The Diverse Mycorrizal Morphology of Rhododendron dauricum, the Fungal Communities Structure and Dynamics from the Mycorrhizosphere
Abstract
1. Introduction
2. Materials and Methods
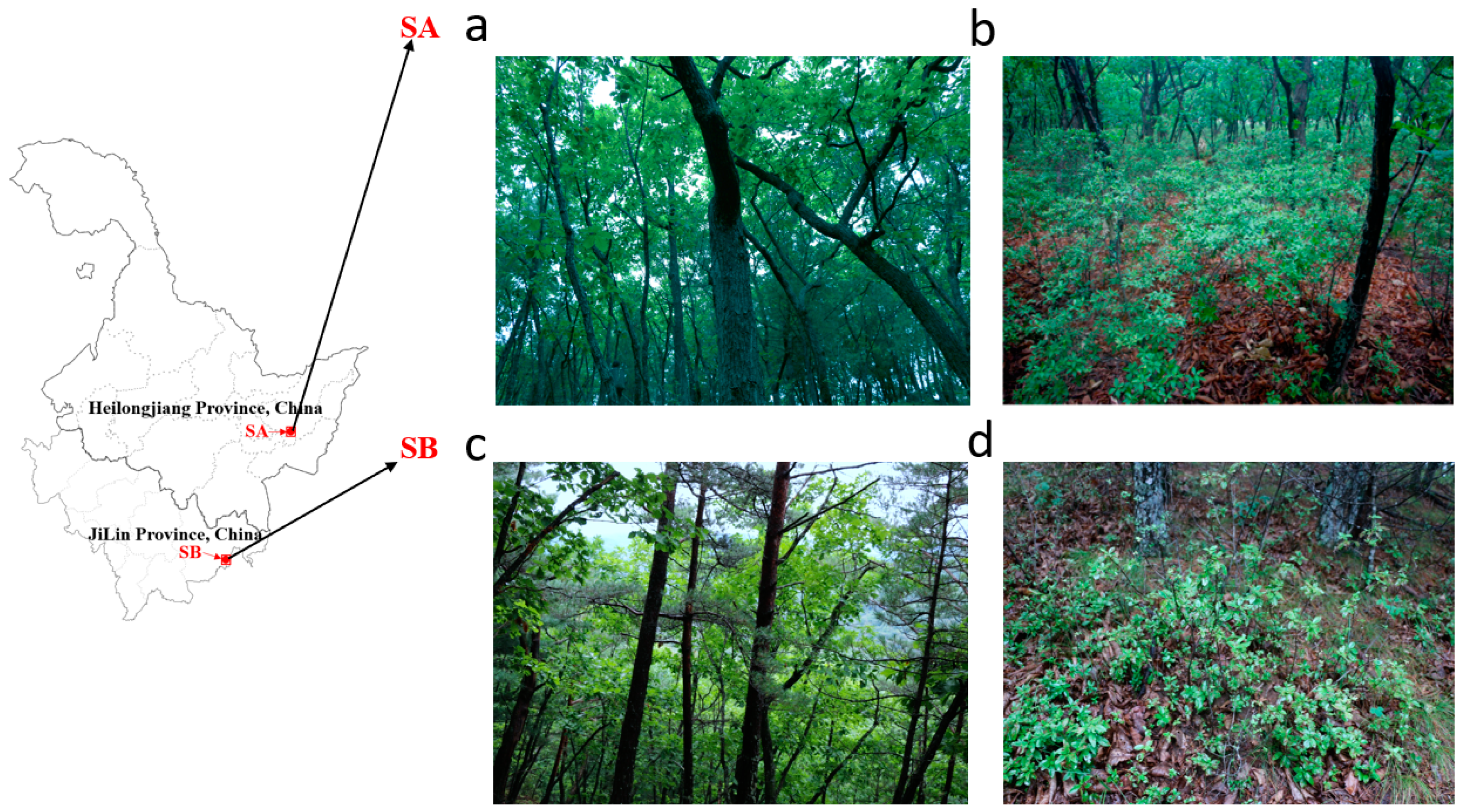
2.1. Sampling Locations
2.2. Collection and Preparation of Mycorrhiza Root Tips’ and Soil Samples
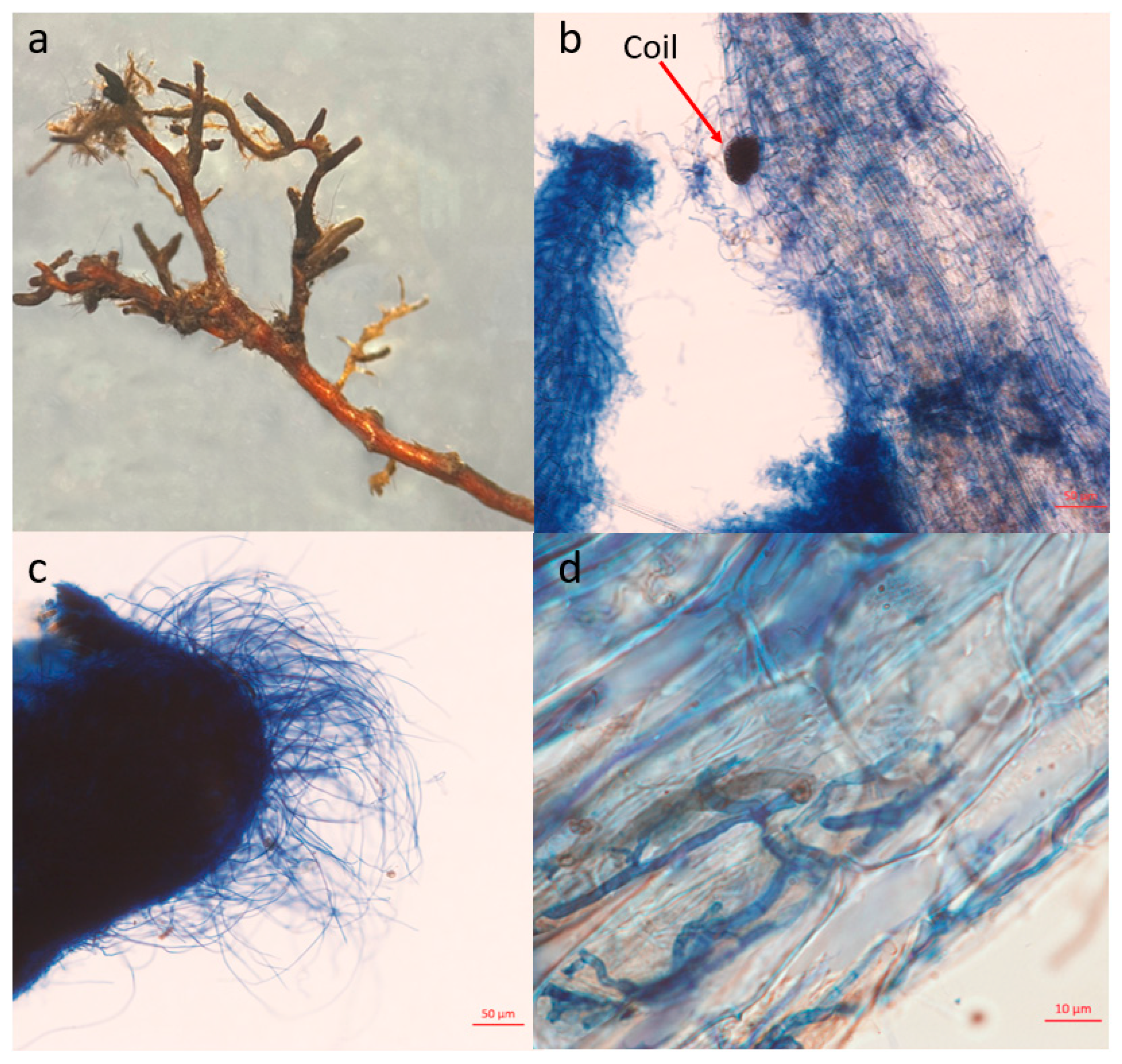
2.3. Collection and Processing of R. dauricum Mycorrhizal Root Tips Samples
2.4. DNA Extraction and Illumina NovaSeq Platform
2.5. Soil Physical and Chemical Properties Analysis
2.6. Climate Elements Collection
2.7. Bioinformatics
2.8. Data Processing
2.9. Fungal Community
3. Results
3.1. Mycorrhizal Ecological Niches of R. dauricum Habitats
3.1.1. R. dauricum Mycorrhizal Morphology
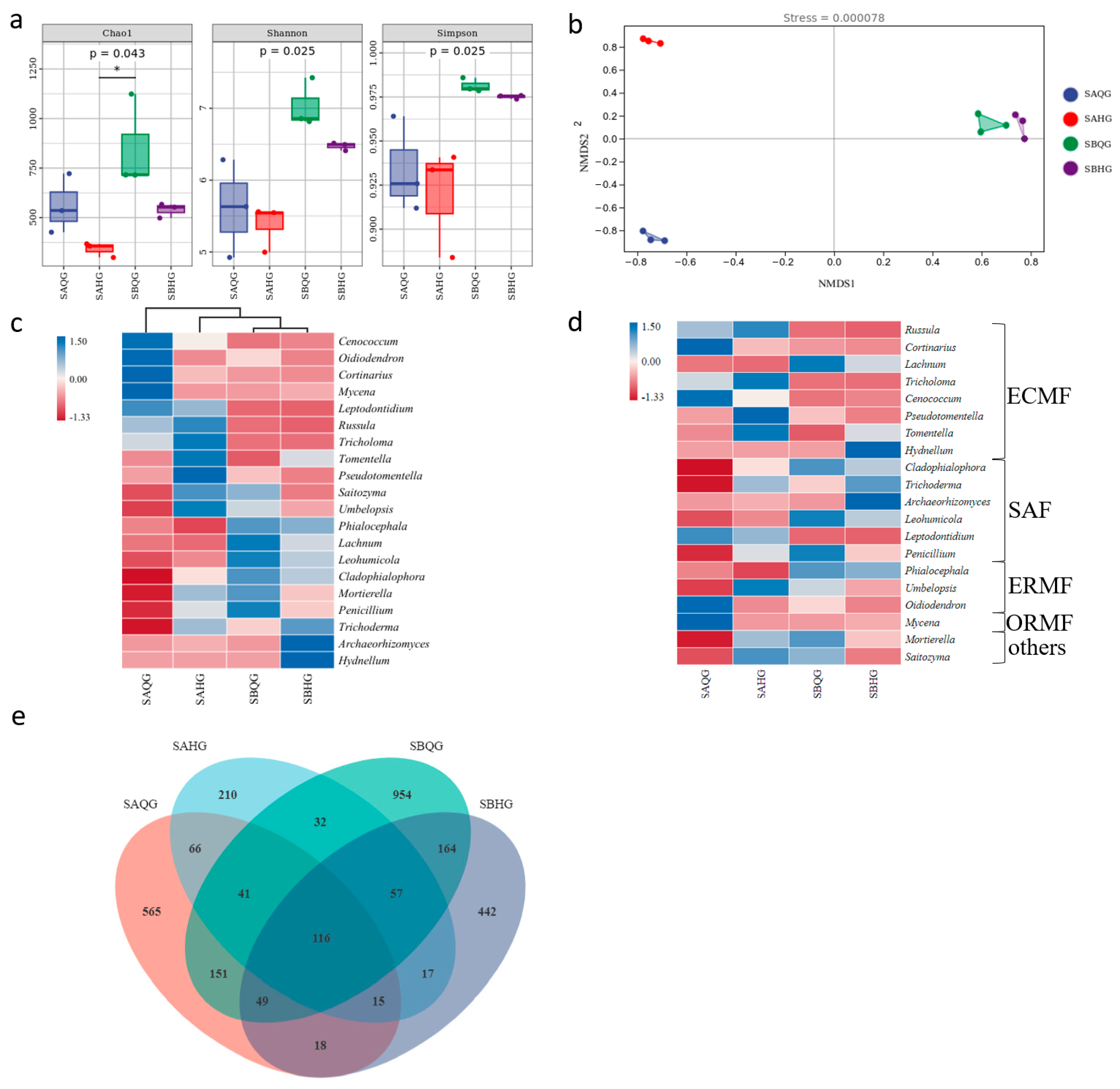
3.1.2. Diversity of Fungi and Mycorrhizal Fungi in Ecological Niches of R. dauricum Habitats
3.2. The Impact of Habitat Soil on the Mycorrhizal Fungal Community in the Habitat of R. dauricum
3.2.1. The Relationship between Abiotic Factors and Mycorrhizal Fungi in Mycorrhizal Samples
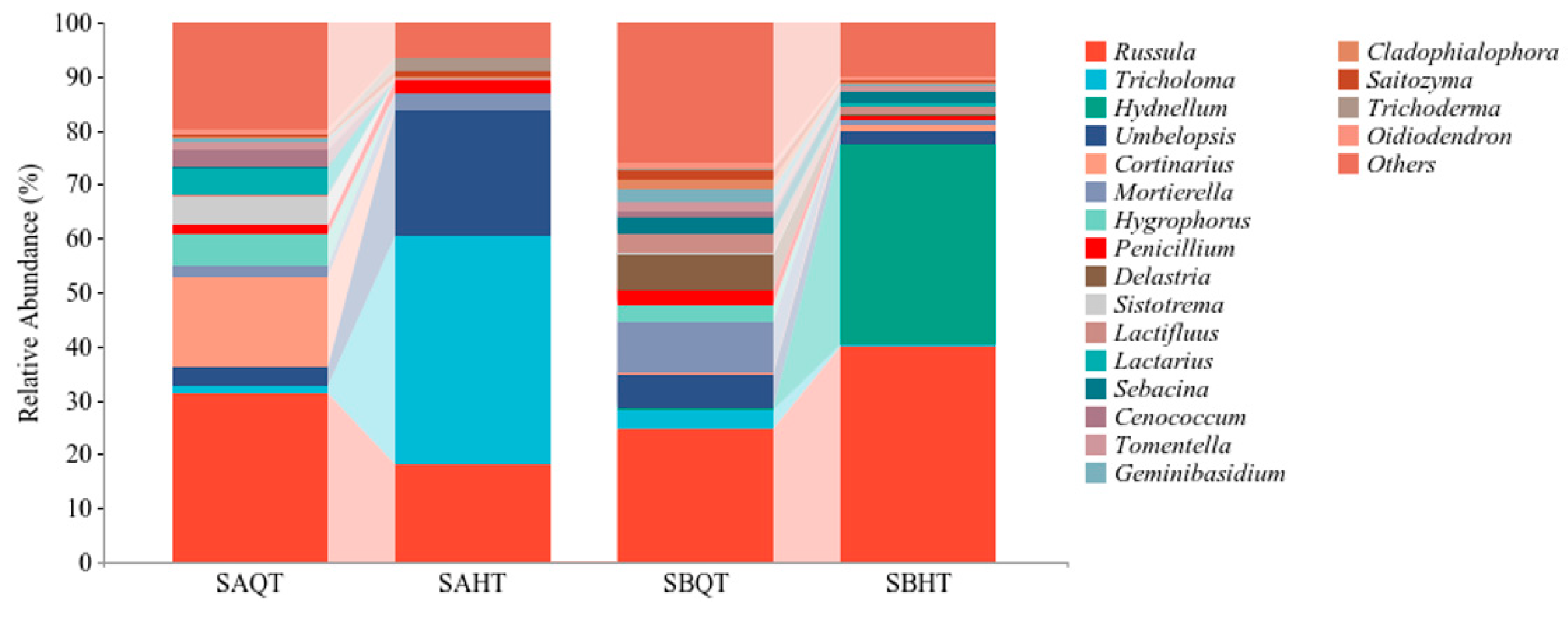
3.2.2. Fungal Species Composition in the Soil of R. dauricum Habitat
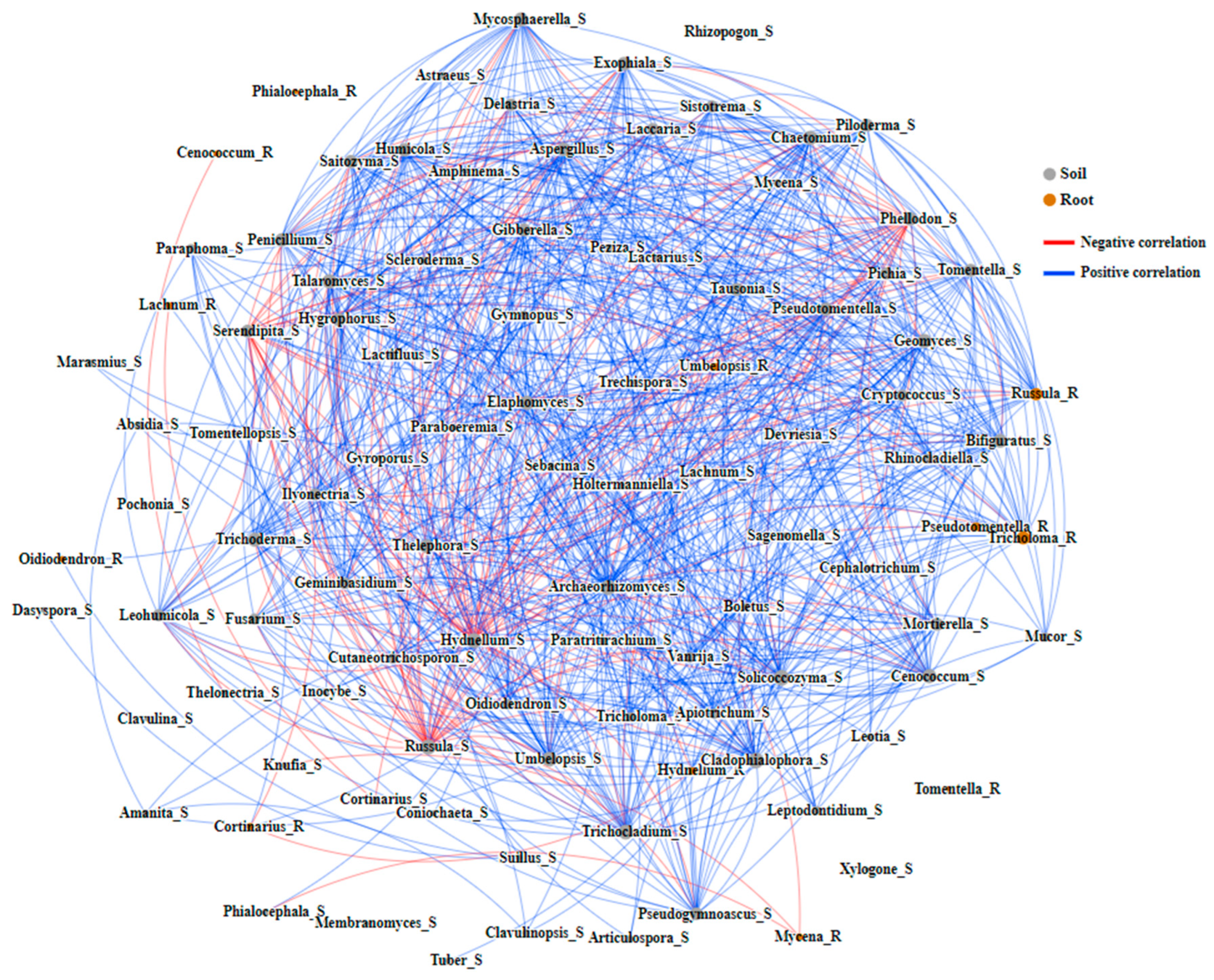
3.2.3. The Relationship between Fungi Community in the Soil and Mycorrhizal Fungi in Mycorrhizal Ecological Niches of the Habitat of R. dauricum
3.3. The Relationship between Climate Factors and Mycorrhizal Fungi in Mycorrhizal Ecological Niches of the Habitat of R. dauricum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liang, Q.; Hu, J.X.; Zhang, X.M.; Xu, W.H. Traditional uses, phytochemistry, pharmacology, toxicology, and quality control of Rhododendron dauricum L. leaves: A comprehensive review. J. Ethnopharmacol. 2023, 305, 116085. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhang, H.; Gao, B.; Zheng, G.; Zha, S.; Yao, G. Highly oxygenated grayanane diterpenoids with structural diversity from the flowers of Rhododendron dauricum and their analgesic activities. Bioorg. Chem. 2023, 132, 106374. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Zhang, X.; Miao, J.; Yuan, H.; Liu, E.; Liang, T.; Li, Q. Farrerol Directly Targets GSK-3 β to Activate Nrf2-ARE Pathway and Protect EA. hy926 Cells against Oxidative Stress-Induced Injuries. Oxidative Med. Cell. Longev. 2020, 2020, 5967434. [Google Scholar] [CrossRef] [PubMed]
- Taura, F.; Iijima, M.; Yamanaka, E.; Takahashi, H.; Kenmoku, H.; Saeki, H.; Morimoto, S.; Asakawa, Y.; Kurosaki, F.; Morita, H. A Novel Class of Plant Type III Polyketide Synthase Involved in Orsellinic Acid Biosynthesis from Rhododendron dauricum. Front. Plant Sci. 2016, 7, 1452. [Google Scholar] [CrossRef] [PubMed]
- Iwata, N.; Wang, N.; Yao, X.; Kitanaka, S. Structures and histamine release inhibitory effects of prenylated orcinol derivatives from Rhododendron dauricum. J. Nat. Prod. 2004, 67, 1106–1109. [Google Scholar] [CrossRef] [PubMed]
- Plassard, C.; Becquer, A.; Garcia, K. Phosphorus Transport in Mycorrhiza: How Far Are We? Trends Plant Sci. 2019, 24, 794–801. [Google Scholar] [CrossRef]
- Wang, Y.H.; Dai, Y.; Kong, W.L.; Zhu, M.L.; Wu, X.Q. Improvement of Sphaeropsis Shoot Blight Disease Resistance by Applying the Ectomycorrhizal Fungus Hymenochaete sp. Rl and Mycorrhizal Helper Bacterium Bacillus pumilus HR10 to Pinus thunbergii. Phytopathology 2022, 112, 1226–1234. [Google Scholar] [CrossRef]
- Yang, Z.; Dong, H.; Zhang, S.; Jiang, J.; Zhu, H.; Yang, H.; Li, L. Isolation and identification of mycorrhizal helper bacteria of Vaccinium uliginosum and their interaction with mycorrhizal fungi. Front. Microbiol. 2023, 14, 1180319. [Google Scholar] [CrossRef]
- Jacott, C.N.; Murray, J.D.; Ridout, C.J. Trade-Offs in Arbuscular Mycorrhizal Symbiosis: Disease Resistance, Growth Responses and Perspectives for Crop Breeding. Agronomy 2017, 7, 75. [Google Scholar] [CrossRef]
- Cairney, J.W.G.; Meharg, A.A. Ericoid mycorrhiza: A partnership that exploits harsh edaphic conditions. Eur. J. Soil Sci. 2010, 54, 735–740. [Google Scholar] [CrossRef]
- Chen, R.J.; Ou, J.; Wang, L.J.; Long, H.Y.; Xiong, X.R. Effects of inoculated different ericoid mycorrhizas strains on physiological characteristics of rhododendron annae under water stress. J. Cent. South Univ. For. Technol. 2017, 37, 43–48. [Google Scholar]
- Kerley, S.J.; Read, D.J. The biology of mycorrhiza in the Ericaceae: XIX. Fungal mycelium as a nitrogen source for the ericoid mycorrhizal fungus Hymenoscyphus ericae and its host plants. New Phytol. 1997, 136, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Perotto, S.; Girlanda, M.; Martino, E. Ericoid Mycorrhizal Fungi: Some New Perspectives on Old Acquaintances; Springer: Dordrecht, The Netherlands, 2002. [Google Scholar]
- Koron, D.; Gogala, N. The use of mycorrhizal fungi in the growing of blueberry plants Vaccinium corymbosum L. In Proceedings of the SHS Acta Horticulturae 525 International Conference on Integrated Fruit Production, Leuven, Belgium, 27 July–1 August 1998. [Google Scholar]
- Strandberg, M.; Johansson, M. Uptake of nutrients in Calluna vulgaris seed plants grown with and without mycorrhiza. For. Ecol. Manag. 1999, 114, 129–135. [Google Scholar] [CrossRef]
- Shi, J.; Wang, X.; Wang, E. Mycorrhizal Symbiosis in Plant Growth and Stress Adaptation: From Genes to Ecosystems. Annu. Rev. Plant Biol. 2023, 74, 569–607. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shi, J.; Xie, Q.; Jiang, Y.; Yu, N.; Wang, E. Nutrient Exchange and Regulation in Arbuscular Mycorrhizal Symbiosis. Molecular plant. 2017, 10, 1147–1158. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, B.; Zheng, S.; Zhang, X.; Wang, X.; Dong, W.; Xie, Q.; Wang, G.; Xiao, Y.; Chen, F.; et al. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 2021, 184, 5527–5540.e18. [Google Scholar] [CrossRef] [PubMed]
- Bledsoe, R.B.; Goodwillie, C.; Peralta, A.L. Long-Term Nutrient Enrichment of an Oligotroph-Dominated Wetland Increases Bacterial Diversity in Bulk Soils and Plant Rhizospheres. mSphere 2020, 5, 10–1128. [Google Scholar] [CrossRef]
- Xiao, C.; Yang, L.; Zhang, L.; Liu, C.; Han, M. Effects of cultivation ages and modes on microbial diversity in the rhizosphere soil of Panax ginseng. J. Ginseng Res. 2016, 40, 28–37. [Google Scholar] [CrossRef]
- Sun, W.; Xiao, E.; Krumins, V.; Häggblom, M.M.; Dong, Y.; Pu, Z.; Li, B.; Wang, Q.; Xiao, T.; Li, F. Rhizosphere Microbial Response to Multiple Metal(loid)s in Different Contaminated Arable Soils Indicates Crop-Specific Metal-Microbe Interactions. Appl. Environ. Microbiol. 2018, 84, e00701-18. [Google Scholar] [CrossRef]
- Wang, Q.Z.; Zhou, L.M.; Han, J.; Cong, B.C.; Yang, H.; Wang, S.M. Investigating the Responses of Microbial Communities to Banana Fusarium Wilt in Suppressive and Conducive Soils Based on Soil Particle-Size Differentiation. Agronomy 2022, 12, 229. [Google Scholar] [CrossRef]
- Yang, Y.; Qiu, K.; Xie, Y.; Li, X.; Zhang, S.; Liu, W.; Huang, Y.; Cui, L.; Wang, S.; Bao, P. Geographical, climatic, and soil factors control the altitudinal pattern of rhizosphere microbial diversity and its driving effect on root zone soil multifunctionality in mountain ecosystems. Sci. Total Environ. 2023, 904, 166932. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Jiang, H.J.; Pan, Y.T.; Lu, F.; Zhu, Q.; Ma, C.Y.; Zhang, A.Y.; Zhou, J.Y.; Zhang, W.; Dai, C.C. Hyphosphere microorganisms facilitate hyphal spreading and root colonization of plant symbiotic fungus in ammonium-enriched soil. ISME J. 2023, 17, 1626–1638. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Xiong, K.; Chi, Y.; He, C.; Fang, J.; He, S. Effect of Cultivated Pastures on Soil Bacterial Communities in the Karst Rocky Desertification Area. Front. Microbiol. 2022, 13, 922989. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Kuehl, J.V.; Chandran, A.; Arkin, A.P. A Simple, Cost-Effective, and Automation-Friendly Direct PCR Approach for Bacterial Community Analysis. mSystems 2021, 6, e0022421. [Google Scholar] [CrossRef] [PubMed]
- McGivern, B.B.; Tfaily, M.M.; Borton, M.A.; Kosina, S.M.; Daly, R.A.; Nicora, C.D.; Purvine, S.O.; Wong, A.R.; Lipton, M.S.; Hoyt, D.W.; et al. Decrypting bacterial polyphenol metabolism in an anoxic wetland soil. Nat. Commun. 2021, 12, 2466. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.; Morris, K.; Vogt, D.J.P.; Gleber, S.C.; Bigalke, M.; Wilcke, W.; Rillig, M.C. Visualizing the dynamics of soil aggregation as affected by arbuscular mycorrhizal fungi. ISME J. 2019, 13, 1639–1646. [Google Scholar] [CrossRef] [PubMed]
- Xing, P.J.; Xu, Y.; Gao, T.T.; Li, G.L.; Zhou, J.J.; Xie, M.L.; Ji, R.Q. The community composition variation of Russulaceae associated with the Quercus mongolica forest during the growing season at Wudalianchi City, China. PeerJ 2020, 8, e8527. [Google Scholar] [CrossRef]
- Phillips, J.M. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 1970, 55, 158-IN18. [Google Scholar] [CrossRef]
- Genre, A.; Lanfranco, L.; Perotto, S.; Bonfante, P. Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 2020, 18, 649–660. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, F.J.R.M.; White, T.; Lee, S.H.; Taylor, L.; Shawetaylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Truog, E. The determination of the readily available phosphorus of soils. Agron. J. 1930, 22, 874–882. [Google Scholar] [CrossRef]
- Soon, Y.K.; Abboud, S. A comparison of some methods for soil organic carbon determination. Communications in Soil. Sci. Plant Anal. 1991, 22, 943–954. [Google Scholar] [CrossRef]
- Wang, W.J.; Li, R.G. Evaluation of soil available nitrogen measurement methods in Hebei Plain. Chin. J. Soil Sci. 1991, 6, 263–266. [Google Scholar]
- Bao, S.D. Soil Agrochemical Analysis, 3rd ed.; China Agricultural Press: Beijing, China, 2000; pp. 1–495. [Google Scholar]
- Ministry of Environmental Protection; Nanjing Institute of Environmental Science; Jiangsu Environmental Monitoring Center. Determination of Soil pH Value Using Potentiometric Method: HJ 962—2018; China Environmental Science Press: Beijing, China, 2018.
- Zhang, S.H.; Huang, M. The Feature Extraction and Data Fusion of RegionalSoil Textures Based on GlS Techniques. Clim. Environ. Res. 2004, 1, 65–79. [Google Scholar]
- China Meteorological Data Network. Available online: http://data.cma.cn/ (accessed on 7 June 2023).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. Available online: https://academic.oup.com/mbe/article/26/7/1641/1128976 (accessed on 7 February 2022). [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory, C.J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. Available online: https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-018-0470-z (accessed on 8 February 2022). [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson, P.J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, W.; Wei, J. Ectomycorrhizal fungal community in the rhizospheric soil of Betula platyphylla in Inner Mongolia. Mycosystema 2018, 37, 294–304. [Google Scholar]
- Liu, R.J.; Chen, Y.L. Mycorrhizalology; Science Press: Beijing, China, 2007. [Google Scholar]
- Yang, X.L.; Yan, W. Isolation and identification for mycorrhizal fungi of Rhododendron dauricum. J. North. Agric. 2016, 44, 54–58. [Google Scholar]
- Cheng, Z.; Wu, S.; Du, J.; Liu, Y.; Sui, X.; Yang, L. Reduced Arbuscular Mycorrhizal Fungi (AMF) Diversity in Light and Moderate Fire Sites in Taiga Forests, Northeast China. Microorganisms 2023, 11, 1836. [Google Scholar] [CrossRef] [PubMed]
- Peterson, T.A.; Mueller, W.C.; Englander, L. Anatomy and ultrastructure of a Rhododendron root–fungus association. Can. J. Bot. 1980, 58, 2421–2433. [Google Scholar] [CrossRef]
- Berch, S.M.; Allen, T.R.; Berbee, M.L. Molecular detection, community structure and phylogeny of ericoid mycorrhizal fungi. Plant Soil 2002, 244, 55–66. [Google Scholar] [CrossRef]
- Allen, T.R.; Miller, T.; Berch, S.M. Culturing and direct DNA extraction find different fungi from the same ericoid mycorrhizal roots. New Phytol. 2003, 160, 255–272. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, Y.; Guo, X.; Ning, D.; Zhou, X.; Feng, J.; Yuan, M.M.; Liu, S.; Guo, J.; Gao, Z.; et al. Reduction of microbial diversity in grassland soil is driven by long-term climate warming. Nat. Microbiol. 2022, 7, 1054–1062. [Google Scholar] [CrossRef]
- Hu, J.; Miller, G.; Shi, W. Abundance, diversity, and composition of root-associated microbial communities varied with tall fescue cultivars under water deficit. Front. Microbiol. 2022, 13, 1078836. [Google Scholar] [CrossRef]
- Shu, X.; He, J.; Zhou, Z.; Xia, L.; Hu, Y.; Zhang, Y.; Zhang, Y.; Luo, Y.; Chu, H.; Liu, W.; et al. Organic amendments enhance soil microbial diversity, microbial functionality and crop yields: A meta-analysis. Sci. Total Environ. 2022, 829, 154627. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, W.; Ma, L.; Ye, X.; Erdenebileg, E.; Wang, R.; Huang, Z.; Indree, T.; Liu, G. Biotic and abiotic drivers of ecosystem multifunctionality: Evidence from the semi-arid grasslands of northern China. Sci. Total Environ. 2023, 887, 164158. [Google Scholar] [CrossRef]
- Wong, S.K.; Cui, Y.; Chun, S.J.; Kaneko, R.; Masumoto, S.; Kitagawa, R.; Mori, A.S.; Lim, A.S.; Uchida, M. Vegetation as a key driver of the distribution of microbial generalists that in turn shapes the overall microbial community structure in the low Arctic tundra. Environ. Microbiome 2023, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Liu, Z.; Cui, H.; Chen, J.; Chen, S.; Gao, H.; Yang, X.; Wang, Y.; Wang, J.; Liu, K.; et al. Contrasting influences of two dominant plants, Dasiphora fruticosa and Ligularia virguarea, on aboveground and belowground communities in an alpine meadow. Front. Microbiol. 2023, 14, 1118789. [Google Scholar] [CrossRef] [PubMed]
- Paries, M.; Gutjahr, C. The good, the bad, and the phosphate: Regulation of beneficial and detrimental plant-microbe interactions by the plant phosphate status. New Phytol. 2023, 239, 29–46. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Yan, Z.; Hao, Y.; Kang, E.; Zhang, X.; Li, M.; Zhang, K.; Yan, L.; Yang, A.; et al. The divergent vertical pattern and assembly of soil bacterial and fungal communities in response to short-term warming in an alpine peatland. Front. Plant Sci. 2022, 13, 986034. [Google Scholar] [CrossRef]
- Soto, M.J.; Dillewijn, P.; Martínez-Abarca, F.; Jiménez-Zurdo, J.I.; Toro, N. Attachment to plant roots and nod gene expression are not affected by pH or calcium in the acid-tolerant alfalfa-nodulating bacteria Rhizobium sp. LPU83. FEMS Microbiol. Ecol. 2004, 48, 71–77. [Google Scholar] [CrossRef]
- Ji, R.Q.; Xu, Y.; Si, Y.J.; Phukhamsakda, C.; Li, Y.; Meng, L.P.; Liu, S.Y.; Xie, M.L. Fungal-Bacterial Networks in the Habitat of SongRong ( Tricholoma matsutake ) and Driving Factors of Their Distribution Rules. J. Fungi 2022, 8, 575. [Google Scholar] [CrossRef]
- Zhang, X.; Song, X.; Wang, T.T.; Huang, L.; Ma, H.; Wang, M.; Tan, D.Y. The responses to long-term nitrogen addition of soil bacterial, fungal, and archaeal communities in a desert ecosystem. Front. Microbiol. 2022, 13, 66–88. [Google Scholar] [CrossRef]
- Carrell, A.A.; Hicks, B.B.; Sidelinger, E.; Johnston, E.R.; Jawdy, S.S.; Clark, M.M.; Klingeman, D.M.; Cregger, M.A. Nitrogen addition alters soil fungal communities, but root fungal communities are resistant to change. Front. Microbiol. 2023, 13, 54–67. [Google Scholar] [CrossRef]
- Lu, Q.W.; Rebecca, A.; Bunn, E.; Whitney, D.; Feng, Y.Y.; DeVetter, L.W.; Tao, H.Y. Arbuscular mycorrhizae influence raspberry growth and soil fertility under conventional and organic fertilization. Front. Microbiol. 2023, 14, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Lu, T.; Qian, H. Pesticide interference and additional effects on plant microbiomes. Sci. Total Environ. 2023, 888, 164149. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Peng, Y.; Li, Z.; Guo, H.; Xia, X.; Li, B.; Yin, W. The Regulation of Nitrate Reductases in Response to Abiotic Stress in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 1202. [Google Scholar] [CrossRef]
- Araki, R.; Kousaka, K.; Namba, K.; Murata, Y.; Murata, J. 2′-Deoxymugineic acid promotes growth of rice (Oryza sativa L.) by orchestrating iron and nitrate uptake processes under high pH conditions. Plant J. Cell Mol. Biol. 2015, 81, 233–246. [Google Scholar] [CrossRef]
- Elhamouly, N.A.; Hewedy, O.A.; Zaitoon, A.; Miraples, A.; Elshorbagy, O.T.; Hussien, S.; El-Tahan, A.; Peng, D. The hidden power of secondary metabolites in plant-fungi interactions and sustainable phytoremediation. Front. Plant Sci. 2022, 13, 1044896. [Google Scholar] [CrossRef] [PubMed]
- Kai, K. Bioorganic chemistry of signaling molecules in microbial communication. J. Pestic. Sci. 2019, 44, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Lopez, E.; Alcazar, P.; Cid, C. Identification of Biomolecules Involved in the Adaptation to the Environment of Cold-Loving Microorganisms and Metabolic Pathways for Their Production. Biomolecules 2021, 11, 1155. [Google Scholar] [CrossRef]
- Chagas, F.O.; Pessotti, R.C.; Caraballo-Rodríguez, A.M.; Pupo, M.T. Chemical signaling involved in plant-microbe interactions. Chem. Soc. Rev. 2018, 47, 1652–1704. [Google Scholar] [CrossRef]
- Yu, J.; Tu, X.; Huang, A.C. Functions and biosynthesis of plant signaling metabolites mediating plant-microbe interactions. Nat. Prod. Rep. 2022, 39, 1393–1422. [Google Scholar] [CrossRef]
- Chang, X.; Kingsley, K.L.; White, J.F. Chemical Interactions at the Interface of Plant Root Hair Cells and Intracellular Bacteria. Microorganisms 2021, 9, 1041. [Google Scholar] [CrossRef]
- Dreher, D.; Baldermann, S.; Schreiner, M.; Hause, B. An arbuscular mycorrhizal fungus and a root pathogen induce different volatiles emitted by Medicago truncatula roots. J. Adv. Res. 2019, 19, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Guasconi, D.; Juhanson, J.; Clemmensen, K.E.; Cousins, S.A.O.; Hugelius, G.; Manzoni, S.; Roth, N.; Fransson, P. Vegetation, topography, and soil depth drive microbial community structure in two Swedish grasslands. FEMS Microbiol. Ecol. 2023, 99, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ma, L.; Zhu, M.; Liu, B.; Liu, X.; Shi, Y. Rhizosphere microbial community assembly and association networks strongly differ based on vegetation type at a local environment scale. Front. Microbiol. 2023, 14, 1129471. [Google Scholar] [CrossRef]
- Carey, C.J.; Beman, J.M.; Eviner, V.T.; Malmstrom, C.M.; Hart, S.C. Soil microbial community structure is unaltered by plant invasion, vegetation clipping, and nitrogen fertilization in experimental semi-arid grasslands. Front. Microbiol. 2015, 6, 466. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Chen, C.; Pang, Z.; Zhang, G.; Wu, J.; Kan, H. Short-Term Vegetation Restoration Enhances the Complexity of Soil Fungal Network and Decreased the Complexity of Bacterial Network. J. Fungi 2022, 8, 1122. [Google Scholar] [CrossRef]
- Hu, L.; Li, Q.; Yan, J.; Liu, C.; Zhong, J. Vegetation restoration facilitates belowground microbial network complexity and recalcitrant soil organic carbon storage in southwest China karst region. Sci. Total Environ. 2022, 820, 153137. [Google Scholar] [CrossRef] [PubMed]
- Kravchenko, I.; Rayko, M.; Tikhonova, E.; Konopkin, A.; Abakumov, E.; Lapidus, A. Agricultural Crops Grown in Laboratory Conditions on Chernevaya Taiga Soil Demonstrate Unique Composition of the Rhizosphere Microbiota. Microorganisms 2022, 10, 2171. [Google Scholar] [CrossRef] [PubMed]
- Bourceret, A.; Guan, R.; Dorau, K.; Mansfeldt, T.; Omidbakhshfard, A.; Medeiros, D.B.; Fernie, A.R.; Hofmann, J.; Sonnewald, U.; Mayer, J.; et al. Maize Field Study Reveals Covaried Microbiota and Metabolic Changes in Roots over Plant Growth. mBio 2022, 13, e0258421. [Google Scholar] [CrossRef]
- Abdelkhalek, A.; Al-Askar, A.A.; Arishi, A.A.; Behiry, S.I. Trichoderma hamatum Strain Th23 Promotes Tomato Growth and Induces Systemic Resistance against Tobacco Mosaic Virus. J. Fungi 2022, 8, 228. [Google Scholar] [CrossRef]
- Wang, K.; Wen, Z.; Asiegbu, F.O. The dark septate endophyte Phialocephala sphaeroides suppresses conifer pathogen transcripts and promotes root growth of Norway spruce. Tree Physiol. 2022, 42, 2627–2639. [Google Scholar] [CrossRef]
- Butler, S.; O’Dwyer, J.P. Stability criteria for complex microbial communities. Nat. Commun. 2018, 9, 2970. [Google Scholar] [CrossRef] [PubMed]
- Angulo, M.T.; Moog, C.H.; Liu, Y.Y. A theoretical framework for controlling complex microbial communities. Nat. Commun. 2019, 10, 1045. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Mahlich, Y.; Miller, M.; Bromberg, Y. fusionDB: Assessing microbial diversity and environmental preferences via functional similarity networks. Nucleic Acids Res. 2018, 46, D535–D541. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, A.; Dennis, P.G.; Forstner, C.; Raghavendra, A.K.H.; Mathesius, U.; Richardson, A.E.; Delhaize, E.; Gilliham, M.; Watt, M.; Ryan, P.R. Manipulating exudate composition from root apices shapes the microbiome throughout the root system. Plant Physiol. 2021, 187, 2279–2295. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Richardson, A.E.; O’Callaghan, M.; DeAngelis, K.M.; Jones, E.E.; Stewart, A.; Firestone, M.K.; Condron, L.M. Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol. Ecol. 2011, 77, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Heuermann, D.; Döll, S.; Schweneker, D.; Feuerstein, U.; Gentsch, N.; von Wirén, N. Distinct metabolite classes in root exudates are indicative for field- or hydroponically-grown cover crops. Front. Plant Sci. 2023, 14, 1122285. [Google Scholar] [CrossRef]
- Xu, Q.; Fu, H.; Zhu, B.; Hussain, H.A.; Zhang, K.; Tian, X.; Duan, M.; Xie, X.; Wang, L. Potassium Improves Drought Stress Tolerance in Plants by Affecting Root Morphology, Root Exudates and Microbial Diversity. Metabolites 2021, 11, 131. [Google Scholar] [CrossRef]
- Song, C.G.; Sun, Y.F.; Wu, D.M.; Gao, N.; Liu, S.; Xu, T.M.; Cui, B.K. Morphology and molecular phylogeny reveal five new species of Hydnellum (Bankeraceae, Thelephorales) from China. Front. Microbiol. 2022, 13, 1049007. [Google Scholar] [CrossRef]
- Wang, S.M.; Han, J.J.; Ma, K.; Jin, T.; Bao, L.; Pei, Y.F.; Liu, H.W. New α-glucosidase inhibitors with p-terphenyl skeleton from the mushroom Hydnellum concrescens. Fitoterapia 2014, 98, 149–155. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; Muñoz-Ucros, J.; Weikl, F.; Pritsch, K.; Goebel, M.; Buckley, D.H.; Bauerle, T.L. The effects of mixed-species root zones on the resistance of soil bacteria and fungi to long-term experimental and natural reductions in soil moisture. Sci. Total Environ. 2023, 873, 162266. [Google Scholar] [CrossRef]
- Li, M.; Wang, H.; Zhao, X.; Feng, W.; Ding, G.; Quan, W. Effect of Ectomycorrhizal Fungi on the Drought Resistance of Pinus massoniana Seedlings. J. Fungi 2023, 9, 471. [Google Scholar] [CrossRef] [PubMed]


| pH | SOM | AK | TN | AP | |
|---|---|---|---|---|---|
| (mg·kg−1) | (mg·kg−1) | (mg·kg−1) | (mg·kg−1) | ||
| F-value | 12.226 | 18.994 | 26.324 | 17.309 | 10.88 |
| p-value | 0.002 | 0.001 | 0 | 0.001 | 0.003 |
| SAQT | 5.14 ± 0.08 b | 79,283.25 ± 2061.76 a | 26.30 ± 1.11 a | 122.45 ± 1.92 a | 7.29 ± 1.78 a |
| SBQT | 5.36 ± 0.03 a | 57,661.96 ± 3500.07 bc | 24.20 ± 1.19 a | 100.83 ± 0.93 b | 5.86 ± 0.13 a |
| SAHT | 4.86 ± 0.07 c | 56,806.44 ± 664.50 c | 25.37 ± 0.49 a | 106.19 ± 5.54 b | 2.05 ± 0.11 b |
| SBHT | 5.22 ± 0.04 ab | 64,972.75 ± 2430.68 b | 16.33 ± 0.50 b | 88.81 ± 3.13 c | 1.01 ± 0.36 b |
| AT (°C) | MP (mm) | ST (°C) | RH (%) | |
|---|---|---|---|---|
| F-value | 3.873 | 53.921 | 1.643 | 4.794 |
| p-value | 0.056 | 0 | 0.255 | 0.034 |
| SAQ | 17.80 ± 1.00 a | 156.77 ± 20.00 b | 17.86 ± 2.00 a | 73.30 ± 1.00 b |
| SAH | 17.71 ± 0.50 a | 219.875 ± 10.00 a | 17.45 ± 1.00 a | 76.69 ± 1.00 a |
| SBQ | 16.12 ± 0.20 b | 130.46 ± 11.99 c | 15.26 ± 2.00 a | 73.87 ± 2.00 b |
| SBH | 17.16 ± 0.80 ab | 94.22 ± 3.05 d | 16.35 ± 1.00 a | 75.55 ± 0.20 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Xu, Y.; Si, Y.-J.; Li, B.-Q.; Chen, P.; Wu, L.-L.; Guo, P.; Ji, R.-Q. The Diverse Mycorrizal Morphology of Rhododendron dauricum, the Fungal Communities Structure and Dynamics from the Mycorrhizosphere. J. Fungi 2024, 10, 65. https://doi.org/10.3390/jof10010065
Liu J, Xu Y, Si Y-J, Li B-Q, Chen P, Wu L-L, Guo P, Ji R-Q. The Diverse Mycorrizal Morphology of Rhododendron dauricum, the Fungal Communities Structure and Dynamics from the Mycorrhizosphere. Journal of Fungi. 2024; 10(1):65. https://doi.org/10.3390/jof10010065
Chicago/Turabian StyleLiu, Jin, Yang Xu, Yan-Ji Si, Bin-Qi Li, Peng Chen, Ling-Ling Wu, Pu Guo, and Rui-Qing Ji. 2024. "The Diverse Mycorrizal Morphology of Rhododendron dauricum, the Fungal Communities Structure and Dynamics from the Mycorrhizosphere" Journal of Fungi 10, no. 1: 65. https://doi.org/10.3390/jof10010065
APA StyleLiu, J., Xu, Y., Si, Y.-J., Li, B.-Q., Chen, P., Wu, L.-L., Guo, P., & Ji, R.-Q. (2024). The Diverse Mycorrizal Morphology of Rhododendron dauricum, the Fungal Communities Structure and Dynamics from the Mycorrhizosphere. Journal of Fungi, 10(1), 65. https://doi.org/10.3390/jof10010065







