Synthetic Biology Tools for Engineering Aspergillus oryzae
Abstract
:1. Introduction
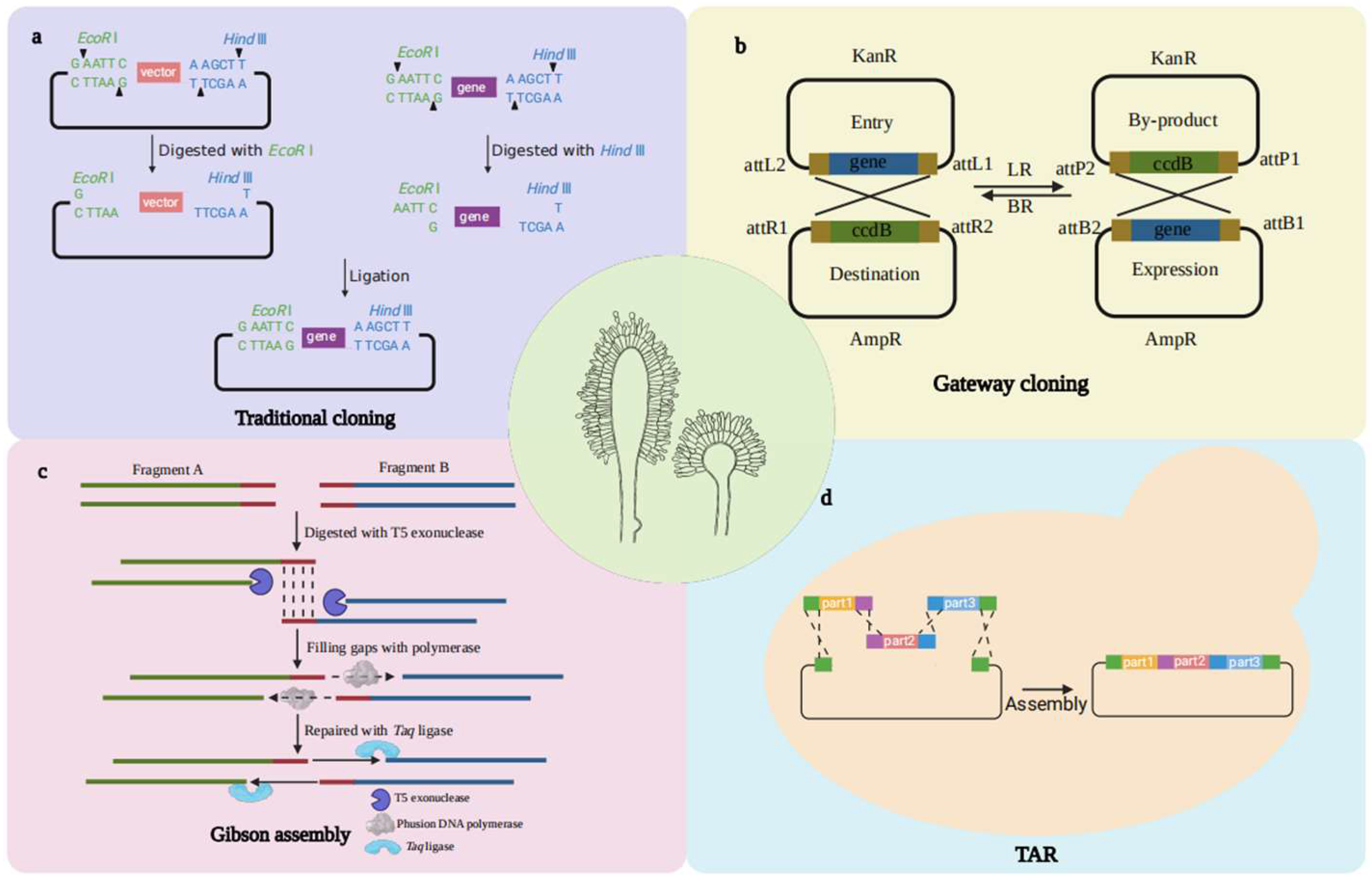
2. DNA Assembly Techniques
2.1. Conventional DNA Manipulation via Restriction/Ligation
2.2. Gateway
2.3. Gibson Assembly
2.4. TAR Technology
3. Gene Expression Regulatory Elements
3.1. Promoter
3.2. Terminator
3.3. Selection Marker
| Host Strain | Origin | Genotypes | Nutritional Sources | References |
|---|---|---|---|---|
| Carboxin-resistance mutant | RIB40 | AosdhB | carboxin | [40] |
| Bleomycin-resistance mutant | RIB40 | Blmb | bleomycin | [41] |
| NS4 | RIB40 | niaD−, SC− | NO2, methionine | [44] |
| NSPID1 | RIB40 | niaD−, SC−, ΔpyrG, ΔligD | NO2, methionine, uracil | [50] |
| niaD300 | RIB40 | niaD− | NO2 | [52] |
| AK2 | AK | ade−, argB− | adenine, arginine | [53] |
| NSR13/NSR1 | RIB40 | niaD−, SC−, adeA− | NO2, methionine, adenine | [49] |
| NSA1 | RIB40 | niaD−, SC−, ΔargB | NO2, methionine, arginine, citrulline | [49] |
| M-2-3 | RIB40 | ΔargB | arginine, citrulline | [54] |
| NSAR1 | RIB40 | niaD−, SC−, ΔargB, adeA− | NO2, methionine, arginine, citrulline, adenine | [49,54,55,56] |
| PTR26 | HL1034 | ptrA | pyrithiamine | [39] |
| 3.042/ΔpyrG | 3.042 | ΔpyrG | uracil, uridine | [57] |
4. Methods of Transformation
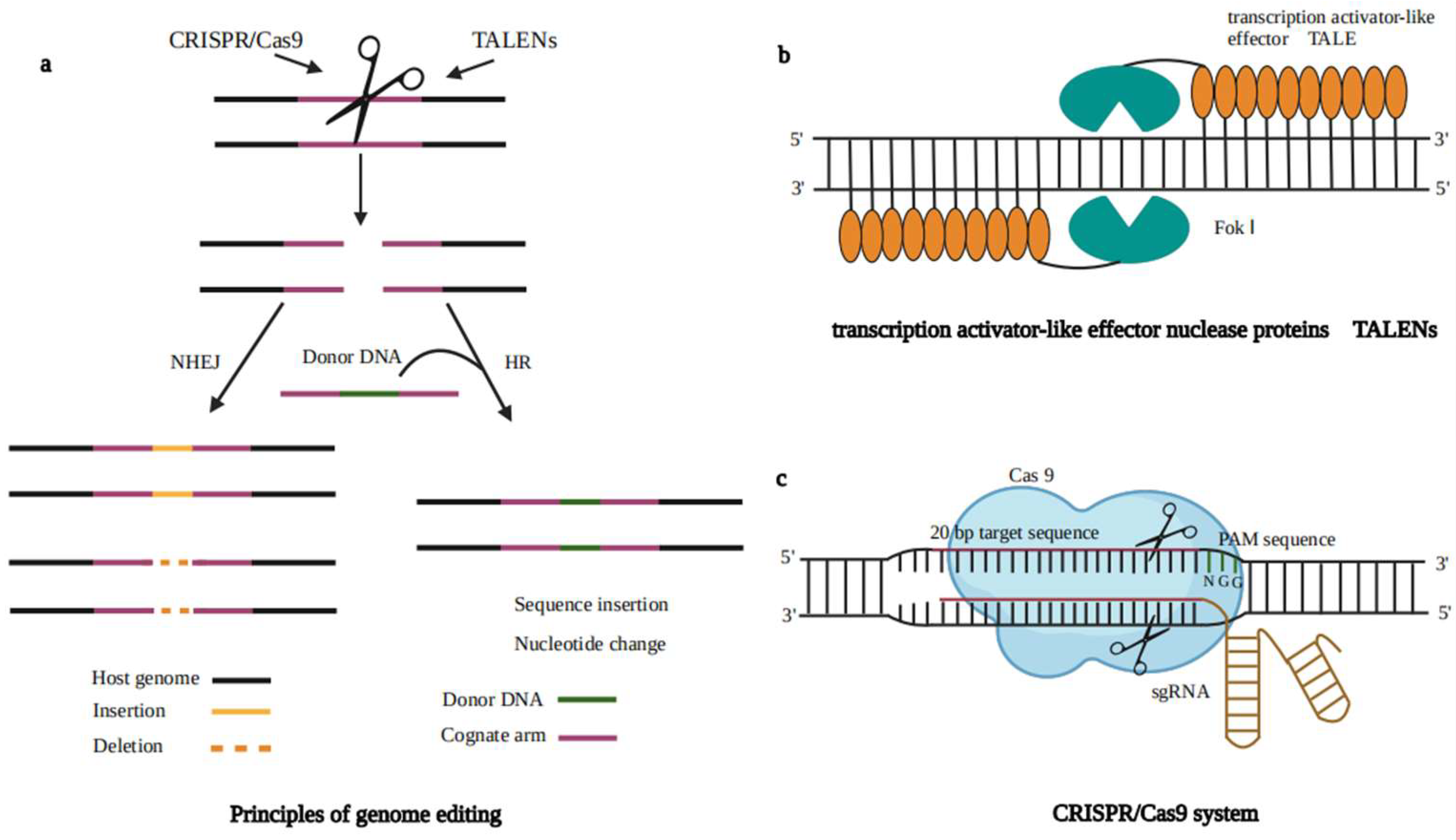
5. Genome Editing Techniques
6. Strategies for Heterologous Protein Production in A. oryzae
6.1. Reduction in Autoprotease Activity in A. oryzae
6.2. Optimization of the Secretion of Heterologous Protein Pathways
6.3. Fusion Expression Strategies
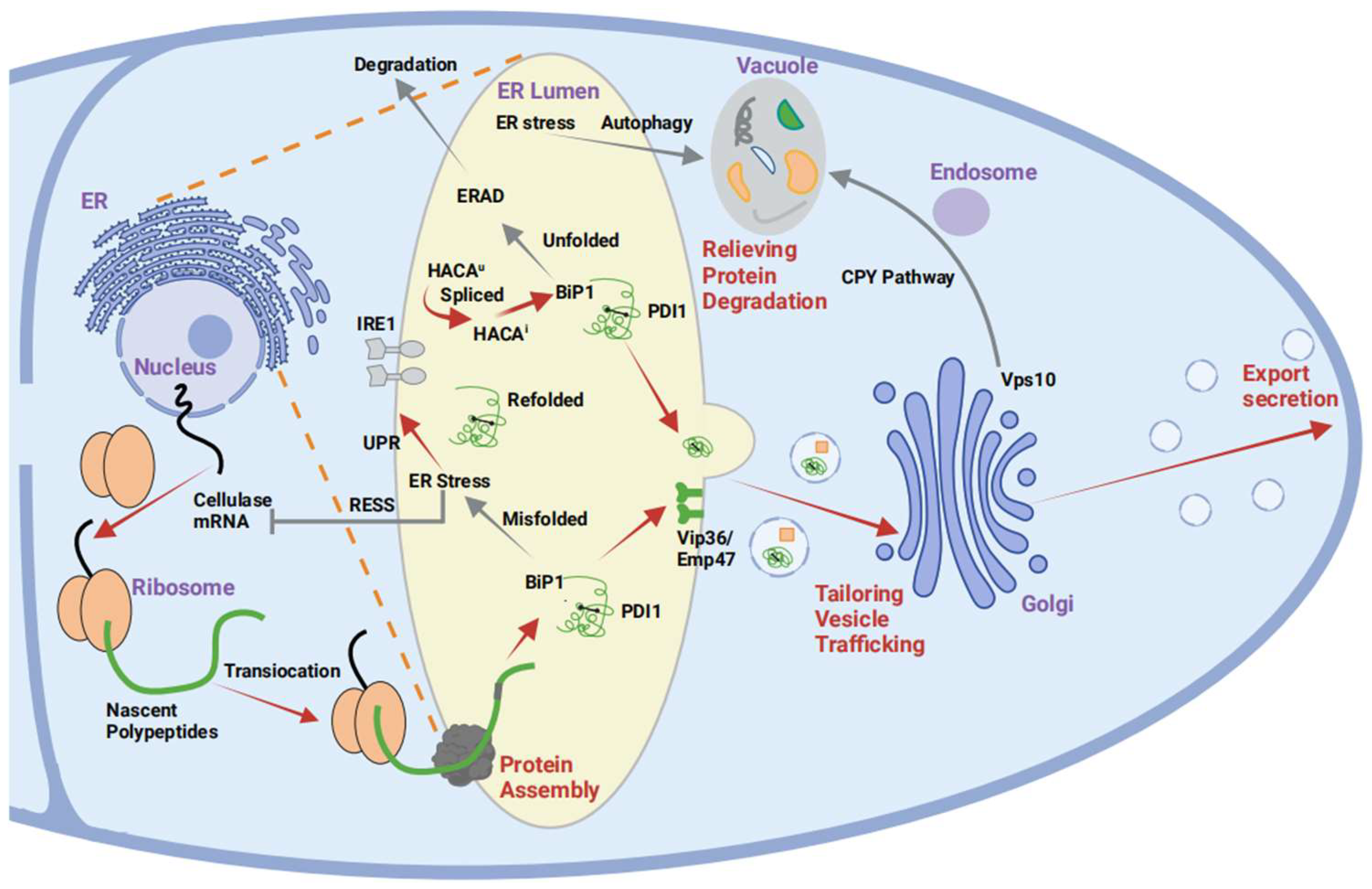
7. Conclusions and Perspectives
- (1)
- DNA assembly is one of the limiting factors for the rapid development of a synthetic biology toolkit for A. oryzae, overcoming limitations associated with long or multiple fragments and scarred DNA after ligation. Enhanced cloning strategies and DNA assembly techniques are essential for the swift construction of gene expression cassettes. Recently, several biotechnology methods for the genetic modification of filamentous fungi, such as the Modular Cloning system or the Golden Braid-based Fungal Braid system, have been used to assemble strains more quickly in a standardized and modular manner. These methods have potential application value in A. oryzae [96,97]. In the future, it is necessary to vigorously develop improved cloning strategies and DNA assembly techniques, to enable faster and more efficient construction of multiple gene expression cassettes.
- (2)
- Promoters and terminators are the most important basic elements in synthetic biology research. To date, several gene expression control elements available for A. oryzae have been identified. However, compared with the model microorganism Saccharomyces cerevisiae, such elements are still rare. It is necessary to construct promoters with shorter sequences, stronger functions, and wider ranges of transcriptional activity to further explore new synthetic biological elements.
- (3)
- Gene editing technology has accomplished rapid editing of A. oryzae with the advantages of simplicity, high efficiency, and high specificity and accelerated the development of engineered strains. CRISPR/Cas9 systems must address off-target effects and improve the efficiency and enhancement of the efficiency of precisely targeted editing, among other aspects. In addition, CRISPR-related derivative technologies based on dCas9 or nCas9 need to be developed for A. oryzae, such as CRISPRa and CRISPRi-mediated gene expression regulation technology and multifunctional CRISPR-mediated combined regulation technology [98,99,100]. The CRISPR system is expected to be rapidly developed and technologically innovated in the future for research on gene function, metabolic pathway reconstruction, precise expression regulation, and high-performance chassis construction in A. oryzae.
- (4)
- In recent years, research on the production capacity of heterologous protease preparations and other useful substances of A. oryzae has greatly improved the production and application of A. oryzae [101,102,103]. Future research could focus on the activity of enzyme extraction, the adverse effects of high concentrations of products on A. oryzae itself, and the utilization of raw materials. This study provides additional effective methods and research space for the breeding of A. oryzae production strains in the future.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kitamoto, K. Cell biology of the Koji mold Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2015, 79, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Gomi, K.; Hasegawa, F.; Machida, M. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia 2006, 162, 143–153. [Google Scholar] [CrossRef]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.-I.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Yao, Y.; Qi, W.; Wang, C.; Hou, L.; Zeng, B.; Cao, X. Draft genome sequence of Aspergillus oryzae strain 3.042. Eukaryot. Cell 2012, 11, 1178. [Google Scholar] [CrossRef] [PubMed]
- Takusagawa, S.; Satoh, Y.; Ohtsu, I.; Dairi, T. Ergothioneine production with Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2019, 83, 181–184. [Google Scholar] [CrossRef]
- Han, H.; Yu, C.; Qi, J.; Wang, P.; Zhao, P.; Gong, W.; Xie, C.; Xia, X.; Liu, C. High-efficient production of mushroom polyketide compounds in a platform host Aspergillus oryzae. Microb. Cell Factories 2023, 22, 60. [Google Scholar] [CrossRef]
- Xiao, Z.-H.; Dong, J.-Y.; Li, A.; Dai, J.-M.; Li, Y.-P.; Hu, Q.-F.; Shao, L.-D.; Matsuda, Y.; Wang, W.-G. Biocatalytic and chemical derivatization of the fungal meroditerpenoid chevalone E. Org. Chem. Front. 2022, 9, 1837–1843. [Google Scholar] [CrossRef]
- Takino, J.; Kozaki, T.; Sato, Y.; Liu, C.; Ozaki, T.; Minami, A.; Oikawa, H. Unveiling Biosynthesis of the Phytohormone Abscisic Acid in Fungi: Unprecedented Mechanism of Core Scaffold Formation Catalyzed by an Unusual Sesquiterpene Synthase. J. Am. Chem. Soc. 2018, 140, 12392–12395. [Google Scholar] [CrossRef]
- Fujii, R.; Minami, A.; Tsukagoshi, T.; Sato, N.; Sahara, T.; Ohgiya, S.; Gomi, K.; Oikawa, H. Total Biosynthesis of Diterpene Aphidicolin, a Specific Inhibitor of DNA Polymerase α: Heterologous Expression of Four Biosynthetic Genes in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2011, 75, 1813–1817. [Google Scholar] [CrossRef]
- Uchida, H.; Arakida, S.; Sakamoto, T.; Kawasaki, H. Expression of Aspergillus oryzae phytase gene in Aspergillus oryzae RIB40 niaD(-). J. Biosci. Bioeng. 2006, 102, 564–567. [Google Scholar] [CrossRef]
- Pahirulzaman, K.A.; Williams, K.; Lazarus, C.M. A toolkit for heterologous expression of metabolic pathways in Aspergillus oryzae. Methods Enzymol. 2012, 517, 241–260. [Google Scholar] [CrossRef] [PubMed]
- Nofiani, R.; de Mattos-Shipley, K.; Lebe, K.E.; Han, L.-C.; Iqbal, Z.; Bailey, A.M.; Willis, C.L.; Simpson, T.J.; Cox, R.J. Strobilurin biosynthesis in Basidiomycete fungi. Nat. Commun. 2018, 9, 3940. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., III; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Fukaya, M.; Ozaki, T.; Minami, A.; Oikawa, H. Biosynthetic machineries of anthraquinones and bisanthraquinones in Talaromyces islandicus. Biosci. Biotechnol. Biochem. 2022, 86, 435–443. [Google Scholar] [CrossRef]
- Kunes, S.; Botstein, D.; Fox, M.S. Transformation of yeast with linearized plasmid DNA. Formation of inverted dimers and recombinant plasmid products. J. Mol. Biol. 1985, 184, 375–387. [Google Scholar] [CrossRef]
- Ma, H.; Kunes, S.; Schatz, P.J.; Botstein, D. Plasmid construction by homologous recombination in yeast. Gene 1987, 58, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Orr-Weaver, T.L.; Szostak, J.W.; Rothstein, R.J. Yeast transformation: A model system for the study of recombination. Proc. Natl. Acad. Sci. USA 1981, 78, 6354–6358. [Google Scholar] [CrossRef] [PubMed]
- Lebe, K.E.; Cox, R.J. Oxidative steps during the biosynthesis of squalestatin S1. Chem. Sci. 2019, 10, 1227–1231. [Google Scholar] [CrossRef]
- Jeennor, S.; Anantayanon, J.; Chutrakul, C.; Panchanawaporn, S.; Laoteng, K. Novel pentose-regulated promoter of Aspergillus oryzae with application in controlling heterologous gene expression. Biotechnol. Rep. 2022, 33, e00695. [Google Scholar] [CrossRef]
- Oda, K.; Terado, S.; Toyoura, R.; Fukuda, H.; Kawauchi, M.; Iwashita, K. Development of a promoter shutoff system in Aspergillus oryzae using a sorbitol-sensitive promoter. Biosci. Biotechnol. Biochem. 2016, 80, 1792–1801. [Google Scholar] [CrossRef]
- Tada, S.; Gomi, K.; Kitamoto, K.; Takahashi, K.; Tamura, G.; Hara, S. Construction of a fusion gene comprising the Taka-amylase A promoter and the Escherichia coli beta-glucuronidase gene and analysis of its expression in Aspergillus oryzae. Mol. Genet. Genom. 1991, 229, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, K.; Tada, S.; Gomi, K.; Kitamoto, K.; Kumagai, C.; Tamura, G. Deletion analysis of the Taka-amylase A gene promoter using a homologous transformation system in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 1992, 56, 1849–1853. [Google Scholar] [CrossRef] [PubMed]
- Hata, Y.; Kitamoto, K.; Gomi, K.; Kumagai, C.; Tamura, G. Functional elements of the promoter region of the Aspergillus oryzae glaA gene encoding glucoamylase. Curr. Genet. 1992, 22, 85–91. [Google Scholar] [CrossRef]
- Minetoki, T.; Gomi, K.; Kitamoto, K.; Kumagai, C.; Tamura, G. Nucleotide sequence and expression of alpha-glucosidase-encoding gene (agdA) from Aspergillus oryzae. Biosci. Biotechnol. Biochem. 1995, 59, 1516–1521. [Google Scholar] [CrossRef] [PubMed]
- Shoji, J.Y.; Maruyama, J.; Arioka, M.; Kitamoto, K. Development of Aspergillus oryzae thiA promoter as a tool for molecular biological studies. FEMS Microbiol. Lett. 2005, 244, 41–46. [Google Scholar] [CrossRef]
- Ishida, H.; Hata, Y.; Kawato, A.; Abe, Y. Improvement of the glaB promoter expressed in solid-state fermentation (SSF) of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2006, 70, 1181–1187. [Google Scholar] [CrossRef]
- Liu, J.; Xie, Z.; Shin, H.D.; Li, J.; Du, G.; Chen, J.; Liu, L. Rewiring the reductive tricarboxylic acid pathway and L-malate transport pathway of Aspergillus oryzae for overproduction of L-malate. J. Biotechnol. 2017, 253, 1–9. [Google Scholar] [CrossRef]
- Kitamoto, N.; Matsui, J.; Kawai, Y.; Kato, A.; Yoshino, S.; Ohmiya, K.; Tsukagoshi, N. Utilization of the TEF1-alpha gene (TEF1) promoter for expression of polygalacturonase genes, pgaA and pgaB, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 1998, 50, 85–92. [Google Scholar] [CrossRef]
- Ishida, H.; Hata, Y.; Kawato, A.; Abe, Y.; Kashiwagi, Y. Isolation of a novel promoter for efficient protein production in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2004, 68, 1849–1857. [Google Scholar] [CrossRef]
- Bando, H.; Hisada, H.; Ishida, H.; Hata, Y.; Katakura, Y.; Kondo, A. Isolation of a novel promoter for efficient protein expression by Aspergillus oryzae in solid-state culture. Appl. Microbiol. Biotechnol. 2011, 92, 561–569. [Google Scholar] [CrossRef]
- Tsuboi, H.; Koda, A.; Toda, T.; Minetoki, T.; Hirotsune, M.; Machida, M. Improvement of the Aspergillus oryzae enolase promoter (P-enoA) by the introduction of cis-element repeats. Biosci. Biotechnol. Biochem. 2005, 69, 206–208. [Google Scholar] [CrossRef] [PubMed]
- Minetoki, T.; Tsuboi, H.; Koda, A.; Ozeki, K. Development of high expression system with the improved promoter using the cis-acting element in Aspergillus species. J. Biol. Macromol. 2003, 3, 89–96. [Google Scholar]
- Ichikawa, K.; Shiono, Y.; Shintani, T.; Watanabe, A.; Kanzaki, H.; Gomi, K.; Koseki, T. Efficient production of recombinant tannase in Aspergillus oryzae using an improved glucoamylase gene promoter. J. Biosci. Bioeng. 2020, 129, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Toda, T.; Sano, M.; Honda, M.; Rimoldi, O.; Yang, Y.; Yamamoto, M.; Takase, K.; Hirozumi, K.; Kitamoto, K.; Minetoki, T.; et al. Deletion analysis of the enolase gene (enoA) promoter from the filamentous fungus Aspegillus oryzae. Curr. Genet. 2001, 40, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Geisberg, J.V.; Moqtaderi, Z.; Fan, X.; Ozsolak, F.; Struhl, K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 2014, 156, 812–824. [Google Scholar] [CrossRef] [PubMed]
- de Mattos-Shipley, K.M.J.; Lazarus, C.M.; Williams, K. Investigating Fungal Biosynthetic Pathways Using Heterologous Gene Expression: Aspergillus oryzae as a Heterologous Host. Methods Mol. Biol. 2022, 2489, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Tagami, K.; Minami, A.; Fujii, R.; Liu, C.; Tanaka, M.; Gomi, K.; Dairi, T.; Oikawa, H. Rapid reconstitution of biosynthetic machinery for fungal metabolites in Aspergillus oryzae: Total biosynthesis of aflatrem. Chembiochem 2014, 15, 2076–2080. [Google Scholar] [CrossRef]
- Nakazawa, T.; Ishiuchi, K.; Praseuth, A.; Noguchi, H.; Hotta, K.; Watanabe, K. Overexpressing transcriptional regulator in Aspergillus oryzae activates a silent biosynthetic pathway to produce a novel polyketide. Chembiochem 2012, 13, 855–861. [Google Scholar] [CrossRef]
- Kubodera, T.; Yamashita, N.; Nishimura, A. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: Cloning, characterization and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 2000, 64, 1416–1421. [Google Scholar] [CrossRef]
- Shima, Y.; Ito, Y.; Kaneko, S.; Hatabayashi, H.; Watanabe, Y.; Adachi, Y.; Yabe, K. Identification of three mutant loci conferring carboxin-resistance and development of a novel transformation system in Aspergillus oryzae. Fungal Genet. Biol. 2009, 46, 67–76. [Google Scholar] [CrossRef]
- Suzuki, S.; Tada, S.; Fukuoka, M.; Taketani, H.; Tsukakoshi, Y.; Matsushita, M.; Oda, K.; Kusumoto, K.-I.; Kashiwagi, Y.; Sugiyama, M. A novel transformation system using a bleomycin resistance marker with chemosensitizers for Aspergillus oryzae. Biochem. Biophys. Res. Commun. 2009, 383, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Todokoro, T.; Bando, H.; Kotaka, A.; Tsutsumi, H.; Hata, Y.; Ishida, H. Identification of a novel pyrithiamine resistance marker gene thiI for genome co-editing in Aspergillus oryzae. J. Biosci. Bioeng. 2020, 130, 227–232. [Google Scholar] [CrossRef] [PubMed]
- van Hartingsveldt, W.; Mattern, I.E.; van Zeijl, C.M.; Pouwels, P.H.; van den Hondel, C.A.M.J.J. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol. Genet. Genom. 1987, 206, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Yamada, O.; Lee, B.R.; Gomi, K. Transformation System for Aspergillus oryzae with Double Auxotrophic Mutations, niaD and sC. Biosci. Biotechnol. Biochem. 1997, 61, 1367–1369. [Google Scholar] [CrossRef]
- Gomi, K.; Iimura, Y.; Hara, S. Integrative Transformation of Aspergillus oryzae with a Plasmid Containing the Aspergillus nidulans argB Gene. Agric. Biol. Chem. 1987, 51, 2549–2555. [Google Scholar] [CrossRef]
- Unkles, S.E.; Campbell, E.I.; de Ruiter-Jacobs, Y.M.J.T.; Broekhuijsen, M.; Macro, J.A.; Carrez, D.; Contreras, R.; van den Hondel, C.A.M.J.J.; Kinghorn, J.R. The development of a homologous transformation system for Aspergillus oryzae based on the nitrate assimilation pathway: A convenient and general selection system for filamentous fungal transformation. Mol. Genet. Genom. MGG 1989, 218, 99–104. [Google Scholar] [CrossRef]
- Jin, F.J.; Maruyama, J.; Juvvadi, P.R.; Arioka, M.; Kitamoto, K. Adenine auxotrophic mutants of Aspergillus oryzae: Development of a novel transformation system with triple auxotrophic hosts. Biosci. Biotechnol. Biochem. 2004, 68, 656–662. [Google Scholar] [CrossRef]
- Gomi, K.; Kitamoto, K.; Kumagai, C. Cloning and molecular characterization of the acetamidase-encoding gene (amdS) from Aspergillus oryzae. Gene 1991, 108, 91–98. [Google Scholar] [CrossRef]
- Jin, F.J.; Maruyama, J.; Juvvadi, P.R.; Arioka, M.; Kitamoto, K. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol. Lett. 2004, 239, 79–85. [Google Scholar] [CrossRef]
- Maruyama, J.; Kitamoto, K. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (DeltaligD) in Aspergillus oryzae. Biotechnol. Lett. 2008, 30, 1811–1817. [Google Scholar] [CrossRef]
- Zhang, S.; Ban, A.; Ebara, N.; Mizutani, O.; Tanaka, M.; Shintani, T.; Gomi, K. Self-excising Cre/mutant lox marker recycling system for multiple gene integrations and consecutive gene deletions in Aspergillus oryzae. J. Biosci. Bioeng. 2017, 123, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Kaieda, M.; Nagayoshi, M.; Hama, S.; Kondo, A.; Fukuda, H. Enantioselective transesterification using immobilized Aspergillus oryzae overexpressing lipase. Appl. Microbiol. Biotechnol. 2004, 65, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Liu, J.; Du, Y.; Pei, X.; Li, M. Aspergillus oryzae Biosynthetic Platform for de Novo Iridoid Production. J. Agric. Food Chem. 2021, 69, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Awakawa, T.; Abe, I. Reconstitution of Polyketide-Derived Meroterpenoid Biosynthetic Pathway in Aspergillus oryzae. J. Fungi 2021, 7, 486. [Google Scholar] [CrossRef]
- Oikawa, H. Reconstitution of biosynthetic machinery of fungal natural products in heterologous hosts. Biosci. Biotechnol. Biochem. 2020, 84, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, H. Heterologous production of fungal natural products: Reconstitution of biosynthetic gene clusters in model host Aspergillus oryzae. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Niu, Y.; He, B.; Ma, L.; Li, G.; Tran, V.-T.; Zeng, B.; Hu, Z. A Dual Selection Marker Transformation System Using Agrobacterium tumefaciens for the Industrial Aspergillus oryzae 3.042. J. Microbiol. Biotechnol. 2019, 29, 230–234. [Google Scholar] [CrossRef]
- Jin, F.J.; Hu, S.; Wang, B.T.; Jin, L. Advances in Genetic Engineering Technology and Its Application in the Industrial Fungus Aspergillus oryzae. Front. Microbiol. 2021, 12, 644404. [Google Scholar] [CrossRef]
- Wang, S.; Chen, H.; Tang, X.; Zhang, H.; Chen, W.; Chen, Y.Q. Molecular tools for gene manipulation in filamentous fungi. Appl. Microbiol. Biotechnol. 2017, 101, 8063–8075. [Google Scholar] [CrossRef]
- Nguyen, K.T.; Ho, Q.N.; Do, L.; Mai, L.T.D.; Pham, D.-N.; Tran, H.T.T.; Le, D.H.; Nguyen, H.Q.; Tran, V.-T. A new and efficient approach for construction of uridine/uracil auxotrophic mutants in the filamentous fungus Aspergillus oryzae using Agrobacterium tumefaciens-mediated transformation. World J. Microbiol. Biotechnol. 2017, 33, 107. [Google Scholar] [CrossRef]
- Nguyen, K.T.; Ho, Q.N.; Pham, T.H.; Phan, T.-N.; Tran, V.-T. The construction and use of versatile binary vectors carrying pyrG auxotrophic marker and fluorescent reporter genes for Agrobacterium-mediated transformation of Aspergillus oryzae. World J. Microbiol. Biotechnol. 2016, 32, 204. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.M.; Lin, T.S.; Wang, C.C.C. Total Heterologous Biosynthesis of Fungal Natural Products in Aspergillus nidulans. J. Nat. Prod. 2022, 85, 2484–2518. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Tang, Y.; Lin, J.; Cai, W. Methods for genetic transformation of filamentous fungi. Microb. Cell Factories 2017, 16, 168. [Google Scholar] [CrossRef] [PubMed]
- Bundock, P.; den Dulk-Ras, A.; Beijersbergen, A.; Hooykaas, P. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995, 14, 3206–3214. [Google Scholar] [CrossRef] [PubMed]
- Sugui, J.A.; Chang, Y.C.; Kwon-Chung, K.J. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: An efficient tool for insertional mutagenesis and targeted gene disruption. Appl. Environ. Microbiol. 2005, 71, 1798–1802. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; He, D.; Li, G.; Gao, S.; Lv, H.; Shan, Q.; Wang, L. An efficient tool for random insertional mutagenesis: Agrobacterium tumefaciens-mediated transformation of the filamentous fungus Aspergillus terreus. J. Microbiol. Methods 2014, 98, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Shao, Q.; Li, C.; Zhao, K.; Jiang, L.; Fan, J.; Jiang, H.; Tao, F. An efficient Agrobacterium-mediated transformation method for aflatoxin generation fungus Aspergillus flavus. J. Microbiol. 2018, 56, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yan, J.N.; Zhang, H.; Liu, T.Q.; Xu, Y.; Zhang, Y.Y.; Li, J. Effect of gpd box copy numbers in the gpdA promoter of Aspergillus nidulans on its transcription efficiency in Aspergillus niger. FEMS Microbiol. Lett. 2018, 365, fny154. [Google Scholar] [CrossRef]
- Maruyama, J.I. Genome Editing Technology and Its Application Potentials in the Industrial Filamentous Fungus Aspergillus oryzae. J. Fungi 2021, 7, 638. [Google Scholar] [CrossRef]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef]
- Mizutani, O.; Arazoe, T.; Toshida, K.; Hayashi, R.; Ohsato, S.; Sakuma, T.; Yamamoto, T.; Kuwata, S.; Yamada, O. Detailed analysis of targeted gene mutations caused by the Platinum-Fungal TALENs in Aspergillus oryzae RIB40 strain and a ligD disruptant. J. Biosci. Bioeng. 2017, 123, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Tanaka, Y.; Okabe, T.; Nakamura, H.; Fujii, W.; Kitamoto, K.; Maruyama, J.-I. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 2016, 38, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Nakamura, H.; Zhang, Y.; Pascal, A.; Fujii, W.; Maruyama, J.-I. Forced Recycling of an AMA1-Based Genome-Editing Plasmid Allows for Efficient Multiple Gene Deletion/Integration in the Industrial Filamentous Fungus Aspergillus oryzae. Appl. Environ. Microbiol. 2019, 85, e01896-18. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Minami, A.; Ozaki, T.; Wu, J.; Kawagishi, H.; Maruyama, J.-I.; Oikawa, H. Efficient Reconstitution of Basidiomycota Diterpene Erinacine Gene Cluster in Ascomycota Host Aspergillus oryzae Based on Genomic DNA Sequences. J. Am. Chem. Soc. 2019, 141, 15519–15523. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, K.G.; Rendsvig, J.K.H.; Jarczynska, Z.D.; Cortes, M.V.d.C.B.; van Esch, A.P.; Morera-Gómez, M.; Contesini, F.J.; Mortensen, U.H. A Mad7 System for Genetic Engineering of Filamentous Fungi. J. Fungi 2022, 9, 16. [Google Scholar] [CrossRef]
- Fleissner, A.; Dersch, P. Expression and export: Recombinant protein production systems for Aspergillus. Appl. Microbiol. Biotechnol. 2010, 87, 1255–1270. [Google Scholar] [CrossRef]
- Liu, D.; Garrigues, S.; de Vries, R.P. Heterologous protein production in filamentous fungi. Appl. Microbiol. Biotechnol. 2023, 107, 5019–5033. [Google Scholar] [CrossRef]
- Daba, G.M.; Mostafa, F.A.; Elkhateeb, W.A. The ancient koji mold (Aspergillus oryzae) as a modern biotechnological tool. Bioresour. Bioprocess. 2021, 8, 52. [Google Scholar] [CrossRef]
- Jin, F.J.; Watanabe, T.; Juvvadi, P.R.; Maruyama, J.-I.; Arioka, M.; Kitamoto, K. Double disruption of the proteinase genes, tppA and pepE, increases the production level of human lysozyme by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2007, 76, 1059–1068. [Google Scholar] [CrossRef]
- Yoon, J.; Maruyama, J.; Kitamoto, K. Disruption of ten protease genes in the filamentous fungus Aspergillus oryzae highly improves production of heterologous proteins. Appl. Microbiol. Biotechnol. 2011, 89, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, T.; Watanabe, T.; Mizogami, Y.; Maruyama, J.-I.; Kitamoto, K. Isolation of Aspergillus oryzae mutants for heterologous protein production from a double proteinase gene disruptant. Appl. Microbiol. Biotechnol. 2009, 82, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.J.; Katayama, T.; Maruyama, J.I.; Kitamoto, K. Comparative genomic analysis identified a mutation related to enhanced heterologous protein production in the filamentous fungus Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2016, 100, 9163–9174. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, N.; Ono, N.; Yoshino-Yasuda, S. Construction of Quintuple Protease and Double Amylase Gene Deletant for Heterologous Protein Production in Aspergillus oryzae KBN616. Food Sci. Technol. Res. 2015, 21, 297–307. [Google Scholar] [CrossRef]
- Kasuya, T.; Nakajima, H.; Kitamoto, K. Cloning and characterization of the bipA gene encoding ER chaperone BiP from Aspergillus oryzae. J. Biosci. Bioeng. 1999, 88, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Aishan, T.; Maruyama, J.; Kitamoto, K. Enhanced production and secretion of heterologous proteins by the filamentous fungus Aspergillus oryzae via disruption of vacuolar protein sorting receptor gene Aovps10. Appl. Environ. Microbiol. 2010, 76, 5718–5727. [Google Scholar] [CrossRef]
- Hoang, H.D.; Maruyama, J.; Kitamoto, K. Modulating endoplasmic reticulum-Golgi cargo receptors for improving secretion of carrier-fused heterologous proteins in the filamentous fungus Aspergillus oryzae. Appl. Environ. Microbiol. 2015, 81, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Maruyama, J.; Kikuma, T.; Arioka, M.; Kitamoto, K. Autophagy delivers misfolded secretory proteins accumulated in endoplasmic reticulum to vacuoles in the filamentous fungus Aspergillus oryzae. Biochem. Biophys. Res. Commun. 2011, 406, 464–470. [Google Scholar] [CrossRef]
- Yoon, J.; Kikuma, T.; Maruyama, J.; Kitamoto, K. Enhanced production of bovine chymosin by autophagy deficiency in the filamentous fungus Aspergillus oryzae. PLoS ONE 2013, 8, e62512. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, C.; Wang, B.; Li, X.; Xiao, J.; Pan, L. Identification of functional cis-elements required for repression of the Taka-amylase A gene under secretion stress in Aspergillus oryzae. Biotechnol. Lett. 2015, 37, 333–341. [Google Scholar] [CrossRef]
- Gouka, R.J.; Punt, P.J.; van den Hondel, C.A. Glucoamylase gene fusions alleviate limitations for protein production in Aspergillus awamori at the transcriptional and (post) translational levels. Appl. Environ. Microbiol. 1997, 63, 488–497. [Google Scholar] [CrossRef]
- Tsuchiya, K.; Nagashima, T.; Yamamoto, Y.; Gomi, K.; Kitamoto, K.; Kumagai, C.; Tamura, G. High level secretion of calf chymosin using a glucoamylase-prochymosin fusion gene in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 1994, 58, 895–899. [Google Scholar] [CrossRef]
- Ohno, A.; Maruyama, J.; Nemoto, T.; Arioka, M.; Kitamoto, K. A carrier fusion significantly induces unfolded protein response in heterologous protein production by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2011, 92, 1197–1206. [Google Scholar] [CrossRef]
- Tanaka, M.; Shintani, T.; Gomi, K. Unfolded protein response is required for Aspergillus oryzae growth under conditions inducing secretory hydrolytic enzyme production. Fungal Genet. Biol. 2015, 85, 1–6. [Google Scholar] [CrossRef]
- Nagamine, S.; Liu, C.; Nishishita, J.; Kozaki, T.; Sogahata, K.; Sato, Y.; Minami, A.; Ozaki, T.; Schmidt-Dannert, C.; Maruyama, J.-I.; et al. Ascomycete Aspergillus oryzae Is an Efficient Expression Host for Production of Basidiomycete Terpenes by Using Genomic DNA Sequences. Appl. Environ. Microbiol. 2019, 85, 15. [Google Scholar] [CrossRef]
- Mózsik, L.; Pohl, C.; Meyer, V.; Bovenberg, R.A.L.; Nygård, Y.; Driessen, A.J.M. Modular Synthetic Biology Toolkit for Filamentous Fungi. ACS Synth. Biol. 2021, 10, 2850–2861. [Google Scholar] [CrossRef]
- Moreno-Giménez, E.; Gandía, M.; Sáez, Z.; Manzanares, P.; Yenush, L.; Orzáez, D.; Marcos, J.F.; Garrigues, S. FungalBraid 2.0: Expanding the synthetic biology toolbox for the biotechnological exploitation of filamentous fungi. Front. Bioeng. Biotechnol. 2023, 11, 1222812. [Google Scholar] [CrossRef]
- Dominguez, A.A.; Lim, W.A.; Qi, L.S. Beyond editing: Repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016, 17, 5–15. [Google Scholar] [CrossRef]
- Lu, A.; Wang, J.; Sun, W.; Huang, W.; Cai, Z.; Zhao, G.; Wang, J. Reprogrammable CRISPR/dCas9-based recruitment of DNMT1 for site-specific DNA demethylation and gene regulation. Cell Discov. 2019, 5, 22. [Google Scholar] [CrossRef]
- Tak, Y.E.; Kleinstiver, B.P.; Nuñez, J.K.; Hsu, J.Y.; E Horng, J.; Gong, J.; Weissman, J.S.; Joung, J.K. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat. Methods 2017, 14, 1163–1166. [Google Scholar] [CrossRef] [PubMed]
- Akasaka, N.; Fujiwara, S. The therapeutic and nutraceutical potential of agmatine, and its enhanced production using Aspergillus oryzae. Amino Acids 2020, 52, 181–197. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, J.; Ding, Q.; Luo, Q.; Liu, L. Morphology engineering of Aspergillus oryzae for l-malate production. Biotechnol. Bioeng. 2019, 116, 2662–2673. [Google Scholar] [CrossRef]
- Zhang, L.; Kang, L.; Xu, Y. Phenotypic, Genomic, and Transcriptomic Comparison of Industrial Aspergillus oryzae Used in Chinese and Japanese Soy Sauce: Analysis of Key Proteolytic Enzymes Produced by Koji Molds. Microbiol. Spectr. 2023, 11, e0083622. [Google Scholar] [CrossRef]
| Promoter | Species | Gene Source | Application | References |
|---|---|---|---|---|
| PxyrA | Aspergillus oryzae | xylose reductase | For the expression of β-glucuronidase | [19] |
| Psor | A. oryzae | sugar transporter-like protein (stl1) and sorbitol dehydrogenase (xyl2) | For eGFP expression | [20] |
| PglaA | Aspergillus niger | glucoamylase | For expression of glucosidase | [23] |
| PthiA | A. oryzae | thiamine thiazole synthase | For eGFP expression | [25] |
| PamyB | A. oryzae | alpha-amylase | For expression of lysozyme | [21,22] |
| PmelO | A. oryzae | tyrosinase | For expression of glucoamylase | [29] |
| PSodM | A. oryzae | manganese superoxide dismutase | For expression of cellulase | [29] |
| PhylA | A. oryzae | hemolysin-like-protein | For the expression of endo-1,4-β-glucanase | [30] |
| PagdA | A. oryzae | α-glucosidase | For the expression of α-glucosidase | [24] |
| PpgkA | A. oryzae | phosphoglycerate kinase | For expression of PEP carboxylase | [27] |
| PenoA | A. oryzae | enolase | For the expression of β-glucuronic acid | [34] |
| Padh | Aspergillus nidulans | alcohol dehydrogenase | For the synthesis of tenellin | [11] |
| PenoA142 | A. oryzae | improved PenoA | For expression of β-Glucuronidase | [31] |
| PglaA142 | A. oryzae | improved PglaA | For the production of tannase | [33] |
| Ptef1 | A. oryzae | translation-elongation factor 1 alpha | For expression of polygalacturonase | [28] |
| PglaB | A. niger | glucose amylase | For expression of β-glucuronidase | [26] |
| PgpdA | A. nidulans | glyceraldehyde-3-phosphate dehydrogenase | For expression of β-galactosidase | [26] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Song, C.; Liu, C.; Wang, P. Synthetic Biology Tools for Engineering Aspergillus oryzae. J. Fungi 2024, 10, 34. https://doi.org/10.3390/jof10010034
Yang H, Song C, Liu C, Wang P. Synthetic Biology Tools for Engineering Aspergillus oryzae. Journal of Fungi. 2024; 10(1):34. https://doi.org/10.3390/jof10010034
Chicago/Turabian StyleYang, Hui, Chaonan Song, Chengwei Liu, and Pengchao Wang. 2024. "Synthetic Biology Tools for Engineering Aspergillus oryzae" Journal of Fungi 10, no. 1: 34. https://doi.org/10.3390/jof10010034
APA StyleYang, H., Song, C., Liu, C., & Wang, P. (2024). Synthetic Biology Tools for Engineering Aspergillus oryzae. Journal of Fungi, 10(1), 34. https://doi.org/10.3390/jof10010034








