Abstract
The plant pathogenic fungus Cytospora is notoriously known for causing woody plant canker diseases, resulting in substantial economic losses to biological forests and fruit trees worldwide. Despite their strong negative ecological impact, the existing and prospective distribution patterns of these plant pathogens in China, according to climate change, have received little attention. In this study, we chose three widely dispersed and seriously damaging species, namely, Cytospora chrysosperma, Cytospora mali, and Cytospora nivea, which are the most common species that damage the Juglans regia, Malus domestica, Eucalyptus, Pyrus sinkiangensis, Populus spp., and Salix spp. in China. We utilized ecological niche modeling to forecast their regional distribution in China under four climate change scenarios (present, SSP 126, SSP 370, and SSP 585). The results show that temperature-related climate factors limit the current distribution ranges of the three species. Currently, the three studied species are highly suitable for northeast, northwest, north, and southwest China. Under future climate scenarios, the distribution ranges of the three species are projected to increase, and the centers of the adequate distribution areas of the three species are expected to shift to high-latitude regions. The three species coexist in China, primarily in the northwest and north regions. The ecological niches of C. chrysosperma and C. nivea are more similar. The distribution range of C. mali can reach the warmer and wetter eastern region, whereas C. chrysosperma and C. nivea are primarily found in drought-prone areas with little rainfall. Our findings can help farmers and planners develop methods to avoid the spread of Cytospora spp. and calculate the costs of applying pesticides to reduce contamination and boost yields.
1. Introduction
Plant pathogens have significant impacts on plant output all around the world [1]. Climate change can directly lead to plant production losses through agricultural meteorological disasters or indirectly through the impacts of plant pathogens [2]. Many studies have found that climate change is a major driver of plant pathogen spatial pattern changes and may improve plant pathogen overwintering survivability [3,4]. The distribution range of many plant pathogens is expected to continue shifting toward high-latitude and high-altitude locations [5,6]. Thus, understanding the spatial distribution patterns, population diversity, and compositions of plant pathogens affected by climate is critical for reducing plant disease losses.
Members of the Cytospora genus are found worldwide and are frequently considered endophytes, saprobes, or phytopathogens that can infect many hosts [7,8]. Cytospora species are mostly connected with woody plant canker diseases such as plant infections. Other diseases, such as Chinese jujube root rot and pomegranate collar rot, have also been observed [9,10]. Wounds caused by biotic (insects and birds) and abiotic (wind, drought, frost, rain, and hail) stress are the primary ways that Cytospora species infect host plants [11,12,13], indicating that harsh circumstances are always favorable for these taxa in China [14]. The genus Cytospora has 110 species, 31 of which have been identified in China [15]. Among the numerous pathogens attacking important economic woody plants, such as Juglans regia [16], Malus domestica [17,18], Eucalyptus [7], Pyrus sinkiangensis [17,18], Populus spp. [19,20], and Salix spp. [13], Cytospora chrysosperma, C. nivea, and C. mali are the most common species and cause severe economic and ecological losses, especially in China [8,13,21]. Their occurrence, development, distribution, and prevalence are vital to the long-term development of China’s forestry industry in the northeast, northwest, north, and southwest [8,13].
Understanding the basic biological features of these three species may help us better appreciate their dangers and the need to anticipate future suitable areas. The first species is C. chrysosperma, which is thought to be the primary cause of Salicaceae canker disease, raising quarantine concerns, particularly in China [8,22]. The species is a typical Cytospora species with a wide host range across Europe, Asia, Africa, Australia, and America [7,14,23]. The optimal growth temperature of C. chrysosperma is 28 °C [24]. The second species is C. mali; like other Cytospora species, it is typically known as an opportunistic pathogen occurring within a relatively narrow host range and mainly attacks economically important fruit crops, such as Malus domestica and Pyrus sinkiangensis [15,25] in China and Japan. C. mali is one of the few well-studied critical pathogens in the genus Cytospora concerning its biology, infection, and populations. Previous research has shown that C. mali can survive in the xylem for five years [26] and that the conidia can maintain their germination potential for 16 days at −15 °C, indicating that the species has strong environmental adaptability [27,28]. The optimal growth temperature of C. mali is 25 °C [24]. The third species is C. nivea, which, like C. chrysosperma, is the second most common pathogenic fungus that mainly harms the host of the Salicaceae family in Asia, Europe, and South America [13,29,30]. Meanwhile, it can also harm Juglans regia [16], Malus domestica [18], and Pyrus sinkiangensis [17]. The optimal growth temperature of C. nivea is 25 °C [24]. From the above, we speculate that the distribution ranges, niche overlap, and climatic preferences may differ between these species, but their geographical distributions and determinants have not yet been addressed. Thus, additional disease management strategies are needed.
Global temperature change is widely acknowledged to significantly impact species distribution [6,31,32] and may increase fungal infections [6,33]. However, few studies [5,34,35] have explored the dispersion of fungi on large spatial and temporal scales due to their distinctive life cycles and development patterns. Determining the potential geographic range of the Cytospora canker is crucial for field management decision making and surveillance. GIS (Geographic information systems) and SDM (spatial data management) present a possible solution to this problem [3]. They will expand across more species, geographic regions, and research topics [5,36].
Currently, niche models like the Maxent (Maximum Entropy) model [37], ENFA (Ecological Niche Factor Analysis) model [38], CLIMEX (Climate Change Experiment) [39], and GARP (Genetic Algorithm for Rule-set Production) model [40] and GLMs (Generalized Linear Models) [41] are widely used to simulate and predict the appropriate areas of fungal diseases [5,42,43,44]. The Maxent modeling approach is widely utilized among these models due to its superior predictive performance [5,45,46]. It is effective even when few occurrence records are known and the association between climatic and environmental elements is unpredictable [6,44,47]. Therefore, we used the Maxent model to analyze the potential distribution of the chosen species.
Given that C. chrysosperma, C. mali, and C. nivea have been linked to woody plant canker diseases, elucidating their ecological and physiological characteristics, such as distribution ranges, habitat shifts, niche overlap, and climatic preferences, would aid in disease prediction and the consideration of their controls. Therefore, the following problems are addressed in this study: (1) determining the key environmental parameters impacting C. chrysosperma, C. mali, and C. nivea distributions; (2) calculating the core distribution for each species to estimate their appropriate areas under present and future climatic scenarios and evaluating their migration propensity; and (3) computing and comparing species’ niche overlap. We hope our findings will be valuable to policymakers in developing measures to combat Cytospora canker disease.
2. Materials and Methods
2.1. Occurrence Records
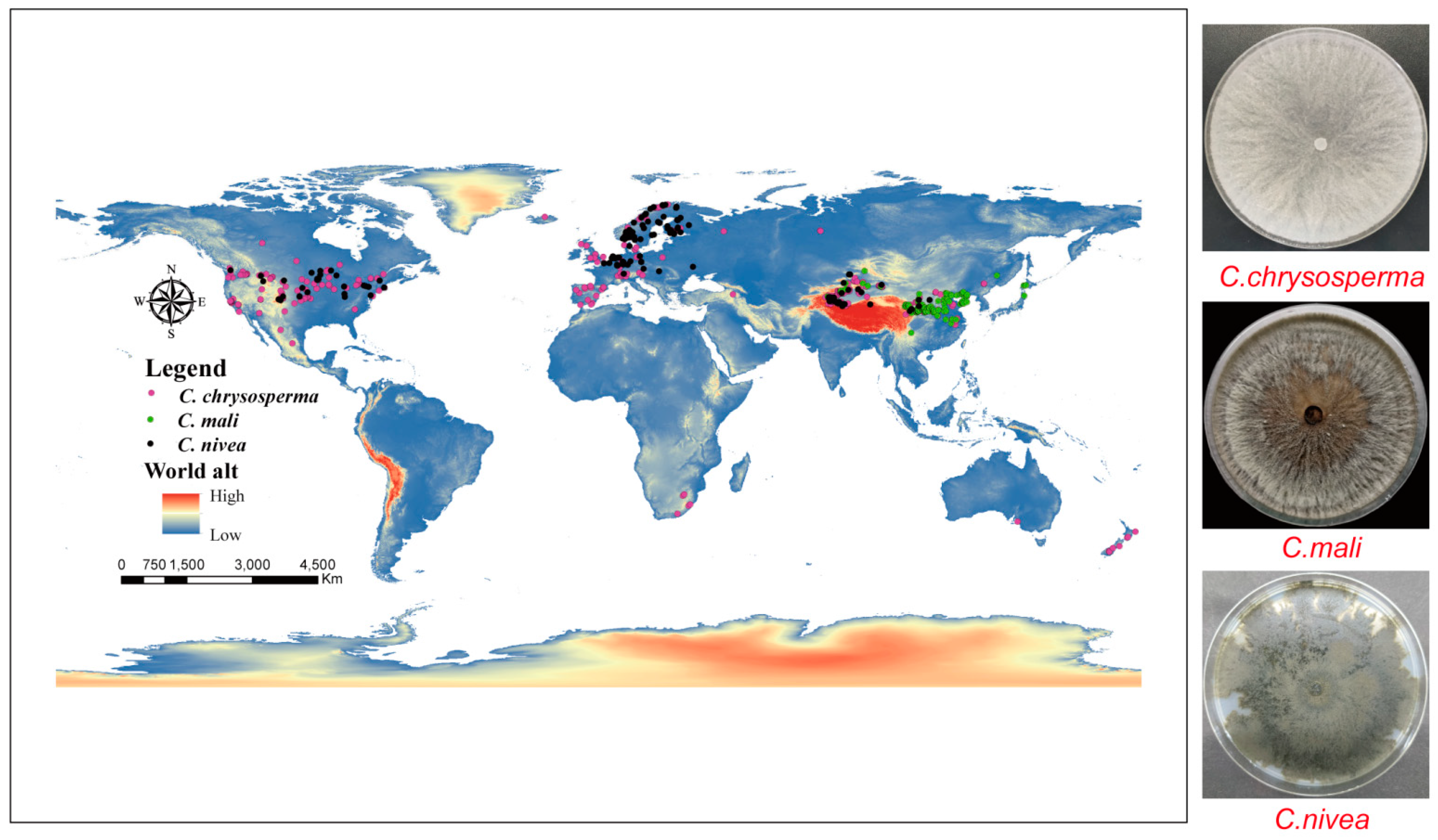
All georeferenced occurrence records in our study were obtained from three different sources: (1) GBIF (The Global Biodiversity Information Facility) (https://www.gbif.org/); (2) relevant articles from the China National Knowledge Infrastructure (CNKI) (https://www.cnki.net/), Web of Science (WOS) (https://www.webofscience.com/), and Google Scholar (https://scholar.google.com.hk); and (3) GPS, which was used to gather 135 C. chrysosperma, 34 C. mali, and 25 C. nivea occurrence points during fieldwork in the Xinjiang Uygur Autonomous Region, China in 2019 and 2023. By ensuring that no two occurrence data points were inside the same raster (~5 km2) [6,48], ENMTools (https://github.com/danlwarren/ENMTools) was utilized to prevent spatial autocorrelation from impairing the model’s performance [48,49]. Ultimately, 374 global occurrence data points for C. chrysosperma, 164 for C. mali, and 166 for C. nivea were retained by our study (Figure 1 and Supplementary Table S1).
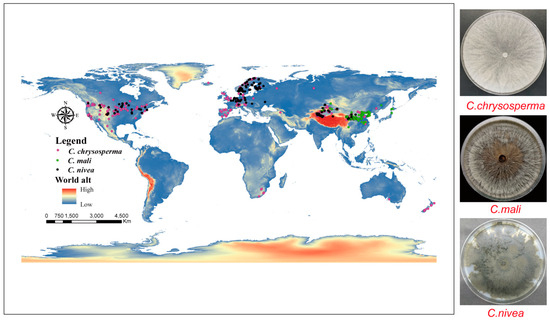
Figure 1.
Global Occurrence records of three studied species. The pink, green and black dots represent C. chrysosperma, C. mali and C. nivea, respectively.
2.2. Environmental Factor Variables
Numerous factors, such as the habitat’s climatic suitability, topography, land cover, and dispersal history, are probably responsible for the spread of numerous fungi [5,21,50]. Since C. chrysosperma, C. mali, and C. nivea typically affect woody plants, we selected soil and vegetation conditions as limiting ecological variables in this study. Supplementary Table S2 displays the selected environmental variables, which include 19 bioclimatic factors, three topographical factors (aspect, slope, and altitude), one global land cover data point (~1 km), and 11 topsoil factors (0–30 cm) with a 2.5 arc-min spatial resolution. These variables were obtained from the World Climate website (http://www.worldclim.org), the Harmonized World Soil Database (http://www.fao.org/soils-portal/), and the Global Maps website (http://globalmaps.github.io). Future climate data were based on BCC-CSM2-MR data in the Coupled Model Intercomparison Project Phase 6 (CMIP6), which is more appropriate for Asia, particularly China [51,52]. To predict the future distributions of C. chrysosperma, C. mali, and C. nivea in 2030, 2050, 2070, and 2090, three independent socioeconomic models driven by CO2 and shared socioeconomic pathways (SSPs) 126, 370, and 585 were chosen in this study.
The species distribution models (SDMs) may overfit due to multicollinearity amongst bioclimatic variables [43]. To eliminate multivariate collinearity, we utilized ENMTools to evaluate the correlation coefficients of the bioclimatic variables. Finally, we kept several meaningful bioclimatic variables for each research species based on the correlation coefficient of the bioclimatic variables (|r| > 0.8) and the contribution of each bioclimatic variable (Supplementary Figure S1) [6,45,48].
2.3. Model Simulation, Optimization, and Evaluation
The regularization multiplier (RM) and feature classes (FCs) are the two most important parameters of the Maxent model, aside from occurrence records and associated bioclimatic variables. A suitable combination of these elements can raise the sensitivity of the model while lowering the risk of overfitting [45,53]. The Maxent model’s parameters were calibrated using the R 3.6.3 “Kuennm” package to determine the ideal model tuning settings for each research species. We examined 31 different FC combinations (L for linear, Q for quadratic, H for hinge, P for product, and T for threshold), and the RM varied progressively throughout a 0.1-unit interval from 0.1 to 4. With delta Akaike minimal information criterion (AICc) values as low as possible (delta AICc = 0 or <2) [54,55], the best Maxent models were identified. Finally, RMs of 0.4, 1.5, and 1.2 were chosen for C. chrysosperma, C. mali, and C. nivea, respectively, along with feature combinations of LQP, LQH, and QT (Table 1).

Table 1.
Results of MaxEnt models with optimized parameters developed for three studied species.
Then, we adjusted the parameters of the Maxent and completed the modeling of the studied species. Specifically, “Create response curves”, “Random seed”, “Do jackknife to measure variable importance”, “Write plot data”, “Write background predictions”, “Replicates 10”, “Replicated run type subsample”, “output file type ‘.asc’”, and “Output format logistic” were the parameters set by the Maxent model. Training data accounted for 75% of the distribution data, with testing data making up the remaining 25%. The default values for the other settings of the software were used. To assess the performance of the model, we used the values of the true skill statistic and area under the receiver operating characteristic (ROC) curve (AUC). AUC values vary from 0 to 1 [56]. A prediction is deemed bad if it is less than 0.5, acceptable if it is between 0.5 and 0.7, good if it is between 0.7 and 0.9, and outstanding if it is between 0.9 and 1 [6,57]. TSS was rated as failed, <0.4; good, 0.4–0.75; and excellent, >0.75 [58,59]. In addition, the efficacy of the model was confirmed in this study by contrasting the results with the field survey data.
2.4. Assessment of Current and Future Distribution Areas
ArcGIS 10.4.1 was used to visualize and further analyze the continuous habitat suitability maps created for each study species. Based on the maximum test sensitivity plus specificity (MTSPS) threshold [6,25,60], binary suitability maps (unsuitable and suitable areas) for each study species were constructed. We developed four climate suitability categories for the three species, unsuitable, low suitable, medium suitable, and high suitable, to interpret the prediction results more easily [6].
Changes in the distribution regions of species can be efficiently reflected by core distribution migration [5]. Therefore, we employed SDMtoolbox (version 2.4) [61] to derive the core distribution migration of three species under present and future concentration scenarios and explain changes in the distribution of the appropriate species areas more clearly. Core distribution migration results were obtained using SDMtoolbox (version 2.4) by importing the “.asc” files produced under various concentration scenarios predicted by the Maxent model into ArcGIS 10.4.1. After that, additional analysis was conducted on the direction and distance of the core distribution migration [61]. Additionally, we calculated the contraction and expansion of the potential range of C. chrysosperma, C. mali, and C. nivea in China with climate change using SDMtoolbox (version 2.4).
2.5. Niche Overlap Analysis
ENMTools (version 1.3) was used to compare the ecological niches of our research species to Maxent projections. Hellinger’s I and Schoener’s D values represented the degree of niche overlap. D values range from 0 (no overlap) to 1 (more than 0.6 indicates significant overlap) [48,62,63]. Additionally, we used ArcGIS 10.4.1 to identify the homologous distribution areas of the three species and mapped the overlapping areas of the species niches under various conditions [43,45].
3. Results
3.1. Model Performance and Variable Contribution
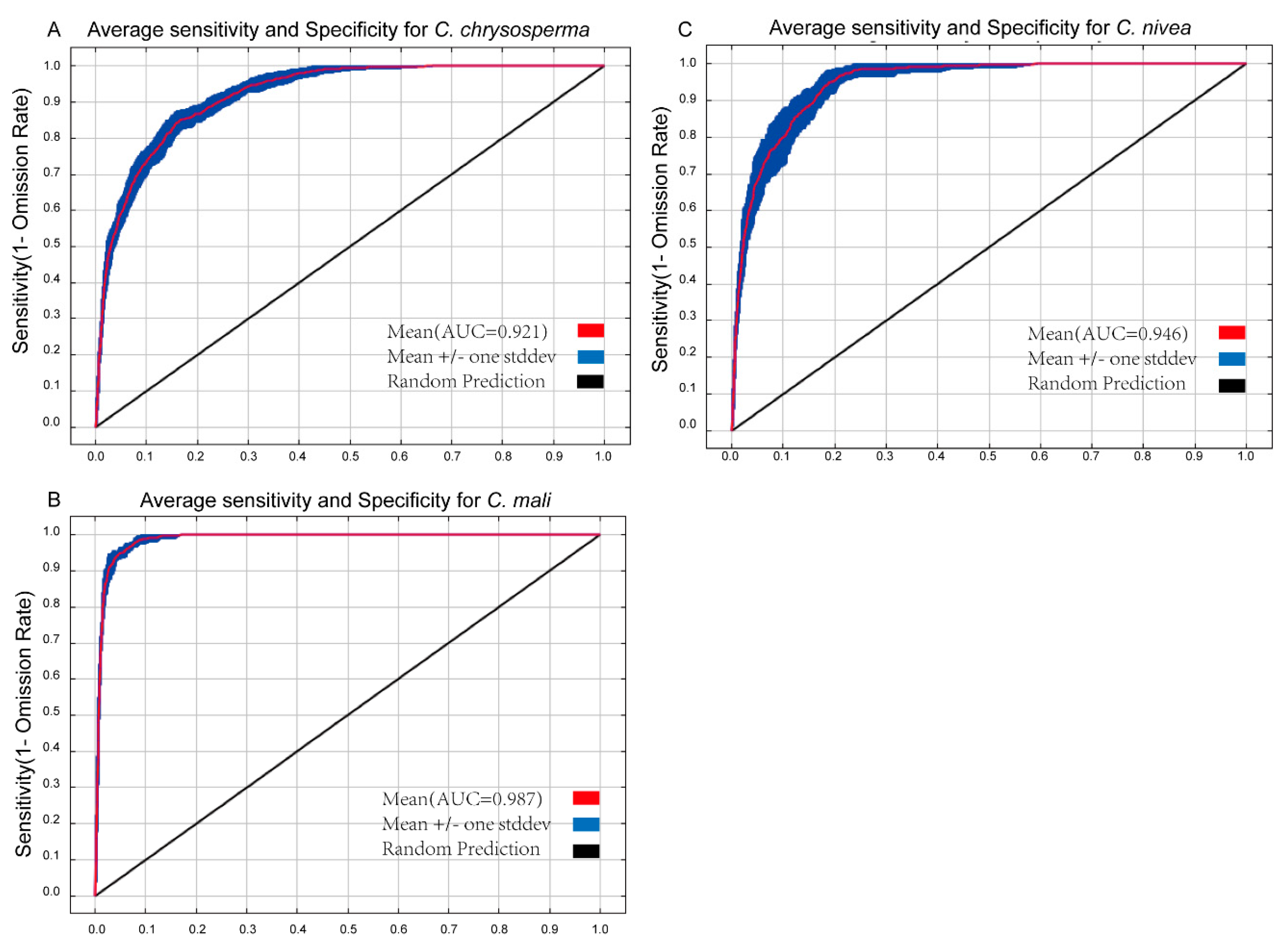
AUC and TSS values have frequently been employed to assess SDM model performance [52,64]. The greater the value, the stronger the relationship between bioclimatic factors and the expected distribution area, and the better the model works [65,66]. The average AUC values for 10 repetitions for C. chrysosperma, C. mali, and C. nivea were 0.921, 0.987, and 0.946 (Figure 2), respectively. Their TSS values were 0.797, 0.898, and 0.802, all exceeding 0.79 (Table 1 and Supplementary Figure S2), indicating that the optimized Maxent model predicted the potential distribution well.
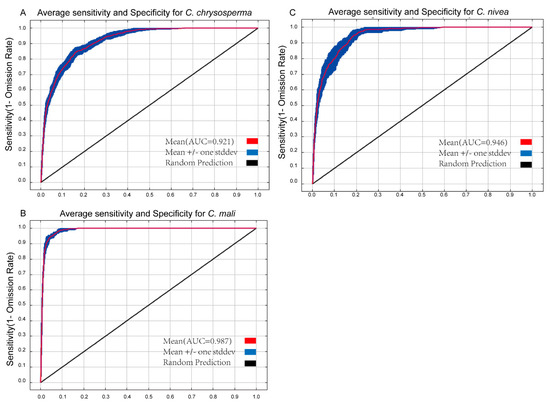
Figure 2.
The receiver operating characteristic curve for target species. (A–C) represent C. chrysosperma, C. mali and C. nivea, respectively.
Based on detailed jackknife testing and a percent contribution study, the main influencing factors for the geographical distribution of the three research species are different (Supplementary Figure S3). The most important bioclimatic variable in the predictions for C. chrysosperma was the annual mean temperature (Bio 1, 64.4%). Other important bioclimatic variables included land cover (8.6%), mean diurnal range (Bio 2, 7.9%), seasonality of precipitation (Bio 15, 7.9%), and topsoil gravel content (T-grave, 7.6%), with a total contribution rate of up to 96.4% (Supplementary Table S3). The mean temperature of the driest quarter (Bio 9, 35.4%) had the highest mean contribution to the C. mali models, followed by precipitation of the coldest quarter (Bio 19, 18.8%), precipitation of the warmest quarter (Bio 18, 11.3%), land cover (11%), and temperature seasonality (Bio 4, 9.5%), with a total contribution rate of up to 86% (Supplemental Table S3). The most significant environmental variable that affected the AUC of the models for C. nivea was the mean temperature of the coldest quarter (Bio 11, 67.7%). This was followed by the precipitation of the wettest month (Bio 13, 9.2%), the mean diurnal range (Bio 2, 7.2%), and the topsoil TEB (5.4%), with a total contribution rate of up to 89.5% (Supplementary Table S3).
For C. chrysosperma, the optimal ranges of the mean diurnal range (Bio 2), annual mean temperature (Bio 1), land cover, and precipitation seasonality (Bio 15) were >1.9 °C, −2.76–17.19 °C, >3.18, and 0–120.14 mm, respectively (Supplementary Figure S3A and Supplementary Table S4). For C. mali, the optimal ranges of the precipitation of the coldest quarter (Bio 19), mean temperature of the driest quarter (Bio 9), precipitation of the warmest quarter (Bio 18), land cover, and temperature seasonality (Bio 4) were >0–46.55 mm, >−10.46–9.17 °C, 8.57–927.25 mm, and 7.08–19.23, respectively (Supplementary Figure S3B and Supplementary Table S4). For C. nivea, the optimal ranges of the precipitation of the wettest month (Bio 13), mean temperature of the coldest quarter (Bio 11), and T-teb were 0–154.09 mm, >−13.64–4.75 °C, and 0–46.58, respectively (Supplementary Figure S3C and Supplementary Table S4). In addition, the range of the maximum probability of factors of the three studied species is shown in Supplementary Table S4.
3.2. Current Distribution Regions of the Three Species
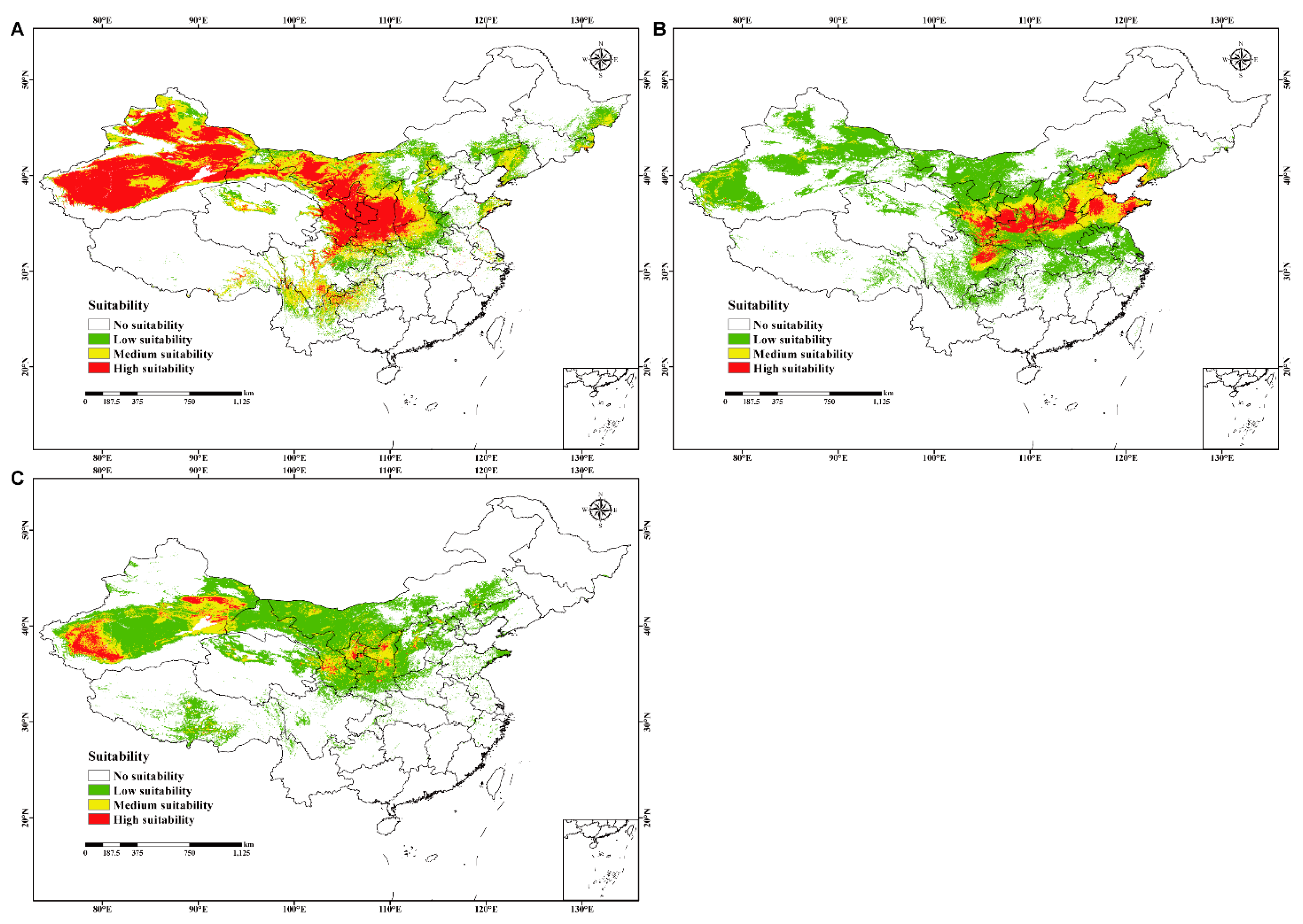
The prediction results of C. chrysosperma, C. mali, and C. nivea in China fit the known distribution in native ranges, reflecting the detection efficiency of our models. Among them, C. chrysosperma had the largest possible distribution area, covering approximately 36.2 × 105 km2, accounting for 37.7% of the Chinese land surface area. The high- and medium-suitability areas primarily included Xinjiang, Gansu, Ningxia, Shaanxi, Sichuan, Midwest Nei Monggol, south-central Shanxi, and north Henan, with sporadic distributions in of Liaoning, Eastern Heilongjiang, Jilin, Shandong, Hebei, Qinghai, Xizang, Yunnan, and Guizhou (Figure 3A and Supplementary Table S5). Meanwhile, C. mali had a relatively narrower distribution range than C. chrysosperma. Specifically, C. mali covered approximately 32.4 × 105 km2, accounting for 33.8% of the Chinese land surface area. The high- and medium-suitability areas primarily involved Gansu, Shaanxi, Shanxi, Hebei, Shandong, Sichuan, Henan, and Tianjin, with sporadic distributions in Ningxia, Liaoning, Beijing, and Xinjiang (Figure 3B and Supplementary Table S6). Moreover, C. nivea covers the smallest possible distribution area of approximately 27.9 × 105 km2, accounting for 29.1%. The high- and medium-suitability areas were mainly in Xinjiang, Gansu, Ningxia, and Shaanxi, with sporadic distributions in Qinghai, Xizang, Shanxi, Hebei, and Nei Monggol (Figure 3C and Supplementary Table S7).
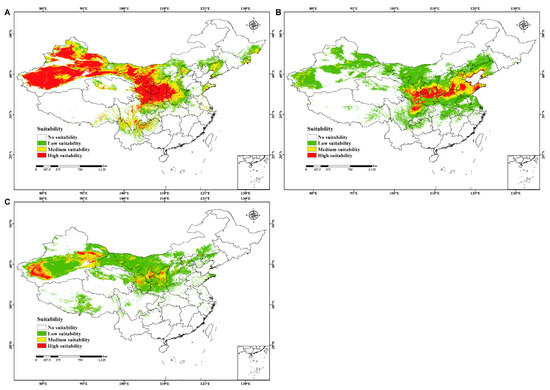
Figure 3.
The current potential geographical distribution for three studied species in China. (A–C) represent C. chrysosperma (no suitability p ≤ 0.2812; low suitability 0.2812 < p ≤ 0.4; medium suitability 0.4 < p ≤ 0.6; high suitability p > 0.6, p = probability), C. mali (no suitability p ≤ 0.0887; low suitability 0.0887 < p ≤ 0.4; medium suitability 0.4 < p ≤ 0.6; high suitability p > 0.6, p = probability) and C. nivea (no suitability p ≤ 0.1184; low suitability 0.1184 < p ≤ 0.4; medium suitability 0.4 < p ≤ 0.6; high suitability p > 0.6, p = probability), respectively.
3.3. Future Distribution Changes of the Three Species
North Qinghai and most of Xinjiang, Gansu, Ningxia, Shaanxi, Nei Monggol, and Sichuan are mainly possible distribution zones for C. chrysosperma with climate change (Supplementary Figure S4). The medium-suitability habitat is predicted to expand as a result of climate change. At the same time, the low- and high-suitability habitats will generally decrease to varying degrees. The average suitable area (39.61%) would increase under the 12 future climatic scenarios compared to the current time (37.75%) (Supplemental Table S5). Although C. mali has a narrower range of acceptable habitats than C. chrysosperma, it may adapt to wetter and warmer climates, such as those in Henan, Shandong, Jiangsu, Hubei, Sichuan, and Guizhou (Supplementary Figure S5). In the 12 future climate scenarios, the high-, medium-, and low-suitability habitats are expected to increase remarkably, and the average suitable areas (44.29%) would increase compared with the current time (33.79%) (Supplementary Table S6). The range of potentially suitable habitats for C. nivea was the smallest, occurring only in Xinjiang, Gansu, Nei Monggol, Ningxia, Xizang, Shaanxi, and Qinghai (Supplementary Figure S6). The high-, medium-, and low-suitability habitats are predicted to expand due to climate change. The average suitable area (34.08%) would increase compared with the current period (29.13%) (Supplementary Table S7).
Supplementary Figures S7–S9 and Tables S8–S10 illustrate the contraction and expansion of the potential range of C. chrysosperma, C. mali, and C. nivea in China with climate change. Although the extent of the possible range changes varied among the SSPs, all the SSPs anticipated northward shifts in the prospective distributions of the three species. C. chrysosperma migrated into high-latitude and high-altitude habitats. At the same time, C. mali and C. nivea migrated to high latitudes and locations, but their tendency to migrate to high altitudes was not great. The expansion area under a high-emission scenario for the three studied species was larger than that under a low-emission scenario compared to the current climatic conditions. The contraction area under a low-emission scenario was lower than that under a high-emission scenario. The results indicate that future climate change will considerably alter the range of appropriate habitats for the three species.
3.4. Comparisons of Overlapping Areas, Distribution Centroids, and Ecological Niches for the Three Studied Species
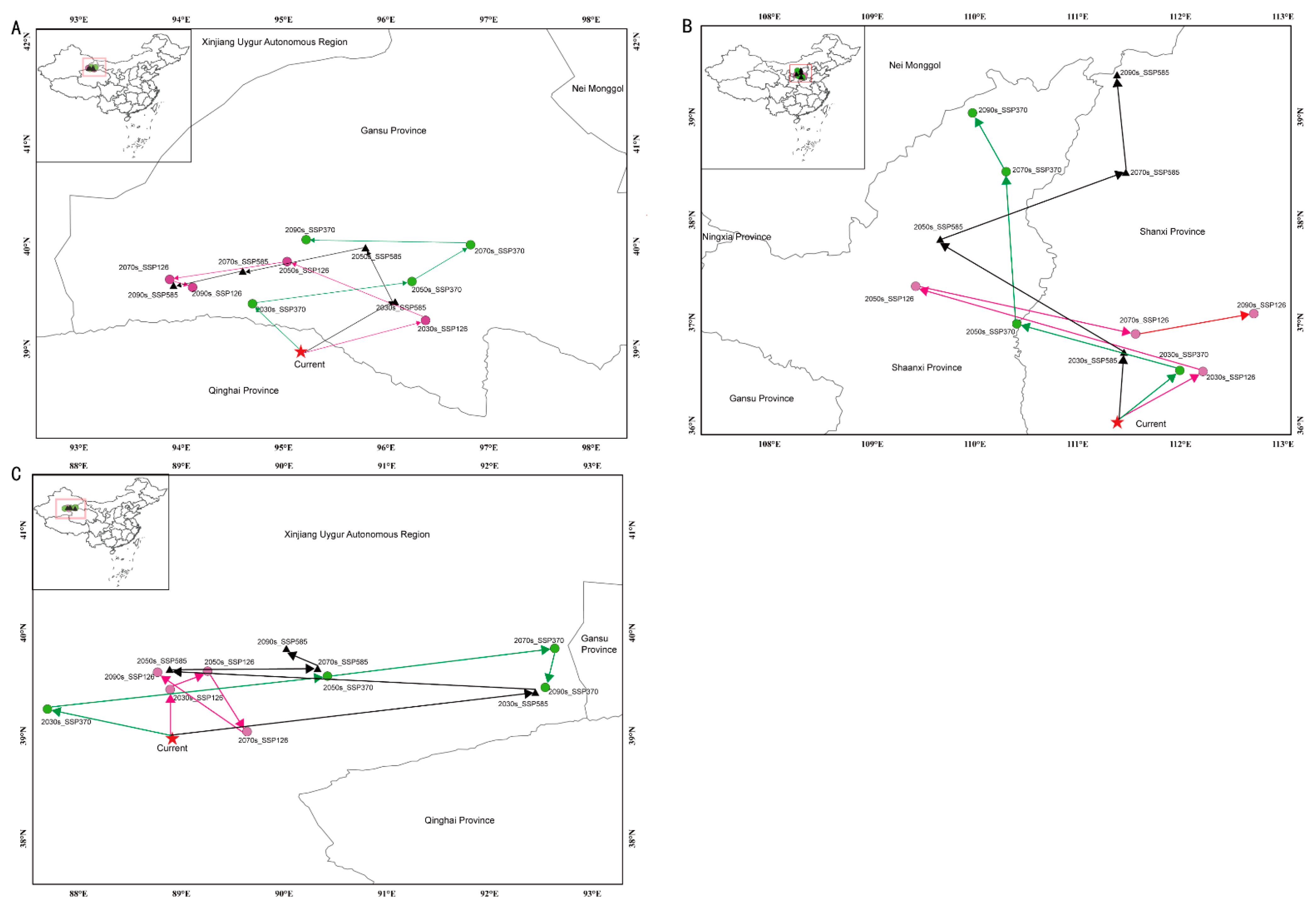
The predictions of the Maxent model under the current climate conditions suggested that the appropriate areas for the three studied species overlapped to varied degrees with the suitable area (Figure 2). The overlapping area between C. chrysosperma and C. nivea was the greatest (28.6 × 104 km2), followed by C. mali (24.9 × 104 km2). The distribution centroids of C. chrysosperma, C. mali, and C. nivea were distributed in Qinghai (95.167233° N, 38.975466° E), Shanxi (111.386901° N, 36.032607° E), and Xinjiang (88.912738° N, 38.976171° E), respectively, under the current climate change scenario (Figure 4 and Table 2). Under various climate change scenarios, the centers of the appropriate habitat areas of the three species were expected to shift to high-latitude regions.
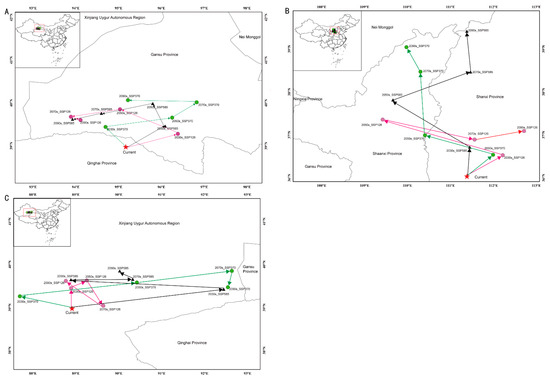
Figure 4.
Centroid shifts of potential suitable area for three studied species under different climatic scenarios in China. Red star indicates the centroids of the suitable habitats of each species under current climate. Dots and triangles represent the centroids of the suitable habitats of each species under different future climate scenarios. (A–C) represent C. chrysosperma, C. mali and C. nivea, respectively.

Table 2.
Centroid migration of suitable areas under future climatic conditions for target species.
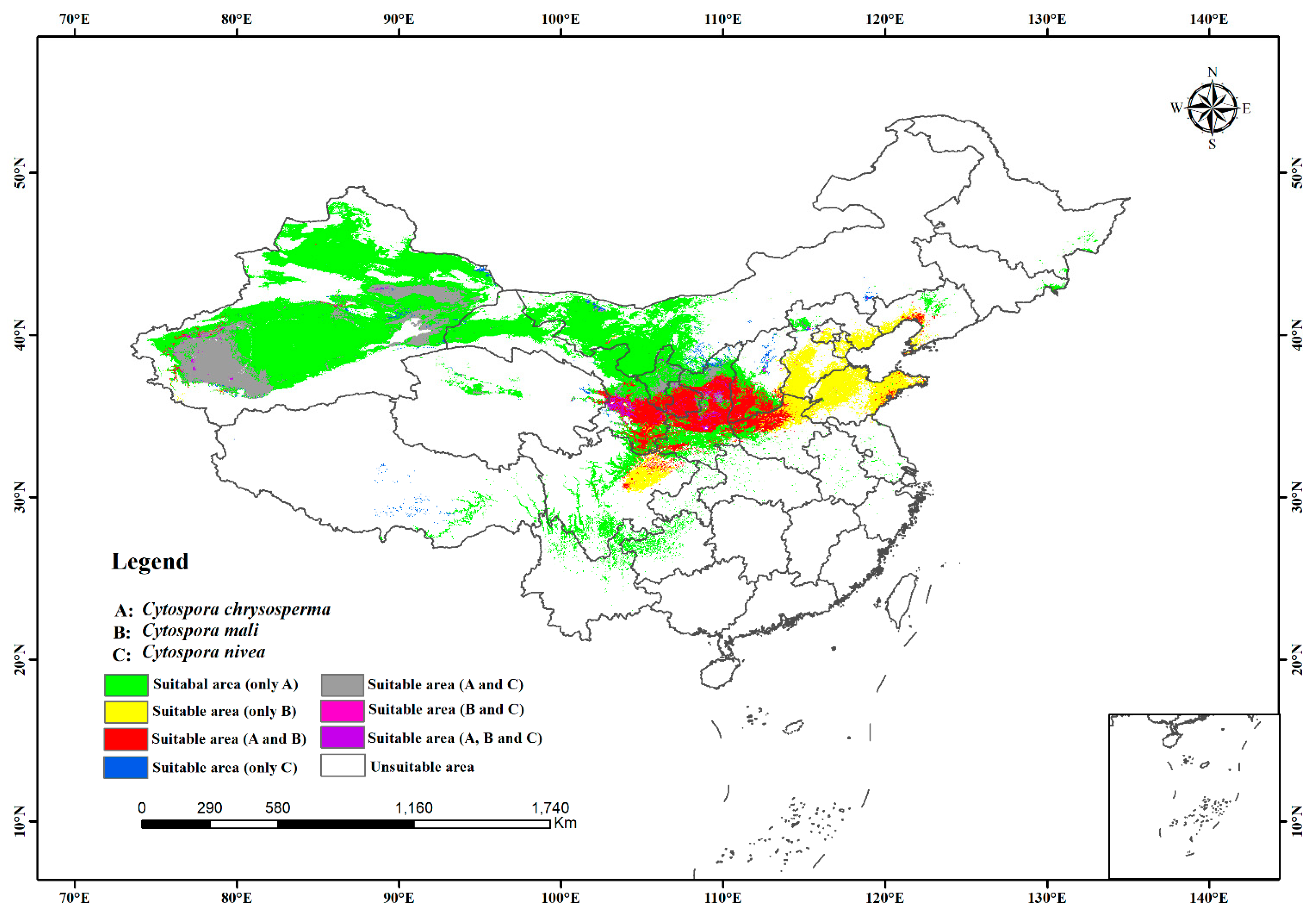
Based on the predicted suitable habitat, we estimated the niche overlap index and mapped the overlap of the potential distribution habitats of the three studied species in China. According to Table 3, there was less niche overlap between C. chrysosperma and C. mali (D = 0.6093, I = 0.8730) than between C. chrysosperma and C. nivea (D = 0.6848, I = 0.8956). However, The range overlap between C. chrysosperma and C. nivea was higher (0.9246) than that between C. chrysosperma and C. mali (0.5285). There was niche overlap and separation for C. mali and C. nivea (D = 0.5488, I = 0.8342) (Table 2). In China, the main niches of C. chrysosperma overlapped with the niches of C. mali and C. nivea. These three species coexist mainly in the northwest and north regions. The niche of C. mali could reach the warm and wet eastern region. In contrast, the niches of C. chrysosperma and C. nivea mainly existed in drought with low rainfall areas in western China (Figure 5).

Table 3.
Niche and range overlap of the potential distribution habitats of the three species.
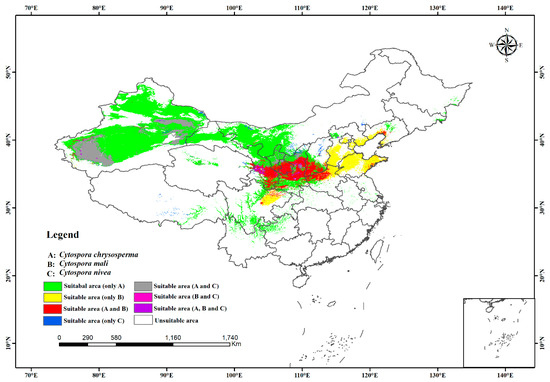
Figure 5.
Niche and range overlap of the potential distribution habitats of the three species.
4. Discussion
Due to the unique growth features of fungi in comparison to other taxa such as animals and plants, species distribution model (SDM) research on fungi began quite late and is scarce [67]. However, with the rapid expansion of disciplines like statistics, computer technology, and geographic information systems in recent years, fungi SDMs have notably increased in the study of pathogenic microfungi, lichens, and macrofungi [5]. Although there are many uses for fungi SDMs, the majority of them are grouped into three main categories: (1) investigating environmental factors that influence occurrence, (2) forecasting occurrence in specific regions, and (3) utilizing fungus as a model organism to investigate ecological or methodological ideas [5]. For example, Ajene et al. [44] employed three species distribution models (BIOCLIM, MaxEnt and Boosted Regression Trees) to predict the current and future potential distribution of Candidatus Liberibacter asiaticus in Africa, and the potential global distribution of Candidatus Liberibacter africanus, using long-term bioclimatic variables. Ejaz et al. [47] utilized four SDMs (GLM, generalized linear model; GAM, generalized additive model; GBM, generalized boosting model; and MaxEnt, maximal entropy) to investigate the suitability of Fusarium spp. disease around the world under climate change.
Currently, there is little information on the prevalence and occurrence of Cytospora canker in large-scale locations. During 2019–2023, our group conducted some work on the epidemic dynamics of Cytospora canker in Xinjiang Uygur Autonomous Region, China, focusing primarily on data gathering and field disease monitoring, laying the fundamental work for constructing prediction models. In this work, we used Maxent to investigate the possible habitat suitability of C. chrysosperma, C. mali, and C. nivea under present and future climatic circumstances. The AUC values all exceeded 0.9, indicating excellent accuracy. The climatic variables, particularly the temperature-related ones, are prominent factors for the studied species when selecting their habitats, such as the annual mean temperature (Bio 1), mean temperature of the coldest quarter (Bio 11), and mean temperature of the driest quarter (Bio 9), followed by the precipitation factors. The topographical factors (altitude, slope, and aspect) have weaker effects on species distribution. This could be as a result of the three investigated species being impacted by environmental factors disproportionately [68].
In general, the effects of terrain and soil factors on species distribution are frequently limited to smaller spatial scales and have a stronger impact on pathogen host plants, whereas climate factors, including temperature and precipitation, have the opposite effect [69]. Previous field investigations have indicated that the three examined species mostly infect host plants via various wounds, temperature is the most important environmental factor influencing the occurrence and prevalence of Cytospora canker disease, and precipitation aids spore dispersal but has a minor impact [70]. Our findings demonstrated that three of the examined species were highly adapted to the regions of north, northeast, and northwest China, implying that the dry and cold circumstances in China are always good for these taxa. The northern area of China, as is well known, has a temperate continental climate and a temperate monsoon climate, with low annual rainfall, cold and dry winters, and high summer temperatures, and wounds are prone to forming in this harsh climate environment [71]. Furthermore, the ideal temperature for the three examined species is 25–28 °C [24], and strains cannot develop when the ambient temperature is below 5 °C or above 35 °C [72]. Therefore, there are often two infection peaks for Cytospora canker each year: one from March to May and another from September to October [24]. Infections from November onward are extremely rare. This is in line with the finding that temperature affects the distribution of the three species under study more so than precipitation. Additionally, the models confirmed that C. chrysosperma was the most widely dispersed species compared to C. mali and C. nivea, implying the greatest potential for harm within the research area, consistent with the findings of previous resource surveys [13,15,19]. However, the predicted potential distributions of the three studied species were much more comprehensive than currently documented. Therefore, additional survey efforts are required to corroborate our findings.
Climate change has been documented as global warming and rising temperatures, which could lead to the redistribution or extinction of species in the future [73,74]. Many studies have shown that habitat migrations are primarily observed and expected to move toward higher latitudes and altitudes under a warming climate [6,32]. Our results showed a similar trend for C. chrysosperma. However, there is a trend for C. mali and C. nivea to shift toward higher latitudes but not toward higher altitudes, suggesting that the altitude either does not affect their distribution or has a minor impact. This may be because each species has unique physiological properties. Thus, adaptations to future climates will differ and depend on the ecological traits of the species. Additionally, the general trend for suitable regions in China is increasing in all of the warming scenarios, and habitat fragmentation is a crucial problem based on the range shift comparison of the present and future (Supplementary Figures S6–S8 and Tables S8–S10). Unsuitable regions may become suitable, whereas moderate and marginal areas may become ideal-suitability areas. This indicates that China must deal with a more serious Cytospora canker crisis.
Niche breadth and niche overlap are essential indicators for describing the actual ecological niche of a species, which, to some extent, reflects the characteristics of plants and their adaptability to the environment [75]. Niche breadth refers to the total utilization of various environmental resources by species. In contrast, niche overlap is the degree of similarity and competition between species in resource utilization [76]. In general, species with wide ecological niches may be more resilient to climate change and more able to adapt to it than species with narrow ecological niches [77]. C. chysosperma had the widest niche breadth among the three species, followed by C. nivea. The niche and range overlaps between C. chysosperma and C. nivea were high. Meanwhile, the geographical diffusion centers of C. chysosperma and C. nivea were also relatively close. However, due to the higher competitive ability of C. chysosperma than C. nivea, the potential diffusion range was significantly higher than that of C. nivea. Although the niche overlap and range overlap of C. chysosperma and C. mali were slightly lower than those of C. chysosperma and C. nivea, the centroids of C. chysosperma and C. mali were far away (Figure 4 and Table 2). With climate change, competition among these species may intensify as their harmful areas in China increase, and spatial overlap is expected to increase.
The three studied species have different potential diffusion zones in northeast, northwest, north, and southwest China. Therefore, it is necessary to develop corresponding prevention and control measures for the potential diffusion characteristics and key diffusion areas of each species to avoid unknown diffusion hazards. The border area between Gansu, Xinjiang, and Qinghai must be given special attention because the current and future core distributions of C. chrysosperma and C. nivea are located in this area. In the future, the monitoring of C. nivea should be strengthened in north Xinjiang, central and east Nei Monggol, central and west Xizang, and north Qinghai. The areas around Bohai Bay, north Loess Plateau, north Qinghai, and north Xinjiang should focus on strengthening the dynamic monitoring and quarantine of C. mali. The dynamic monitoring and quarantine of C. chrysosperma should be strengthened in the western loess plateau, eastern, northeast, and western Sichuan plateau, western Xizang, Tarim Basin, and Junggar Basin.
There are indeed some limitations to our study. First, the accuracy of species occurrence data, especially from published sources, increases the forecasting uncertainty. Due to the lack of latitude and longitude coordinates, precise distribution locations were determined by searching for place names with coordinate positioning software, which may have resulted in geographical mistakes. Second, the model was based on an ideal niche and did not consider the effects of additional elements, such as the species self-diffusion ability, sampling biases, uncertainties in identification and taxonomy, host conditions, species interactions, human activities, variety type, medication frequency, and socioeconomic structure. Third, prediction precision still has some limitations, although the individual SDM model in this study demonstrated great prediction accuracy. The accuracy and performance levels of SDMs vary considerably between approaches and species. Some research has shown that approaches that integrate numerous individual models produce robust estimates of the possible distributions of species, which can be used to increase the model prediction accuracy. The causes mentioned above may result in discrepancies between the expected and actual distributions. Therefore, future research must address these issues that impact the precision of model predictions.
5. Conclusions
Based on the worldwide distribution records and bioclimatic data in this study, we used the maximum entropy model (Maxent) to predict the potentially suitable areas of C. chrysosperma, C. mali, and C. nivea in China. The findings revealed that temperature-related climate factors limit the three species’ current distribution ranges. The three studied species were highly suitable for northeast, northwest, north, and southwest China. Under future climate scenarios, the distribution areas of the three species are expected to increase, and the centers of the appropriate habitat areas of the three species will shift to high-latitude regions. The three species coexist in China, primarily in the northwest and north regions. The ecological niches of C. chrysosperma, C. nivea, and C. mali are more similar. The distribution range of C. mali can reach the warmer and wetter eastern region, whereas C. chrysosperma and C. nivea are primarily found in drought-prone areas with little rainfall. Our findings can help farmers and planners develop methods to avoid the spread of Cytospora spp. and calculate the costs of applying pesticides to reduce contamination and boost yields.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/jof10010038/s1: Figure S1 Correlation analysis of various environmental factors. Figure S2 Jackknife test of variable importance Regularized training gain. Figure S3 Response curves for predictors in MaxEne model. Figure S4 The future potential geographical distributions of C. chrysosperma in China under future climatic conditions. Figure S5 The future potential geographical distributions of C. mali in China under future climatic conditions. Figure S6 The future potential geographical distributions of C. nivea in China under future climatic conditions. Figure S7 Adaptability changes in C. chrysosperma under different future climate scenarios. Figure S8 Adaptability changes in C. mali under different future climate scenarios. Figure S9 Adaptability changes in C. nivea under different future climate scenarios. Table S1 Geographical distributions of C. chrysosperma, C. mali and C. nivea species sampled in this study. Table S2 Environmental variables for current period model analysis. Table S3 List of selected environmental variables for each species. Table S4 Key climatic factors influencing habitat distributions of three species. Table S5 Dynamics of changes in distribution area of C. chrysosperma under different climate scenarios. Table S6 Dynamics of changes in distribution area of C. mali under different climate scenarios. Table S7 Dynamics of changes in distribution area of C. nivea under different climate scenarios. Table S8 Percentage of distribution changes of C. chrysosperma between current and future climate-change scenarios. Table S9 Percentage of distribution changes of C. mali between current and future climate-change scenarios. Table S10 Percentage of distribution changes of C. nivea between current and future climate-change scenarios.
Author Contributions
Conceptualization, C.Y., H.H. and L.W.; data curation, C.Y. and H.H.; formal analysis, C.Y. and H.H; funding acquisition, Z.K., L.H., L.W. and H.F.; investigation, C.Y., H.H., Z.W. and S.S.; methodology, C.Y. and H.H.; project administration, L.W. and H.F.; software, C.Y.; supervision, L.W. and H.F.; visualization, C.Y. and H.H.; writing—original draft, C.Y.; writing—review and editing, C.Y., H.H. and L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Joint Funds of the National Natural Science Foundation of China (Grant No. U1903206), the Bingtuan Science and Technology Program (2020DB006) and the Project of Scientific Research and Innovation for postgraduates in Tarim University (TDBSCX202105).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Fones, H.N.; Bebber, D.P.; Chaloner, T.M.; Kay, W.T.; Steinberg, G.; Gurr, S.J. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 2020, 1, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Chaloner, T.M.; Gurr, S.J.; Bebber, D.P. Plant pathogen infection risk tracks global crop yields under climate change. Nat. Clim. Change 2021, 11, 710–715. [Google Scholar] [CrossRef]
- Guo, Y.L.; Li, X.; Zhao, Z.F.; Nawaz, Z. Predicting the impacts of climate change, soils and vegetation types on the geographic distribution of Polyporus umbellatus in China. Sci. Total Environ. 2019, 648, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.S.; Zhang, W.; Peng, Y.; Zhao, F.; Chang, X.Q.; Xing, K.; Zhu, L.; Ma, G.; Yang, H.P.; Rudolf, V.H.W. Climate warming promotes pesticide resistance through expanding overwintering range of a global pest. Nat. Commun. 2021, 12, 10. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.X.; Guillera-Arroita, G.; May, T.W.; Lahoz-Monfort, J.J.; Elith, J. Using Species Distribution Models For Fungi. Fungal Biol. Rev. 2020, 34, 74–88. [Google Scholar] [CrossRef]
- Yan, C.C.; Hao, H.T.; Wang, Z.; Sha, S.S.; Zhang, Y.W.; Wang, Q.P.; Kang, Z.S.; Huang, L.L.; Wang, L.; Feng, H.Z. Prediction of Suitable Habitat Distribution of Cryptosphaeria pullmanensis in the World and China under Climate Change. J. Fungi 2023, 9, 739. [Google Scholar] [CrossRef] [PubMed]
- Adams, G.C.; Wingfield, M.J.; Common, R.; Roux, H. Phylogenetic relationships and morphology of Cytospora species and related teleomorphs (Ascomycota, Diaporthales, Valsaceae) from Eucalyptus. Stud Mycol. Stud. Mycol. 2004, 52, 1–144. [Google Scholar]
- Fan, X.L.; Bezerra, J.D.P.; Tian, C.M.; Crous, P.W. Cytospora (Diaporthales) in China. Persoonia 2020, 45, 1–45. [Google Scholar] [CrossRef]
- Palavouzis, S.C.; Tzamos, S.; Paplomatas, E.; Thomidis, T. First report of cytospora punicae isolated from pomegranate plants with symptom of collar rot in northern Greece. J. Plant Pathol. 2015, 97, 216. [Google Scholar]
- Du, Q.; Zhao, S.F.; Wu, C.L.; Kong, L.; Zhang, P. Root Rot of Chinese Jujube (Ziziphus jujuba) Caused by Cytospora sacculus in China. Plant Dis. 2015, 97, 1661. [Google Scholar] [CrossRef]
- Guyon, J.; Hoffman, J. Survey of aspen disease in the intermountain region. USDA For. Serv. For. Health Prot. Ogden Utah. 2011. Available online: https://digitalcommons.usu.edu/aspen_bib/7040/ (accessed on 1 December 2023).
- Worrall, J.J.; Rehfeldt, G.E.; Hamann, A.; Hogg, E.H.; Marchetti, S.B.; Michaelian, M.; Gray, L.K. Recent declines of Populus tremuloides in North America linked to climate. For. Ecol. Manag. 2013, 299, 35–51. [Google Scholar] [CrossRef]
- Rong, M. Taxonomy and Phylogeny of Cytospora in Xinjiang, Northwest of China; Beijing Forestry University: Beijing, China, 2017. [Google Scholar]
- Fan, X.L.; Tian, C.M.; Yang, Q.; Liang, Y.M.; You, C.J.; Zhang, Y.B. Cytospora from Salix in northern China. Mycotaxon 2014, 129, 303–315. [Google Scholar] [CrossRef]
- Xinlei, F. Phylogeny and Taxonomy of Cytospora in Yellow River Region of China; Beijing Forestry University: Beijing, China, 2016. [Google Scholar]
- Shasha, X. The Occurrence, Pathogenic Structural Composition and Genetic Diversity of Walnut Canker Disease in Xinjiang; Tarim University: Alar, China, 2022. [Google Scholar]
- Zhe, W. Pathogen Population Composition of Fragrant Pear Valsa Canker in Fragrant Pear Trees and Transcriptome Analysis of Dominant Species Cytospora pyri; Tarim University: Alar, China, 2023. [Google Scholar]
- Sha, S. Population Structure Analysis of Apple Valsa Canker Fungus and Evaluation of the Biocontrol Potential of Bacillus velezensis SY01 Surrounding the Tarim Basin; Tarim University: Alar, China, 2023. [Google Scholar]
- Yinming, L. Study of Ascomycetous Species on Branches Disease from Desert Plants in Xinjiang; Xinjiang Agricultural University: Urumqi, China, 2017. [Google Scholar]
- Jia, W.; Ma, Q.; Su, J.; Li, Z.; Liu, Y.; Song, P. Pathogen Identification of Poplar Stem Rot in Hohhot of InnerMongolia. J. Northwest For. Univ. 2022, 37, 210–215+237. [Google Scholar]
- Wang, Y.; Xie, S.; Cao, J.; Zhao, H.; Yin, X.; Guo, Y.; Xu, C.; Guo, L.; Wu, H.; Zhang, M. Lasiodiplodia regiae sp. nov.: A new species causing canker and dieback of fruit trees in China. Phytopathology 2023, 113, 1210–1221. [Google Scholar] [CrossRef]
- Ayse, K.; Semra, K.; Gulsan, T.S.; Esin, S.; Takashi, S.; Gerard, A.; Gerard, V.; Richard, S. Fulminating fungal sinusitis caused by Valsa sordida, a plant pathogen, in a patient immunocompromised by acute myeloid leukemia. Med. Mycol. 2006, 44, 531–539. [Google Scholar] [CrossRef]
- Spielman, L.J. A monograph of Valsa on hardwoods in North America. Can. J. Bot. 1985, 63, 1355–1378. [Google Scholar] [CrossRef]
- Yawei, Z. The Species Distribution, Biological Characteristics and Pathogenicity of Cytospora on Populus and Salix in Xinjiang; Xinjiang Agricultural University: Urumqi, China, 2022. [Google Scholar]
- Xu, W.; Sun, H.; Jin, J.; Cheng, J. Predicting the potential distribution of apple canker pathogen (Valsa mali) in China under climate change. Forests 2020, 11, 1126. [Google Scholar] [CrossRef]
- Ke, X.; Huang, L.; Han, Q.; Gao, X.; Kang, Z. Histological and cytological investigations of the infection and colonization of apple bark by Valsa mali var. mali. Australas. Plant Pathol. 2013, 42, 85–93. [Google Scholar] [CrossRef]
- Kamiri, L.K. Epidemiology of Cytospora Canker Caused in Colorado Blue Spruce by Valsa kunzei. Phytopathology 1981, 71, 941. [Google Scholar] [CrossRef]
- Wang, S.; Hu, T.; Wang, Y.; Luo, Y.; Michailides, T.J.; Cao, K. New understanding on infection processes of Valsa canker of apple in China. Eur. J. Plant Pathol. 2016, 146, 531–540. [Google Scholar] [CrossRef]
- Fan, X.; Hyde, K.; Liu, M.; Tian, C. Cytospora species associated with walnut canker disease in China, with description of a new species C. gigalocus. Fungal Biol. 2015, 119, 310–319. [Google Scholar] [CrossRef]
- Kepley, J.B.; Reeves, F.B.; Jacobi, W.R.; Adams, G.C. Species associated with cytospora canker on Populus tremuloides. Mycotaxon 2015, 130, 783–805. [Google Scholar] [CrossRef]
- Bebber, D.P.; Ramotowski, M.A.; Gurr, S.J. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Change 2013, 3, 985–988. [Google Scholar] [CrossRef]
- McKay, A. Pathogens in a warming world. Nat. Ecol. Evol. 2023, 7, 2. [Google Scholar] [CrossRef]
- La Porta, N.; Capretti, P.; Thomsen, I.M.; Kasanen, R.; Hietala, A.M.; Von Weissenberg, K. Forest pathogens with higher damage potential due to climate change in Europe. Can. J. Plant Pathol. 2008, 30, 177–195. [Google Scholar] [CrossRef]
- Guo, Y.; Li, X.; Zhao, Z.; Wei, H.; Gao, B.; Gu, W. Prediction of the potential geographic distribution of the ectomycorrhizal mushroom Tricholoma matsutake under multiple climate change scenarios. Sci. Rep. 2017, 7, 46221. [Google Scholar] [CrossRef]
- Zhu, Y.; Yao, K.; Ma, M.; Cui, Y.; Xu, J.; Chen, W.; Yang, R.; Wu, C.; Gong, G. Occurrence Regionalization of Kiwifruit Brown Spot in Sichuan. J. Fungi 2023, 9, 899. [Google Scholar] [CrossRef]
- Andrew, C.; Diez, J.; James, T.Y.; Kauserud, H. Fungarium specimens: A largely untapped source in global change biology and beyond. Philos. Trans. R. Soc. B Biol. Sci. 2018, 374, 20170392. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Hirzel, A.H.; Hausser, J.; Chessel, D.; Perrin, N. cological-niche factor analysis: How to compute habitat-suitability maps without absence data? Ecology 2002, 83, 2027–2036. [Google Scholar] [CrossRef]
- Pattison, R.R.; Mack, R.N. Potential distribution of the invasive tree Triadica sebifera (Euphorbiaceae) in the United States: Evaluating CLIMEX predictions with field trials. Glob. Change Biol. 2008, 14, 813–826. [Google Scholar] [CrossRef]
- Bell, J.F. Tree-based methods. In Machine Learning Methods for Ecological Applications; Springer: Boston, MA, USA, 1999; pp. 89–105. [Google Scholar]
- Yee, T.W.; Mitchell, N.D. Generalized additive models in plant ecology. J. Veg. Sci. 1991, 2, 587–602. [Google Scholar] [CrossRef]
- Bai, R.; Wang, J.; Li, N. Climate change increases the suitable area and suitability degree of rubber tree powdery mildew in China. Ind. Crops Prod. 2022, 189, 115888. [Google Scholar] [CrossRef]
- Zhao, H.X.; Xian, X.Q.; Yang, N.W.; Zhang, Y.J.; Liu, H.; Wan, F.H.; Guo, J.Y.; Liu, W.X. Insights from the biogeographic approach for biocontrol of invasive alien pests: Estimating the ecological niche overlap of three egg parasitoids against Spodoptera frugiperda in China. Sci. Total Environ. 2023, 862, 13. [Google Scholar] [CrossRef]
- Ajene, I.J.; Khamis, F.; van Asch, B.; Pietersen, G.; Rasowo, B.A.; Ekesi, S.; Mohammed, S. Habitat suitability and distribution potential of Liberibacter species (“Candidatus Liberibacter asiaticus“ and “Candidatus Liberibacter africanus“) associated with citrus greening disease. Divers. Distrib. 2020, 26, 575–588. [Google Scholar] [CrossRef]
- Ding, W.C.; Li, H.Y.; Wen, J.B. Response of the invasive plant Ailanthus altissima (Mill.) Swingle and its two important natural enemies (Eucryptorrhynchus scrobiculatus (Motschulsky) and E. brandti (Harold)) to climate change. Ecol. Indic. 2022, 143, 14. [Google Scholar] [CrossRef]
- Cohen, S.D. Estimating the Climate Niche of Sclerotinia sclerotiorum Using Maximum Entropy Modeling. J. Fungi 2023, 9, 892. [Google Scholar] [CrossRef]
- Ejaz, M.R.; Jaoua, S.; Ahmadi, M.; Shabani, F. An examination of how climate change could affect the future spread of Fusarium spp. around the world, using correlative models to model the changes. Environ. Technol. Innov. 2023, 31, 15. [Google Scholar] [CrossRef]
- Warren, D.L.; Glor, R.E.; Turelli, M.J.E. ENMTools: A toolbox for comparative studies of environmental niche models. Ecography 2010, 33, 607–611. [Google Scholar] [CrossRef]
- Marchioro, C.A.; Krechemer, F.S. Prevention is better than cure: Integrating habitat suitability and invasion threat to assess global biological invasion risk by insect pests under climate change. Pest Manag. Sci. 2021, 77, 4510–4520. [Google Scholar] [CrossRef] [PubMed]
- Guisan, A.; Thuiller, W.; Zimmermann, N.E. Habitat Suitability and Distribution Models: With Applications in R; Cambridge University Press: Cambridge, UK, 2017. [Google Scholar]
- Lu, Y.; Wu, T.; Li, Y.; Yang, B. Mitigation of the double ITCZ syndrome in BCC-CSM2-MR through improving parameterizations of boundary-layer turbulence and shallow convection. Geosci. Model Dev. 2021, 14, 5183–5204. [Google Scholar] [CrossRef]
- Zhou, Y.R.; Lu, X.; Zhang, G.F. Potentially differential impacts on niche overlap between Chinese endangered Zelkova schneideriana and its associated tree species under climate change. Front. Ecol. Evol. 2023, 11. [Google Scholar] [CrossRef]
- Morales, N.S.; Fernández, I.C.; Baca-González, V. MaxEnt’s parameter configuration and small samples: Are we paying attention to recommendations? A systematic review. Peerj 2017, 5, e3093. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Dudík, M.; Schapire, R.E.; Blair, M.E. Opening the black box: An open-source release of Maxent. Ecography 2017, 40, 887–893. [Google Scholar] [CrossRef]
- Cobos, M.E.; Peterson, A.T.; Barve, N.; Osorio-Olvera, L.J.P. kuenm: An R package for detailed development of ecological niche models using Maxent. PeerJ 2019, 7, e6281. [Google Scholar] [CrossRef]
- Hosni, E.M.; Nasser, M.G.; Al-Ashaal, S.A.; Rady, M.H.; Kenawy, M.A.J.S.R. Modeling current and future global distribution of Chrysomya bezziana under changing climate. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Hu, X.; Wu, F.; Guo, W.; Liu, N.J.S.S.S. Identification of potential cultivation region for Santalum album in China by the MaxEnt ecologic niche model. Sci. Silvae Sin. 2014, 50, 27–33. [Google Scholar]
- Rezaei, S.; Mohammadi, A.; Shadloo, S.; Ranaie, M.; Wan, H.Y. Climate change induces habitat shifts and overlaps among carnivores in an arid and semi-arid ecosystem. Ecol. Inform. 2023, 77, 9. [Google Scholar] [CrossRef]
- Eskildsen, A.; le Roux, P.C.; Heikkinen, R.K.; Hoye, T.T.; Kissling, W.D.; Pöyry, J.; Wisz, M.S.; Luoto, M. Testing species distribution models across space and time: High latitude butterflies and recent warming. Glob. Ecol. Biogeogr. 2013, 22, 1293–1303. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, H.; Yue, J.; Xie, T.; Xu, Y.; Tian, Y.J.F. Predicting shifts in the suitable climatic distribution of walnut (Juglans regia L.) in China: Maximum entropy model paves the way to forest management. Forests. 2018, 9, 103. [Google Scholar] [CrossRef]
- Brown, J.L. SDM toolbox: A python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods Ecol. Evol. 2014, 5, 694–700. [Google Scholar] [CrossRef]
- Hyseni, C.; Garrick, R.C. Ecological Drivers of Species Distributions and Niche Overlap for Three Subterranean Termite Species in the Southern Appalachian Mountains, USA. Insects 2019, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Rödder, D.; Engler, J.O. Quantitative metrics of overlaps in Grinnellian niches: Advances and possible drawbacks. Glob. Ecol. Biogeogr. 2011, 20, 915–927. [Google Scholar] [CrossRef]
- Yan, C.; Hao, H.; Feng, H.; Wang, Z.; Sha, S.; Li, M.; Wang, L.; Kang, Z. Whole genome sequence of Cryptosphaeria pullmanensis, an important pathogenic fungus potentially threatening crop and forestry production. Genomics 2023, 115, 110576. [Google Scholar] [CrossRef] [PubMed]
- Moya, W.; Jacome, G.; Yoo, C. Past, current, and future trends of red spiny lobster based on PCA with MaxEnt model in Galapagos Islands, Ecuador. Ecol. Evol. 2017, 7, 4881–4890. [Google Scholar] [CrossRef]
- Ye, X.; Zhang, M.; Yang, Q.; Ye, L.; Liu, Y.; Zhang, G.; Chen, S.; Lai, W.; Wen, G.; Zheng, S. Prediction of suitable distribution of a critically endangered plant Glyptostrobus pensilis. Forests 2022, 13, 257. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species Distribution Models: Ecological Explanation and Prediction Across Space and Time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Zhang, J.; Nielsen, S.E.; Chen, Y.; Georges, D.; Qin, Y.; Wang, S.S.; Svenning, J.C.; Thuiller, W. Extinction risk of North American seed plants elevated by climate and land-use change. J. Appl. Ecol. 2017, 54, 303–312. [Google Scholar] [CrossRef]
- Yu, F.Y.; Wang, T.J.; Groen, T.A.; Skidmore, A.K.; Yang, X.F.; Ma, K.P.; Wu, Z.F. Climate and land use changes will degrade the distribution of Rhododendrons in China. Sci. Total Environ. 2019, 659, 515–528. [Google Scholar] [CrossRef]
- Chen, C.; Li, B.H.; Dong, X.L.; Wang, C.X.; Lian, S.; Liang, W.X. Effects of Temperature, Humidity, and Wound Age on Valsa mali Infection of Apple Shoot Pruning Wounds. Plant Dis. 2016, 100, 2394–2401. [Google Scholar] [CrossRef]
- Min, X.J.; Zang, Y.X.; Sun, W.; Ma, J.Y. Contrasting water sources and water-use efficiency in coexisting desert plants in two saline-sodic soils in northwest China. Plant Biol. 2019, 21, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.R. Biological Characteristics of the Pathogens from Different Host Plants Caused Canker and Screening Fungicide against Pear Valsa; Huazhong Agriculture University: Wuhan, China, 2015. [Google Scholar]
- Zhao, Q.; Li, R.; Gao, Y.Y.; Yao, Q.; Guo, X.Q.; Wang, W.G. Modeling impacts of climate change on the geographic distribution of medicinal plant Fritillaria cirrhosa D. Don. Plant Biosyst. 2018, 152, 349–355. [Google Scholar] [CrossRef]
- Ljubicic, I.; Varga, F.; Bogdanovic, S.; Sklepic, L.; Britvec, M.; Temunovic, M. Comparative assessment of habitat suitability and niche overlap of three medicinal and melliferous Satureja L. species (Lamiaceae) from the eastern Adriatic region: Exploring potential for cultivation. Ecol. Inform. 2023, 76, 13. [Google Scholar] [CrossRef]
- Wang, W.T.; Gao, S.Y.; Wang, S.F. Predictive studies of potential invasive areas for four poisonous weeds in Gansu grassland. Acta Ecol. Sin. 2019, 39, 5301–5307. [Google Scholar]
- Silvertown, J.W. The distribution of plants in limestone pavement-tests of species interaction and niche separation against null hypotheses. J. Ecol. 1983, 71, 819–828. [Google Scholar] [CrossRef]
- Abolmaali, S.M.R.; Tarkesh, M.; Bashari, H. MaxEnt modeling for predicting suitable habitats and identifying the effects of climate change on a threatened species, Daphne mucronata, in central Iran. Ecol. Inform. 2018, 43, 116–123. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).