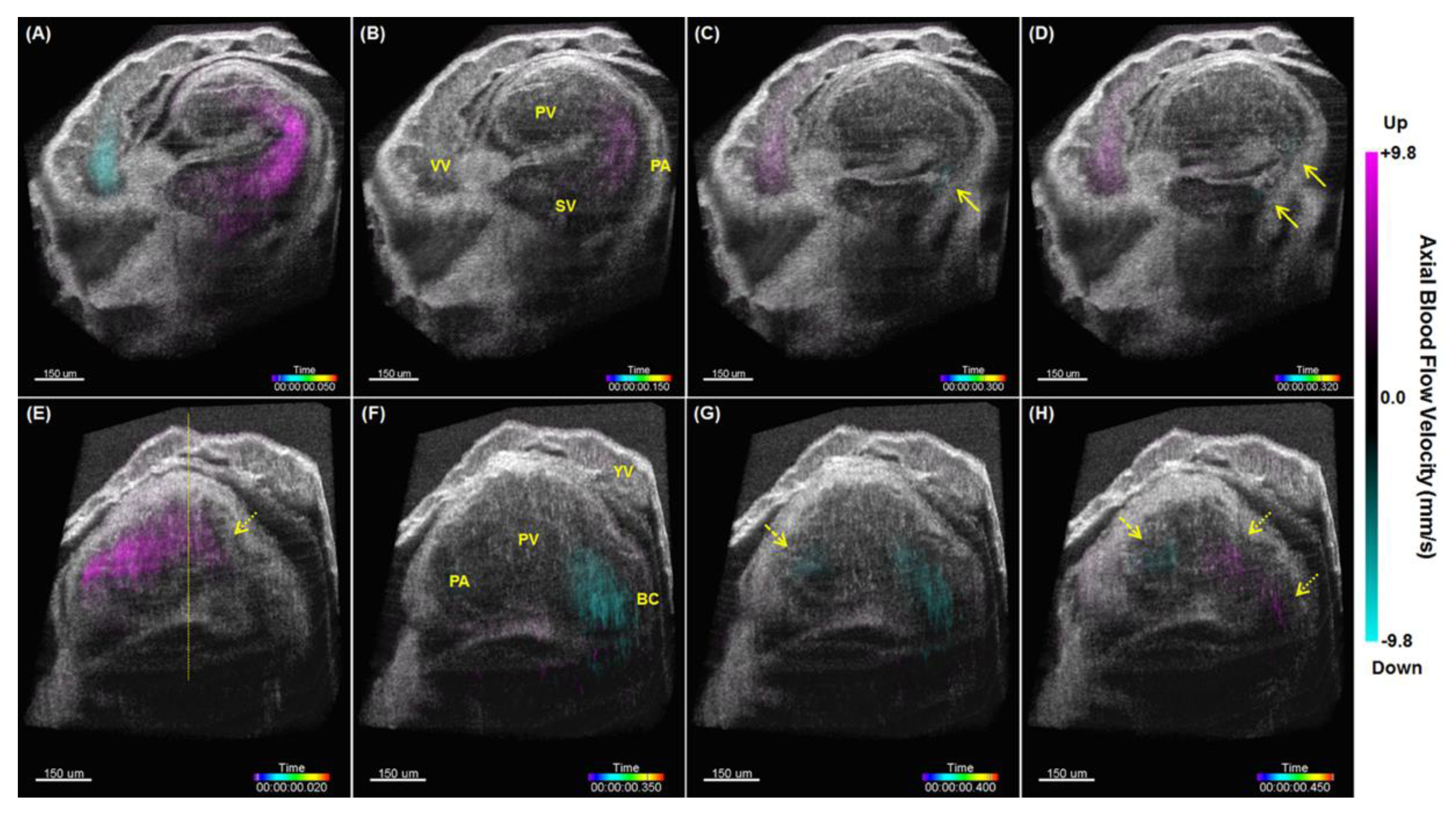
5. Hemodynamic Imaging with Doppler OCT
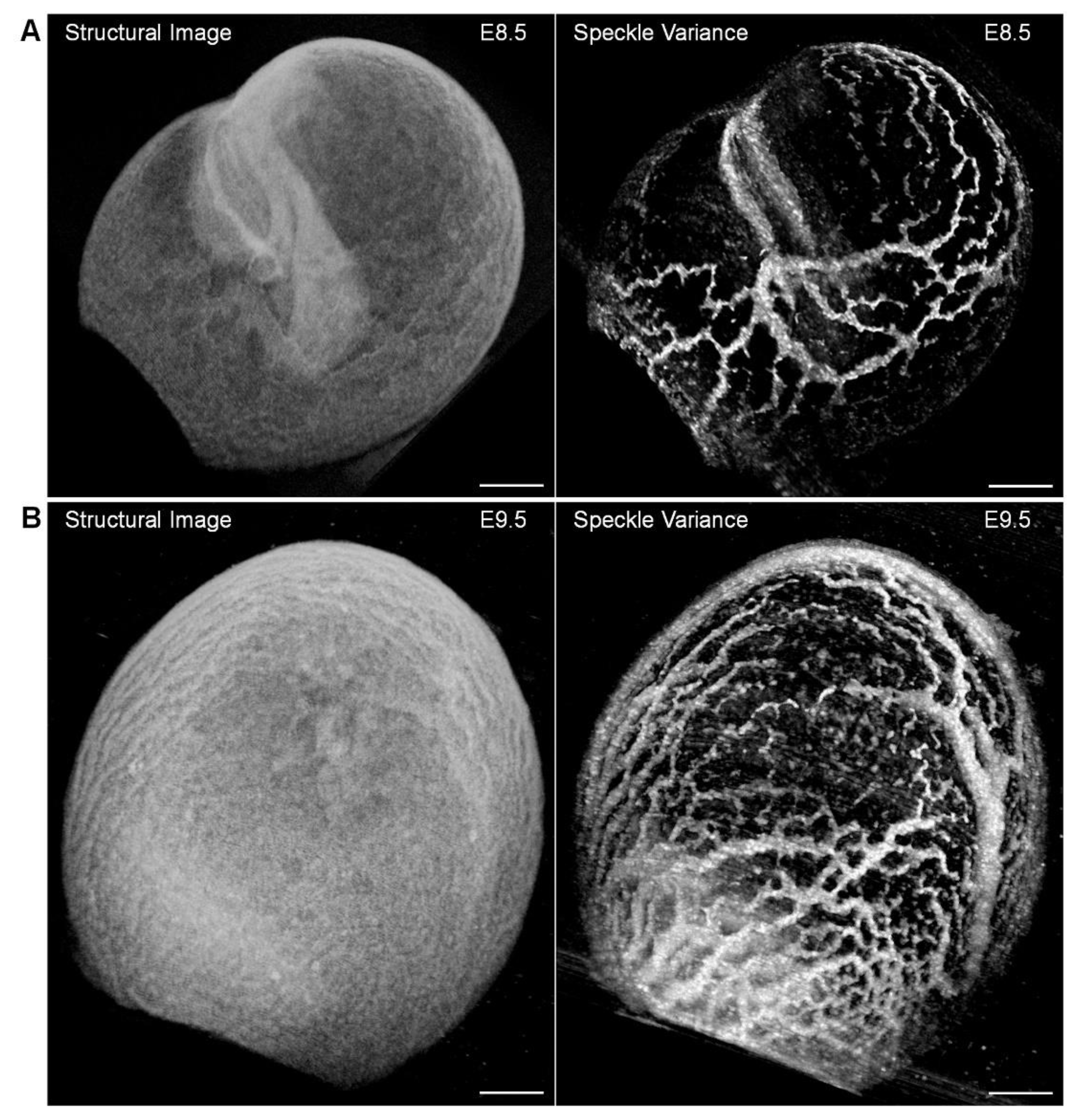
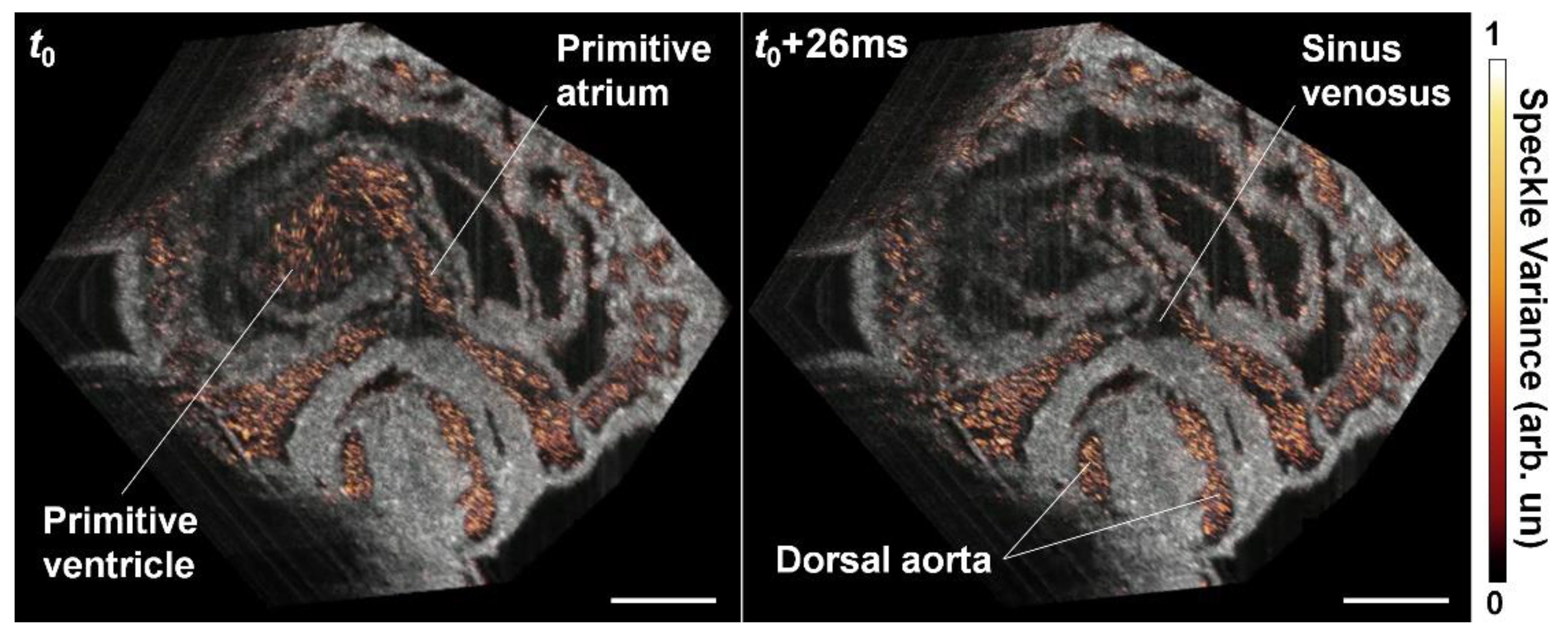
6. OCT Angiography Approach for Cardiovascular Imaging
7. Perspective on Future Developments
Funding
Acknowledgments
Conflicts of Interest
References
- Bondue, A.; Blanpain, C. Mesp1 A Key Regulator of Cardiovascular Lineage Commitment. Circ. Res. 2010, 107, 1414–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.M. Mesp1 at the heart of mesoderm lineage specification. Cell Stem Cell 2008, 3, 1–2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Liang, X.; Najafi, N.; Cass, M.; Lin, L.; Cai, C.-L.; Chen, J.; Evans, S.M. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev. Biol. 2007, 304, 286–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leach, J.P.; Heallen, T.; Zhang, M.; Rahmani, M.; Morikawa, Y.; Hill, M.C.; Segura, A.; Willerson, J.T.; Martin, J.F. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 2017, 550, 260–264. [Google Scholar] [CrossRef] [Green Version]
- Heallen, T.; Zhang, M.; Wang, J.; Bonilla-Claudio, M.; Klysik, E.; Johnson, R.L.; Martin, J.F. Hippo Pathway Inhibits Wnt Signaling to Restrain Cardiomyocyte Proliferation and Heart Size. Science 2011, 332, 458–461. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.-J.; Lin, C.-Y.; Chen, C.-H.; Zhou, B.; Chang, C.-P. Partitioning the heart: Mechanisms of cardiac septation and valve development. Development 2012, 139, 3277–3299. [Google Scholar] [CrossRef] [Green Version]
- Furtado, M.B.; Costa, M.W.; Rosenthal, N.A. The cardiac fibroblast: Origin, identity and role in homeostasis and disease. Differentiation 2016, 92, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Acharya, A.; Baek, S.; Huang, G.; Eskiocak, B.; Goetsch, S.; Sung, C.Y.; Banfi, S.; Sauer, M.F.; Olsen, G.S.; Duffield, J.S.; et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 2012, 139, 2139–2149. [Google Scholar] [CrossRef] [Green Version]
- Van Weerd, J.H.; Christoffels, V.M. The formation and function of the cardiac conduction system. Development 2016, 143, 197–210. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Wang, G.; Lin, L.; Lowe, J.; Zhang, Q.; Bu, L.; Chen, Y.; Chen, J.; Sun, Y.; Evans, S.M. HCN4 Dynamically Marks the First Heart Field and Conduction System Precursors. Circ. Res. 2013, 113, 399–407. [Google Scholar] [CrossRef]
- Srivastava, D.; Olson, E.N. A genetic blueprint for cardiac development. Nature 2000, 407, 35025190. [Google Scholar] [CrossRef] [PubMed]
- Karunamuni, G.H.; Gu, S.; Ford, M.R.; Peterson, L.M.; Ma, P.; Wang, Y.T.; Rollins, A.M.; Jenkins, M.W.; Watanabe, M. Capturing structure and function in an embryonic heart with biophotonic tools. Front. Physiol. 2014, 5, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yelbuz, T.M.; Choma, M.A.; Thrane, L.; Kirby, M.L.; Izatt, J.A. Optical Coherence Tomography. Circulation 2002, 106, 2771–2774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, M.W.; Patel, P.; Deng, H.; Montano, M.M.; Watanabe, M.; Rollins, A.M. Phenotyping transgenic embryonic murine hearts using optical coherence tomography. Appl. Opt. 2007, 46, 1776–1781. [Google Scholar] [CrossRef] [PubMed]
- Garita, B.; Jenkins, M.W.; Han, M.; Zhou, C.; VanAuker, M.; Rollins, A.M.; Watanabe, M.; Fujimoto, J.G.; Linask, K.K. Blood flow dynamics of one cardiac cycle and relationship to mechanotransduction and trabeculation during heart looping. Am. J. Physiol.-Heart Circ. Physiol. 2011, 300, H879–H891. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.; McLachlan, J.C. Looping of chick embryo hearts in vitro. J. Anat. 1990, 168, 257–263. [Google Scholar] [PubMed]
- Lopez, A.L.; Garcia, M.D.; Dickinson, M.E.; Larina, I.V. Live confocal microscopy of the developing mouse embryonic yolk sac vasculature. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2015; Volume 1214, pp. 163–172. [Google Scholar] [CrossRef]
- Parasoglou, P.; Berrios-Otero, C.A.; Nieman, B.J.; Turnbull, D.H. High-resolution MRI of early-stage mouse embryos. NMR Biomed. 2013, 26, 224–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Wu, D.; Turnbull, D.H. In Utero MRI of Mouse Embryos. Methods Mol. Biol. 2018, 1718, 285–296. [Google Scholar] [CrossRef]
- Srinivasan, S.; Baldwin, H.S.; Aristizabal, O.; Kwee, L.; Labow, M.; Artman, M.; Turnbull, D.H. Noninvasive, In Utero Imaging of Mouse Embryonic Heart Development With 40-MHz Echocardiography. Circulation 1998, 98, 912–918. [Google Scholar] [CrossRef]
- Phoon, C.K.L.; Aristizabal, O.; Turnbull, D.H. 40 MHz Doppler characterization of umbilical and dorsal aortic blood flow in the early mouse embryo. Ultrasound Med. Biol. 2000, 26, 1275–1283. [Google Scholar] [CrossRef]
- Wang, S.; Larina, I.V.; Larin, K.V. Label-free optical imaging in developmental biology [Invited]. Biomed. Opt. Express 2020, 11, 2017–2040. [Google Scholar] [CrossRef] [PubMed]
- Larina, I.V.; Larin, K.V.; Justice, M.J.; Dickinson, M.E. Optical Coherence Tomography for live imaging of mammalian development. Curr. Opin. Genet. Dev. 2011, 21, 579–584. [Google Scholar] [CrossRef] [Green Version]
- Raghunathan, R.; Singh, M.; Dickinson, M.E.; Larin, K.V. Optical coherence tomography for embryonic imaging: A review. J. Biomed. Opt. 2016, 21, 050902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boppart, S.A.; Brezinski, M.E.; Bouma, B.E.; Tearney, G.J.; Fujimoto, J.G. Investigation of developing embryonic morphology using optical coherence tomography. Dev. Biol. 1996, 177, 54–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boppart, S.A.; Bouma, B.E.; Brezinski, M.E.; Tearney, G.J.; Fujimoto, J.G. Imaging developing neural morphology using optical coherence tomography. J. Neurosci. Methods 1996, 70, 65–72. [Google Scholar] [CrossRef]
- Boppart, S.A.; Tearney, G.J.; Bouma, B.E.; Southern, J.F.; Brezinski, M.E.; Fujimoto, J.G. Noninvasive assessment of the developing Xenopus cardiovascular system using optical coherence tomography. Proc. Natl. Acad. Sci. USA 1997, 94, 4256–4261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Boer, J.F.; Leitgeb, R.; Wojtkowski, M. Twenty-five years of optical coherence tomography: The paradigm shift in sensitivity and speed provided by Fourier domain OCT [Invited]. Biomed. Opt. Express 2017, 8, 3248–3280. [Google Scholar] [CrossRef] [Green Version]
- Larin, K.V.; Larina, I.V.; Liebling, M.; Dickinson, M.E. Live Imaging of Early Developmental Processes in Mammalian Embryos with Optical Coherence Tomography. J. Innov. Opt. Health Sci. 2009, 2, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Brown, W.; Maher, J.R.; Levinson, H.; Wax, A. Functional optical coherence tomography: Principles and progress. Phys. Med. Biol. 2015, 60, R211–R237. [Google Scholar] [CrossRef]
- Larina, I.V.; Sudheendran, N.; Ghosn, M.; Jiang, J.; Cable, A.; Larin, K.V.; Dickinson, M.E. Live imaging of blood flow in mammalian embryos using Doppler swept-source optical coherence tomography. J. Biomed. Opt. 2008, 13, 060506. [Google Scholar] [CrossRef] [Green Version]
- Sudheendran, N.; Syed, S.H.; Dickinson, M.E.; Larina, I.V.; Larin, K.V. Speckle variance OCT imaging of the vasculature in live mammalian embryos. Laser Phys. Lett. 2011, 8, 247–252. [Google Scholar] [CrossRef]
- Jenkins, M.W.; Rothenberg, F.; Roy, D.; Nikolski, V.P.; Hu, Z.; Watanabe, M.; Wilson, D.L.; Efimov, I.R.; Rollins, A.M. 4D embryonic cardiography using gated optical coherence tomography. Opt. Express 2006, 14, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Marks, D.L.; Ralston, T.S.; Boppart, S.A. Three-dimensional optical coherence tomography of the embryonic murine cardiovascular system. J. Biomed. Opt. 2006, 11, 021014. [Google Scholar] [CrossRef]
- Hammond, J. Recovery and Culture of Tubal Mouse Ova. Nature 1949, 163, 28–29. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.; Rieger, D. Historical Background of Gamete and Embryo Culture. In Embryo Culture, Methods and Protocols; Thomas, B., Pool, J.E.S., Gary, D.S., Eds.; Humana Press: London, UK, 2012. [Google Scholar] [CrossRef]
- Udan, R.S.; Vadakkan, T.J.; Dickinson, M.E. Dynamic responses of endothelial cells to changes in blood flow during vascular remodeling of the mouse yolk sac. Development 2013, 140, 4041–4050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, E.A.V.; Crotty, D.; Kulesa, P.M.; Waters, C.W.; Baron, M.H.; Fraser, S.E.; Dickinson, M.E. Dynamic in vivo imaging of postimplantation mammalian embryos using whole embryo culture. Genesis 2002, 34, 228–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tam, P.P. Postimplantation mouse development: Whole embryo culture and micro-manipulation. Int. J. Dev. Biol. 1998, 42, 895–902. [Google Scholar]
- Wang, S.; Garcia, M.D.; Lopez, A.L.; Overbeek, P.A.; Larin, K.V.; Larina, I.V. Dynamic imaging and quantitative analysis of cranial neural tube closure in the mouse embryo using optical coherence tomography. Biomed. Opt. Express 2017, 8, 407–419. [Google Scholar] [CrossRef]
- Lopez, A.L.; Wang, S.; Larin, K.V.; Overbeek, P.A.; Larina, I.V. Live four-dimensional optical coherence tomography reveals embryonic cardiac phenotype in mouse mutant. J. Biomed. Opt. 2015, 20, 090501. [Google Scholar] [CrossRef] [Green Version]
- Lopez, R.A.L.; Larina, I.V. Dynamic Imaging of Mouse Embryos and Cardiodynamics in Static Culture. Methods Mol. Biol. 2018, 1752, 41. [Google Scholar]
- Mariampillai, A.; Standish, B.A.; Munce, N.R.; Randall, C.; Liu, G.; Jiang, J.Y.; Cable, A.E.; Vitkin, I.A.; Yang, V.X. Doppler optical cardiogram gated 2D color flow imaging at 1000 fps and 4D in vivo visualization of embryonic heart at 45 fps on a swept source OCT system. Opt. Express 2007, 15, 1627–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebling, M.; Forouhar, A.S.; Gharib, M.; Fraser, S.E.; Dickinson, M.E. Four-dimensional cardiac imaging in living embryos via postacquisition synchronization of nongated slice sequences. J. Biomed. Opt. 2005, 10, 054001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebling, M.; Forouhar, A.S.; Wolleschensky, R.; Zimmermann, B.; Ankerhold, R.; Fraser, S.E.; Gharib, M.; Dickinson, M.E. Rapid three-dimensional imaging and analysis of the beating embryonic heart reveals functional changes during development. Dev. Dyn. 2006, 235, 2940–2948. [Google Scholar] [CrossRef] [PubMed]
- Grishina, O.A.; Wang, S.; Larina, I.V. Speckle variance optical coherence tomography of blood flow in the beating mouse embryonic heart. J. Biophotonics 2017, 10, 735–743. [Google Scholar] [CrossRef]
- Larina, I.V.; Larin, K.V.; Dickinson, M.E.; Liebling, M. Sequential Turning Acquisition and Reconstruction (STAR) method for four-dimensional imaging of cyclically moving structures. Biomed. Opt. Express 2012, 3, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Bhat, S.; Larina, I.V.; Larin, K.V.; Dickinson, M.E.; Liebling, M. 4D reconstruction of the beating embryonic heart from two orthogonal sets of parallel optical coherence tomography slice-sequences. IEEE Trans. Med. Imaging 2013, 32, 578–588. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Singh, M.; Lopez, A.L., 3rd; Wu, C.; Raghunathan, R.; Schill, A.; Li, J.; Larin, K.V.; Larina, I.V. Direct four-dimensional structural and functional imaging of cardiovascular dynamics in mouse embryos with 1.5 MHz optical coherence tomography. Opt. Lett. 2015, 40, 4791–4794. [Google Scholar] [CrossRef]
- Yoo, J.; Larina, I.V.; Larin, K.V.; Dickinson, M.E.; Liebling, M. Increasing the field-of-view of dynamic cardiac OCT via post-acquisition mosaicing without affecting frame-rate or spatial resolution. Biomed. Opt. Express 2011, 2, 2614–2622. [Google Scholar] [CrossRef] [Green Version]
- Bhat, S.; Larina, I.V.; Larin, K.V.; Dickinson, M.E.; Liebling, M. Multiple-cardiac-cycle noise reduction in dynamic optical coherence tomography of the embryonic heart and vasculature. Opt. Lett. 2009, 34, 3704–3706. [Google Scholar] [CrossRef] [Green Version]
- Lopez, A.; Wang, S.; Larin, K.; Larina, I. Live dynamic analysis of the developing cardiovascular system in mice. SPIE BiOS 2017, 10043, 100430Q. [Google Scholar]
- Chen, Z.; Zhang, J. Doppler Optical Coherence Tomography. In Optical Coherence Tomography: Technology and Applications; Drexler, W., Fujimoto, J.G., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 1289–1320. [Google Scholar] [CrossRef]
- Wang, X.J.; Milner, T.E.; Nelson, J.S. Characterization of fluid flow velocity by optical Doppler tomography. Opt. Lett. 1995, 20, 1337–1339. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Milner, T.E.; Srinivas, S.; Wang, X.; Malekafzali, A.; van Gemert, M.J.C.; Nelson, J.S. Noninvasive imaging of in vivo blood flow velocity using optical Doppler tomography. Opt. Lett. 1997, 22, 1119–1121. [Google Scholar] [CrossRef] [PubMed]
- Izatt, J.A.; Kulkarni, M.D.; Yazdanfar, S.; Barton, J.K.; Welch, A.J. In vivo bidirectional color Doppler flow imaging of picoliter blood volumes using optical coherence tomography. Opt. Lett. 1997, 22, 1439–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Chen, Z.; Ding, Z.; Ren, H.; Nelson, J.S. Real-time phase-resolved functional optical coherence tomography by use of optical Hilbert transformation. Opt. Lett. 2002, 27, 98–100. [Google Scholar] [CrossRef] [PubMed]
- Westphal, V.; Yazdanfar, S.; Rollins, A.M.; Izatt, J.A. Real-time, high velocity-resolution color Doppler optical coherence tomography. Opt. Lett. 2002, 27, 34–36. [Google Scholar] [CrossRef] [PubMed]
- Yang, V.; Gordon, M.; Qi, B.; Pekar, J.; Lo, S.; Seng-Yue, E.; Mok, A.; Wilson, B.; Vitkin, I. High speed, wide velocity dynamic range Doppler optical coherence tomography (Part I): System design, signal processing, and performance. Opt. Express 2003, 11, 794–809. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Lin, A.J.; Tromberg, B.J.; Chen, Z. A comparison of Doppler optical coherence tomography methods. Biomed. Opt. Express 2012, 3, 2669–2680. [Google Scholar] [CrossRef]
- Yazdanfar, S.; Kulkarni, M.D.; Izatt, J.A. High resolution imaging of in vivo cardiac dynamics using color Doppler optical coherence tomography. Opt. Express 1997, 1, 424–431. [Google Scholar] [CrossRef]
- Yang, V.X.D.; Gordon, M.L.; Seng-Yue, E.; Lo, S.; Qi, B.; Pekar, J.; Mok, A.; Wilson, B.C.; Vitkin, I.A. High speed, wide velocity dynamic range Doppler optical coherence tomography (Part II): Imaging in vivo cardiac dynamics of Xenopus laevis. Opt. Express 2003, 11, 1650–1658. [Google Scholar] [CrossRef] [Green Version]
- Choma, M.A.; Suter, M.J.; Vakoc, B.J.; Bouma, B.E.; Tearney, G.J. Heart wall velocimetry and exogenous contrast-based cardiac flow imaging in Drosophila melanogaster using Doppler optical coherence tomography. J. Biomed. Opt. 2010, 15, 056020. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Jenkins, M.W.; Peterson, L.M.; Doughman, Y.-Q.; Rollins, A.M.; Watanabe, M. Optical coherence tomography captures rapid hemodynamic responses to acute hypoxia in the cardiovascular system of early embryos. Dev. Dyn. 2012, 241, 534–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, M.W.; Peterson, L.M.; Gu, S.; Gargesha, M.; Wilson, D.L.; Watanabe, M.; Rollins, A.M. Measuring hemodynamics in the developing heart tube with four-dimensional gated Doppler optical coherence tomography. J. Biomed. Opt. 2010, 15, 066022. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Yin, X.; Shi, L.; Li, P.; Thornburg, K.L.; Wang, R.; Rugonyi, S. Biomechanics of the Chick Embryonic Heart Outflow Tract at HH18 Using 4D Optical Coherence Tomography Imaging and Computational Modeling. PLoS ONE 2012, 7, e40869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, L.M.; Jenkins, M.W.; Gu, S.; Barwick, L.; Watanabe, M.; Rollins, A.M. 4D shear stress maps of the developing heart using Doppler optical coherence tomography. Biomed. Opt. Express 2012, 3, 3022–3032. [Google Scholar] [CrossRef] [Green Version]
- Ford, S.M.; McPheeters, M.T.; Wang, Y.T.; Ma, P.; Gu, S.; Strainic, J.; Snyder, C.; Rollins, A.M.; Watanabe, M.; Jenkins, M.W. Increased regurgitant flow causes endocardial cushion defects in an avian embryonic model of congenital heart disease. Congenit. Heart Dis. 2017, 12, 322–331. [Google Scholar] [CrossRef] [Green Version]
- Midgett, M.; Goenezen, S.; Rugonyi, S. Blood flow dynamics reflect degree of outflow tract banding in Hamburger–Hamilton stage 18 chicken embryos. J. R. Soc. Interface 2014, 11, 20140643. [Google Scholar] [CrossRef]
- Larina, I.V.; Ivers, S.; Syed, S.; Dickinson, M.E.; Larin, K.V. Hemodynamic measurements from individual blood cells in early mammalian embryos with Doppler swept source OCT. Opt. Lett. 2009, 34, 986–988. [Google Scholar] [CrossRef]
- Wang, S.; Lakomy, D.S.; Garcia, M.D.; Lopez, A.L.; Larin, K.V.; Larina, I.V. Four-dimensional live imaging of hemodynamics in mammalian embryonic heart with Doppler optical coherence tomography. J. Biophotonics 2016, 9, 837–847. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Larina, I.V. Live mechanistic assessment of localized cardiac pumping in mammalian tubular embryonic heart. J. Biomed. Opt. 2020, 25, 086001. [Google Scholar] [CrossRef]
- Mahmud, M.S.; Cadotte, D.W.; Vuong, B.; Sun, C.; Luk, T.W.; Mariampillai, A.; Yang, V.X. Review of speckle and phase variance optical coherence tomography to visualize microvascular networks. J. Biomed. Opt. 2013, 18, 50901. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-L.; Wang, R.K. Optical coherence tomography based angiography [Invited]. Biomed. Opt. Express 2017, 8, 1056–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Y.; Tan, O.; Tokayer, J.; Potsaid, B.; Wang, Y.; Liu, J.J.; Kraus, M.F.; Subhash, H.; Fujimoto, J.G.; Hornegger, J.; et al. Split-spectrum amplitude-decorrelation angiography with optical coherence tomography. Opt. Express 2012, 20, 4710–4725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Huang, Z.; Qiu, B.; Meng, X.; You, Y.; Liu, X.; Liu, G.; Zhou, C.; Yang, K.; Maier, A.; et al. Comparative study of deep learning models for optical coherence tomography angiography. Biomed. Opt. Express 2020, 11, 1580–1597. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lopez, A.; Larina, I. Functional optical coherence tomography for live dynamic analysis of mouse embryonic cardiogenesis. SPIE BiOS 2018, 10493, 104930C. [Google Scholar]
- Kulkarni, P.M.; Rey-Villamizar, N.; Merouane, A.; Sudheendran, N.; Wang, S.; Garcia, M.; Larina, I.V.; Roysam, B.; Larin, K.V. Algorithms for improved 3-D reconstruction of live mammalian embryo vasculature from optical coherence tomography data. Quant. Imaging Med. Surg. 2015, 5, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, R.; Wu, C.; Singh, M.; Liu, C.-H.; Miranda, R.C.; Larin, K.V. Evaluating the effects of maternal alcohol consumption on murine fetal brain vasculature using optical coherence tomography. J. Biophotonics 2018, 11, e201700238. [Google Scholar] [CrossRef]
- Raghunathan, R.; Liu, C.-H.; Kouka, A.; Singh, M.; Miranda, R.C.; Larin, K.V. Assessing the acute effects of prenatal synthetic cannabinoid exposure on murine fetal brain vasculature using optical coherence tomography. J. Biophotonics 2019, 12, e201900050. [Google Scholar] [CrossRef]
- Raghunathan, R.; Liu, C.-H.; Ambekar, Y.S.; Singh, M.; Miranda, R.C.; Larin, K.V. Optical coherence tomography angiography to evaluate murine fetal brain vasculature changes caused by prenatal exposure to nicotine. Biomed. Opt. Express 2020, 11, 3618–3632. [Google Scholar] [CrossRef]
- Jenkins, M.W.; Watanabe, M.; Rollins, A.M. Longitudinal Imaging of Heart Development With Optical Coherence Tomography. IEEE J. Sel. Top. Quantum Electron. 2012, 18, 1166–1175. [Google Scholar] [CrossRef] [Green Version]
- Piliszek, A.; Kwon, G.S.; Hadjantonakis, A.-K. Ex utero culture and live imaging of mouse embryos. Methods Mol. Biol. 2011, 770, 243–257. [Google Scholar] [CrossRef] [Green Version]
- Nagy, A.; Gertsenstein, M.; Vintersten, K.; Behringer, R. Roller Culture of Postimplantation Embryos. Cold Spring Harb. Protoc. 2006, 2006, pdb-prot4371. [Google Scholar] [CrossRef] [PubMed]
- Mattison, S.P.; Kim, W.; Park, J.; Applegate, B.E. Molecular Imaging in Optical Coherence Tomography. Curr. Mol. Imaging 2014, 3, 88–105. [Google Scholar] [CrossRef] [PubMed]
- Joos, K.M.; Shen, J.-H. Miniature real-time intraoperative forward-imaging optical coherence tomography probe. Biomed. Opt. Express 2013, 4, 1342–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Furtmüller, G.J.; Tong, D.; Zhu, S.; Lee, W.P.A.; Brandacher, G.; Kang, J.U. MEMS-Based Handheld Fourier Domain Doppler Optical Coherence Tomography for Intraoperative Microvascular Anastomosis Imaging. PLoS ONE 2014, 9, e114215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, L.; Qin, X.; He, Y.; Sang, X.; Pan, J.; Xu, T.; Men, J.; Tanzi, R.E.; Li, A.; Ma, Y.; et al. Segmentation of Drosophila heart in optical coherence microscopy images using convolutional neural networks. J. Biophotonics 2018, 11, e201800146. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Men, J.; Yang, Z.; Jerwick, J.; Li, A.; Tanzi, R.E.; Zhou, C. FlyNet 2.0: Drosophila heart 3D (2D + time) segmentation in optical coherence microscopy images using a convolutional long short-term memory neural network. Biomed. Opt. Express 2020, 11, 1568–1579. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Menon, P.G.; Kowalski, W.; Pekkan, K. Time-resolved OCT-μPIV: A new microscopic PIV technique for noninvasive depth-resolved pulsatile flow profile acquisition. Exp. Fluids 2012, 54, 1426. [Google Scholar] [CrossRef]
- Larin, K.V.; Sampson, D.D. Optical coherence elastography - OCT at work in tissue biomechanics [Invited]. Biomed. Opt. Express 2017, 8, 1172–1202. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Larin, K.V. Optical coherence elastography for tissue characterization: A review. J. Biophotonics 2015, 8, 279–302. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Yin, X.; Shi, L.; Liu, A.; Rugonyi, S.; Wang, R.K. Measurement of Strain and Strain Rate in Embryonic Chick Heart In Vivo Using Spectral Domain Optical Coherence Tomography. IEEE Trans. Biomed. Eng. 2011, 58, 2333–2338. [Google Scholar]
- Peng, L.; Aiping, L.; Liang, S.; Xin, Y.; Sandra, R.; Ruikang, K.W. Assessment of strain and strain rate in embryonic chick heart in vivo using tissue Doppler optical coherence tomography. Phys. Med. Biol. 2011, 56, 7081. [Google Scholar]
- Li, P.; Wang, R.K. Optical coherence tomography provides an ability to assess mechanical property of cardiac wall of developing outflow tract in embryonic heart in vivo. J. Biomed. Opt. 2012, 17, 120502. [Google Scholar] [CrossRef] [Green Version]
- Ko, Z.Y.G.; Mehta, K.; Jamil, M.; Yap, C.H.; Chen, N. A method to study the hemodynamics of chicken embryo’s aortic arches using optical coherence tomography. J. Biophotonics 2017, 10, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dur, O.; Patrick, M.J.; Tinney, J.P.; Tobita, K.; Keller, B.B.; Pekkan, K. Aortic Arch Morphogenesis and Flow Modeling in the Chick Embryo. Ann. Biomed. Eng. 2009, 37, 1069–1081. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, W.J.; Dur, O.; Wang, Y.; Patrick, M.J.; Tinney, J.P.; Keller, B.B.; Pekkan, K. Critical Transitions in Early Embryonic Aortic Arch Patterning and Hemodynamics. PLoS ONE 2013, 8, e60271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, L.M.; Gu, S.; Karunamuni, G.; Jenkins, M.W.; Watanabe, M.; Rollins, A.M. Embryonic aortic arch hemodynamics are a functional biomarker for ethanol-induced congenital heart defects [Invited]. Biomed. Opt. Express 2017, 8, 1823–1837. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Maurer, B.; Hermann, B.; Zabihian, B.; Sandrian, M.G.; Unterhuber, A.; Baumann, B.; Zhang, E.Z.; Beard, P.C.; Weninger, W.J.; et al. Dual modality optical coherence and whole-body photoacoustic tomography imaging of chick embryos in multiple development stages. Biomed. Opt. Express 2014, 5, 3150–3159. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Huang, Y.; Qiu, Q.; Wang, K.; Li, Z.; Yao, Y.; Liu, G.; Zhao, Q.; Chen, X. Three-dimensional label-free imaging of mammalian yolk sac vascular remodeling with optical resolution photoacoustic microscopy. Photoacoustics 2020, 17, 100152. [Google Scholar] [CrossRef]
- Laufer, J.; Norris, F.C.; Cleary, J.O.; Zhang, E.Z.; Treeby, B.E.; Cox, B.T.; Johnson, S.P.; Scambler, P.; Lythgoe, M.F.; Beard, P.C. In vivo photoacoustic imaging of mouse embryos. J. Biomed. Opt. 2012, 17, 061220. [Google Scholar] [CrossRef] [Green Version]
- Raghunathan, R.; Zhang, J.; Wu, C.; Rippy, J.; Singh, M.; Larin, K.V.; Scarcelli, G. Evaluating biomechanical properties of murine embryos using Brillouin microscopy and optical coherence tomography. J. Biomed. Opt. 2017, 22, 1–6. [Google Scholar] [CrossRef]
- Zhang, J.; Raghunathan, R.; Rippy, J.; Wu, C.; Finnell, R.H.; Larin, K.V.; Scarcelli, G. Tissue biomechanics during cranial neural tube closure measured by Brillouin microscopy and optical coherence tomography. Birth Defects Res. 2019, 111, 991–998. [Google Scholar] [CrossRef]
- Prevedel, R.; Diz-Muñoz, A.; Ruocco, G.; Antonacci, G. Brillouin microscopy: An emerging tool for mechanobiology. Nat. Methods 2019, 16, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Ambekar, Y.S.; Singh, M.; Zhang, J.; Nair, A.; Aglyamov, S.R.; Scarcelli, G.; Larin, K.V. Multimodal quantitative optical elastography of the crystalline lens with optical coherence elastography and Brillouin microscopy. Biomed. Opt. Express 2020, 11, 2041–2051. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Sudheendran, N.; Singh, M.; Larina, I.V.; Dickinson, M.E.; Larin, K.V. Rotational imaging optical coherence tomography for full-body mouse embryonic imaging. J. Biomed. Opt. 2016, 21, 26002. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Le, H.; Ran, S.; Singh, M.; Larina, I.V.; Mayerich, D.; Dickinson, M.E.; Larin, K.V. Comparison and combination of rotational imaging optical coherence tomography and selective plane illumination microscopy for embryonic study. Biomed. Opt. Express 2017, 8, 4629–4639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed, S.H.; Coughlin, A.J.; Garcia, M.D.; Wang, S.; West, J.L.; Larin, K.V.; Larina, I.V. Optical coherence tomography guided microinjections in live mouse embryos: High-resolution targeted manipulation for mouse embryonic research. J. Biomed. Opt. 2015, 20, 051020. [Google Scholar] [CrossRef]
- Hoog, T.G.; Fredrickson, S.J.; Hsu, C.-W.; Senger, S.M.; Dickinson, M.E.; Udan, R.S. The effects of reduced hemodynamic loading on morphogenesis of the mouse embryonic heart. Dev. Biol. 2018, 442, 127–137. [Google Scholar] [CrossRef]
- Lopez, A.L., 3rd; Wang, S.; Larina, I.V. Optogenetic cardiac pacing in cultured mouse embryos under imaging guidance. J. Biophotonics 2020, e202000223. [Google Scholar] [CrossRef]
- Dickinson, M.E.; Flenniken, A.M.; Ji, X.; Teboul, L.; Wong, M.D.; White, J.K.; Meehan, T.F.; Weninger, W.J.; Westerberg, H.; Adissu, H.; et al. High-throughput discovery of novel developmental phenotypes. Nature 2016, 537, 508–514. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez, A.L., III; Wang, S.; Larina, I.V. Embryonic Mouse Cardiodynamic OCT Imaging. J. Cardiovasc. Dev. Dis. 2020, 7, 42. https://doi.org/10.3390/jcdd7040042
Lopez AL III, Wang S, Larina IV. Embryonic Mouse Cardiodynamic OCT Imaging. Journal of Cardiovascular Development and Disease. 2020; 7(4):42. https://doi.org/10.3390/jcdd7040042
Chicago/Turabian StyleLopez, Andrew L., III, Shang Wang, and Irina V. Larina. 2020. "Embryonic Mouse Cardiodynamic OCT Imaging" Journal of Cardiovascular Development and Disease 7, no. 4: 42. https://doi.org/10.3390/jcdd7040042
APA StyleLopez, A. L., III, Wang, S., & Larina, I. V. (2020). Embryonic Mouse Cardiodynamic OCT Imaging. Journal of Cardiovascular Development and Disease, 7(4), 42. https://doi.org/10.3390/jcdd7040042




