Molecular Signaling and Metabolic Responses during the Interaction between Human Keratinocytes (HaCaT) and the Dermatophyte Trichophyton rubrum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
2.2. Co-Culture of Human Keratinocytes and T. rubrum
2.3. RNA Extraction and cDNA Synthesis
2.4. Real-Time Quantitative Polymerase Chain Reaction
2.5. Glucose Quantification
2.6. Lactate Quantification
2.7. Enzymatic Dosage of Lactate Dehydrogenase (LDH)
2.8. Statistical Analysis
3. Results
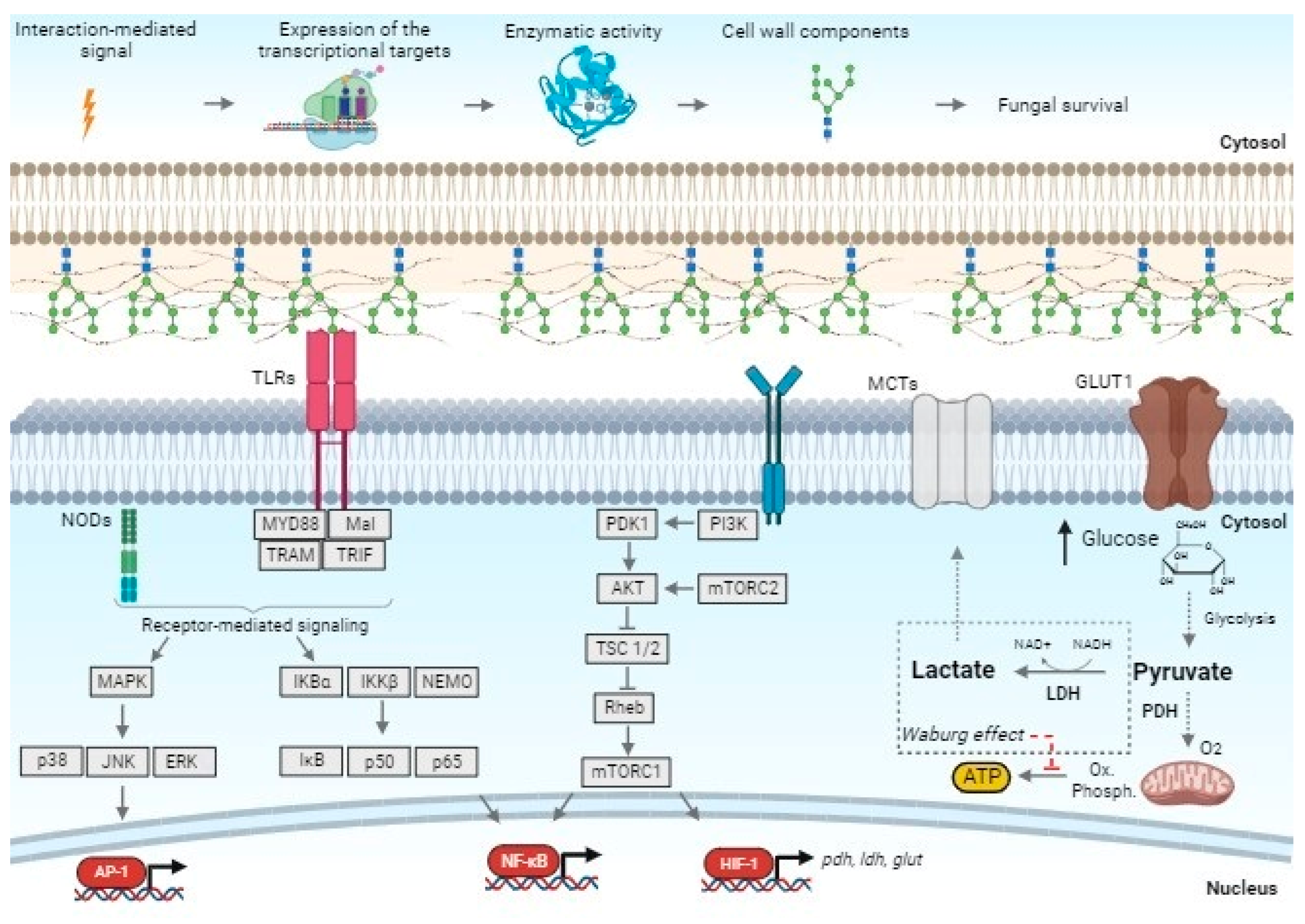
3.1. Transcriptional Response of Keratinocyte PRR Genes
3.2. Overexpression of Human MTOR, HIF-1α, and the Glycolytic Genes (PDHA, GLUT1, and LDHA)
3.3. Intracellular Glucose Levels Increase in HaCaT Cells
3.4. Over-Induction of Lactate Levels in the Cell Supernatant
3.5. Intracellular LDH Activity Decreases in Co-Culture, Whereas the Extracellular Activity Boosts in 24 h
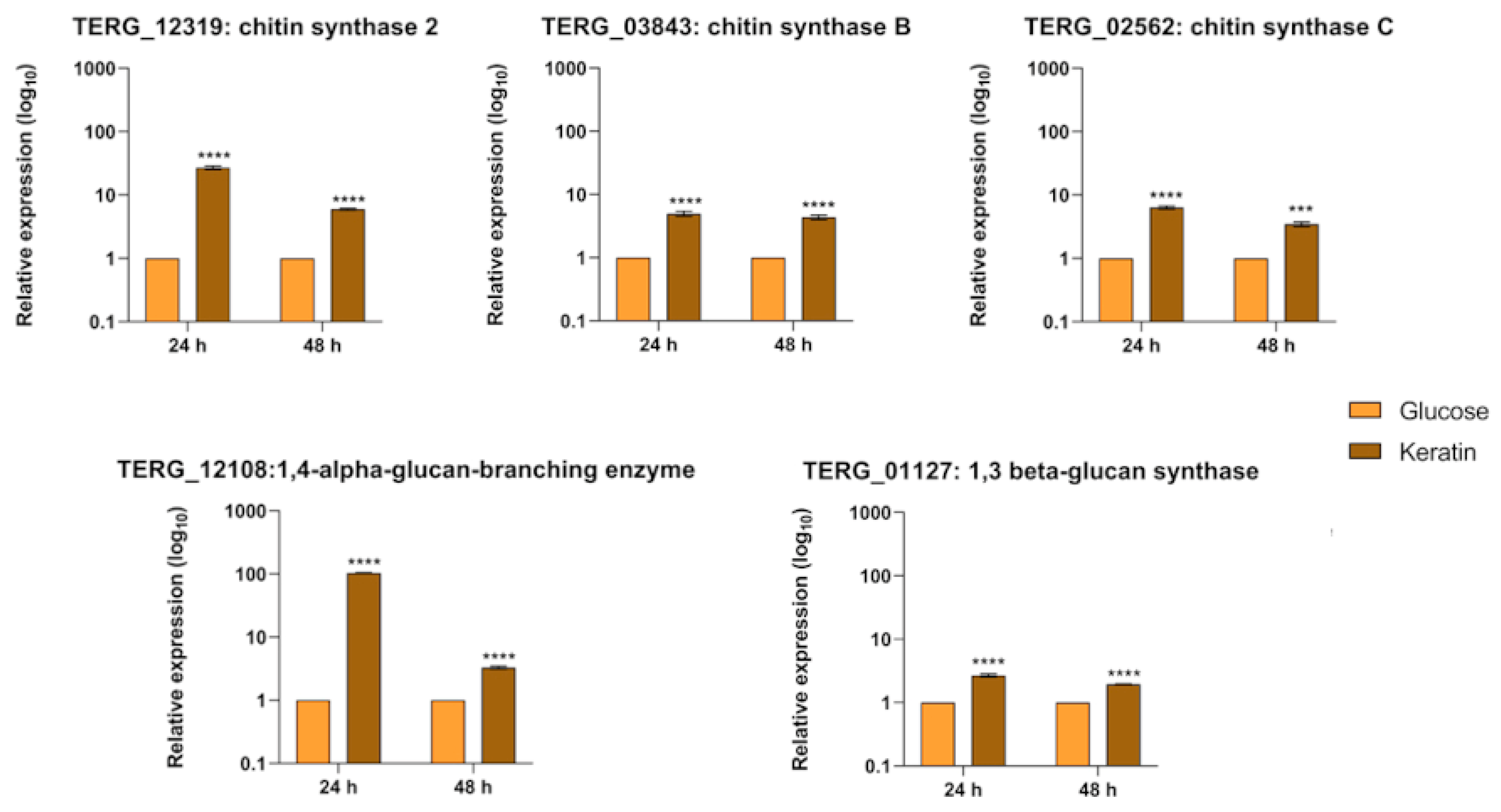
3.6. Transcript Levels of Cell-Wall-Related Genes Are Boosted during Fungal Growth in Keratin but Show Different Regulation Patterns in Co-Culture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petrucelli, M.F.; de Abreu, M.H.; Cantelli, B.A.M.; Segura, G.G.; Nishimura, F.G.; Bitencourt, T.A.; Marins, M.; Fachin, A.L. Epidemiology and diagnostic perspectives of dermatophytoses. J. Fungi 2020, 6, 310. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rossi, N.M.; Peres, N.T.A.; Bitencourt, T.A.; Martins, M.P.; Rossi, A. State-of-the-Art dermatophyte infections: Epidemiology aspects, pathophysiology, and resistance mechanisms. J. Fungi 2021, 7, 629. [Google Scholar] [CrossRef]
- Ciesielska, A.; Kawa, A.; Kanarek, K.; Soboń, A.; Szewczyk, R. Metabolomic analysis of Trichophyton rubrum and Microsporum canis during keratin degradation. Sci. Rep. 2021, 11, 3959. [Google Scholar] [CrossRef]
- Cruz, A.H.S.; Santos, R.S.; Martins, M.P.; Peres, N.T.A.; Trevisan, G.L.; Mendes, N.S.; Martinez-Rossi, N.M.; Rossi, A. Relevance of nutrient-sensing in the pathogenesis of Trichophyton rubrum and Trichophyton interdigitale. Front. Fungal Biol. 2022, 3, 858968. [Google Scholar] [CrossRef] [PubMed]
- Martins-Santana, L.; Petrucelli, M.F.; Sanches, P.R.; Martinez-Rossi, N.M.; Rossi, A. Peptidase regulation in Trichophyton rubrum is mediated by the synergism between alternative splicing and StuA-dependent transcriptional mechanisms. Front. Microbiol. 2022, 13, 2251. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rossi, N.M.; Persinoti, G.F.; Peres, N.T.A.; Rossi, A. Role of pH in the pathogenesis of dermatophytoses. Mycoses 2012, 55, 381–387. [Google Scholar] [CrossRef]
- Martins, M.P.; Rossi, A.; Sanches, P.R.; Bortolossi, J.C.; Martinez-Rossi, N.M. Comprehensive analysis of the dermatophyte Trichophyton rubrum transcriptional profile reveals dynamic metabolic modulation. Biochem. J. 2020, 477, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Nozawa, M.S.; Nozawa, S.R.; Martinez-Rossi, N.M.; Rossi, A. The dermatophyte Trichophyton rubrum secretes an EDTA-sensitive alkaline phosphatase on high-phosphate medium. Braz. J. Microbiol. 2003, 34, 161–164. [Google Scholar] [CrossRef]
- Hatinguais, R.; Willment, J.A.; Brown, G.D. PAMPs of the fungal cell wall and mammalian PRRs. Curr. Top. Microbiol. Immunol. 2020, 425, 187–223. [Google Scholar] [CrossRef]
- Ward, R.A.; Vyas, J.M. The first line of defense: Effector pathways of anti-fungal innate immunity. Curr. Opin. Microbiol. 2020, 58, 160–165. [Google Scholar] [CrossRef]
- Piipponen, M.; Li, D.; Xu Landén, N. The immune functions of keratinocytes in skin wound healing. Int. J. Mol. Sci. 2020, 21, 2–26. [Google Scholar] [CrossRef] [PubMed]
- Weerasinghe, H.; Traven, A. Immunometabolism in fungal infections: The need to eat to compete. Curr. Opin. Microbiol. 2020, 58, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Llibre, A.; Grudzinska, F.S.; O’Shea, M.K.; Duffy, D.; Thickett, D.R.; Mauro, C.; Scott, A. Lactate cross-talk in host-pathogen interactions. Biochem. J. 2021, 478, 3157–3178. [Google Scholar] [CrossRef]
- Wickersham, M.; Wachtel, S.; Wong Fok Lung, T.; Soong, G.; Jacquet, R.; Richardson, A.; Parker, D.; Prince, A. Metabolic stress drives keratinocyte defenses against Staphylococcus aureus Infection. Cell Rep. 2017, 18, 2742–2751. [Google Scholar] [CrossRef]
- Gu, Z.; Zhang, H.; Guo, X.; Cao, Y. Enhanced glycogen metabolism supports the survival and proliferation of HPV-infected keratinocytes in Condylomata acuminata. J. Investig. Dermatol. 2020, 140, 1513–1523.e5. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.P.; Silva, L.G.; Rossi, A.; Sanches, P.R.; Souza, L.D.R.; Martinez-Rossi, N.M. Global analysis of cell wall genes revealed putative virulence factors in the dermatophyte Trichophyton rubrum. Front. Microbiol. 2019, 10, 2168. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Cove, D.J. The Induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta (BBA) Enzymol. Biol. Oxid. 1966, 113, 51–56. [Google Scholar] [CrossRef]
- Petrucelli, M.F.; Peronni, K.; Sanches, P.R.; Komoto, T.T.; Matsuda, J.B.; da Silva Junior, W.A.; Beleboni, R.O.; Martinez-Rossi, N.M.; Marins, M.; Fachin, A.L. Dual RNA-Seq analysis of Trichophyton rubrum and HacaT keratinocyte co-culture highlights important genes for fungal-host interaction. Genes 2018, 9, 362. [Google Scholar] [CrossRef]
- Jacobson, L.S.; McIntyre, L.; Mykusz, J. Assessment of Real-Time PCR cycle threshold values in Microsporum canis culture-positive and culture-negative cats in an animal shelter: A field study. J. Feline Med. Surg. 2018, 20, 108–113. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ma, R.; Zhang, D.; Hu, P.-C.; Li, Q.; Lin, C.-Y. HOXB7-S3 inhibits the proliferation and invasion of MCF-7 human breast cancer cells. Mol. Med. Rep. 2015, 12, 4901–4908. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Ma, X.; Kang, H.; Gao, J.; Min, W.; Guan, H.; Diao, Y.; Lu, W.; Wang, X. Antitumor activity of the selective cyclooxygenase-2 inhibitor, celecoxib, on breast cancer in vitro and in vivo. Cancer Cell Int. 2012, 12, 53. [Google Scholar] [CrossRef]
- Jacob, T.R.; Peres, N.T.A.; Persinoti, G.F.; Silva, L.G.; Mazucato, M.; Rossi, A.; Martinez-Rossi, N.M. Rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med. Mycol. 2012, 50, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Fu, L.; He, L.; Han, J.; Zhao, M.; Yu, Z.; Yang, Q.; He, M.; Gu, C.; Xiao, W. Landscape of keratinocytes transcriptome alterations in response to Trichophyton mentagrophytes infection. Microb. Pathog. 2022, 164, 105426. [Google Scholar] [CrossRef] [PubMed]
- Burstein, V.L.; Beccacece, I.; Guasconi, L.; Mena, C.J.; Cervi, L.; Chiapello, L.S. Skin immunity to dermatophytes: From experimental infection models to human disease. Front. Immunol. 2020, 11, 605644. [Google Scholar] [CrossRef]
- Netea, M.G.; Gow, N.A.R.; Munro, C.A.; Bates, S.; Collins, C.; Ferwerda, G.; Hobson, R.P.; Bertram, G.; Hughes, H.B.; Jansen, T.; et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and toll-like receptors. J. Clin. Investig. 2006, 116, 1642–1650. [Google Scholar] [CrossRef]
- Fuchs, K.; Gloria, Y.C.; Wolz, O.; Herster, F.; Sharma, L.; Dillen, C.A.; Täumer, C.; Dickhöfer, S.; Bittner, Z.; Dang, T.; et al. The fungal ligand chitin directly binds TLR2 and triggers inflammation dependent on oligomer size. EMBO Rep. 2018, 19, e201846065. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yi, J.; Yin, S.; Li, M.; Ye, C.; Lai, W.; Chen, J. Trichophyton rubrum conidia modulate the expression and transport of toll-like receptor 2 in HacaT cell. Microb. Pathog. 2015, 83–84, 1–5. [Google Scholar] [CrossRef]
- Li, Y.; Chen, J.; Wan, M.J.; Lai, W.; Zheng, Y.; Li, M.R.; Chen, R.Z.; Li, X.X. The immune response of human keratinocytes to Trichophyton rubrum conidia is partially mediated by toll-like receptor-2, 4, dectin-1 and cytokines. J. South. Med. Univ. 2011, 31, 678–681. [Google Scholar]
- Netea, M.G.; Van De Veerdonk, F.; Verschueren, I.; Van Der Meer, J.W.M.; Kullberg, B.J. Role of TLR1 and TLR6 in the host defense against disseminated candidiasis. FEMS Immunol. Med. Microbiol. 2007, 52, 118–123. [Google Scholar] [CrossRef]
- Trindade, B.C.; Chen, G.Y. NOD1 and NOD2 in inflammatory and infectious diseases. Immunol. Rev. 2020, 297, 139–161. [Google Scholar] [CrossRef]
- Danis, J.; Mellett, M.; Wertz, P.W. Molecular sciences nod-like receptors in host defence and disease at the epidermal barrier. Int. J. Mol. Sci. 2021, 22, 4677. [Google Scholar] [CrossRef] [PubMed]
- Seiti, F.; Yoshikawa, Y.; Ferreira, L.G.; Rogério De Almeida, S.; Abio Seiti, F.; Rog Erio De Almeida, S. IL-1 signaling inhibits Trichophyton rubrum conidia development and modulates the IL-17 response in vivo. Virulence 2015, 6, 449–457. [Google Scholar] [CrossRef]
- Petrucelli, M.F.; Cantelli, B.A.M.; Marins, M.; Fachin, A.L. The transcriptional regulation of genes involved in the immune innate response of keratinocytes co-cultured with Trichophyton rubrum reveals important roles of cytokine GM-CSF. J. Fungi 2022, 8, 1151. [Google Scholar] [CrossRef] [PubMed]
- García-Madrid, L.A.; Huizar-López, M.D.R.; Flores-Romo, L.; Islas-Rodríguez, A.E. Trichophyton rubrum manipulates the innate immune functions of human keratinocytes. Cent. Eur. J. Biol. 2011, 6, 902–910. [Google Scholar] [CrossRef]
- Saijo, S.; Iwakura, Y. Dectin-1 and Dectin-2 in innate immunity against fungi. Int. Immunol. 2011, 23, 467–472. [Google Scholar] [CrossRef]
- Wang, X.-X.; Sun, H.-Y.; Yang, Q.-Z.; Guo, B.; Sai, Y.; Zhang, J. Hypoxia-inducible factor-1α and glucose transporter 1 in the malignant transformation of oral lichen planus. Int. J. Clin. Exp. Pathol. 2017, 10, 8369–8376. [Google Scholar]
- da Silva-Ferreira, S.; Duarte-Oliveira, C.; Antunes, D.; Barbosa-Matos, C.; Mendes-Frias, A.; Torrado, E.; Costa, S.; Silvestre, R.; Cunha, C.; Carvalho, A. Hypoxia inducible-factor 1 alpha regulates neutrophil recruitment during fungal-elicited granulomatous inflammation. Front. Cell Infect. Microbiol. 2022, 12, 1005839. [Google Scholar] [CrossRef]
- Wei, S.; Fan, Q.; Liang, Y.; Xiaodong, Z.; Ma, Y.; Zhihong, Z.; Hua, X.; Su, D.; Sun, H.; Li, H.; et al. Promotion of glycolysis by HOTAIR through GLUT1 upregulation via MTOR signaling. Oncol. Rep. 2017, 38, 1902–1908. [Google Scholar] [CrossRef]
- Pollizzi, K.N.; Powell, J.D. Integrating canonical and metabolic signalling programmes in the regulation of t cell responses. Nat. Rev. Immunol. 2014, 14, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Jiang, Y.; Zhang, M. Crosstalk between glucose metabolism, lactate production and immune response modulation. Cytokine Growth Factor. Rev. 2022, 68, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, I.; Prasad, P.D.; Thangaraju, M.; Manicassamy, S. Lactate-dependent regulation of immune responses by dendritic cells and macrophages. Front. Immunol. 2021, 12, 691134. [Google Scholar] [CrossRef] [PubMed]
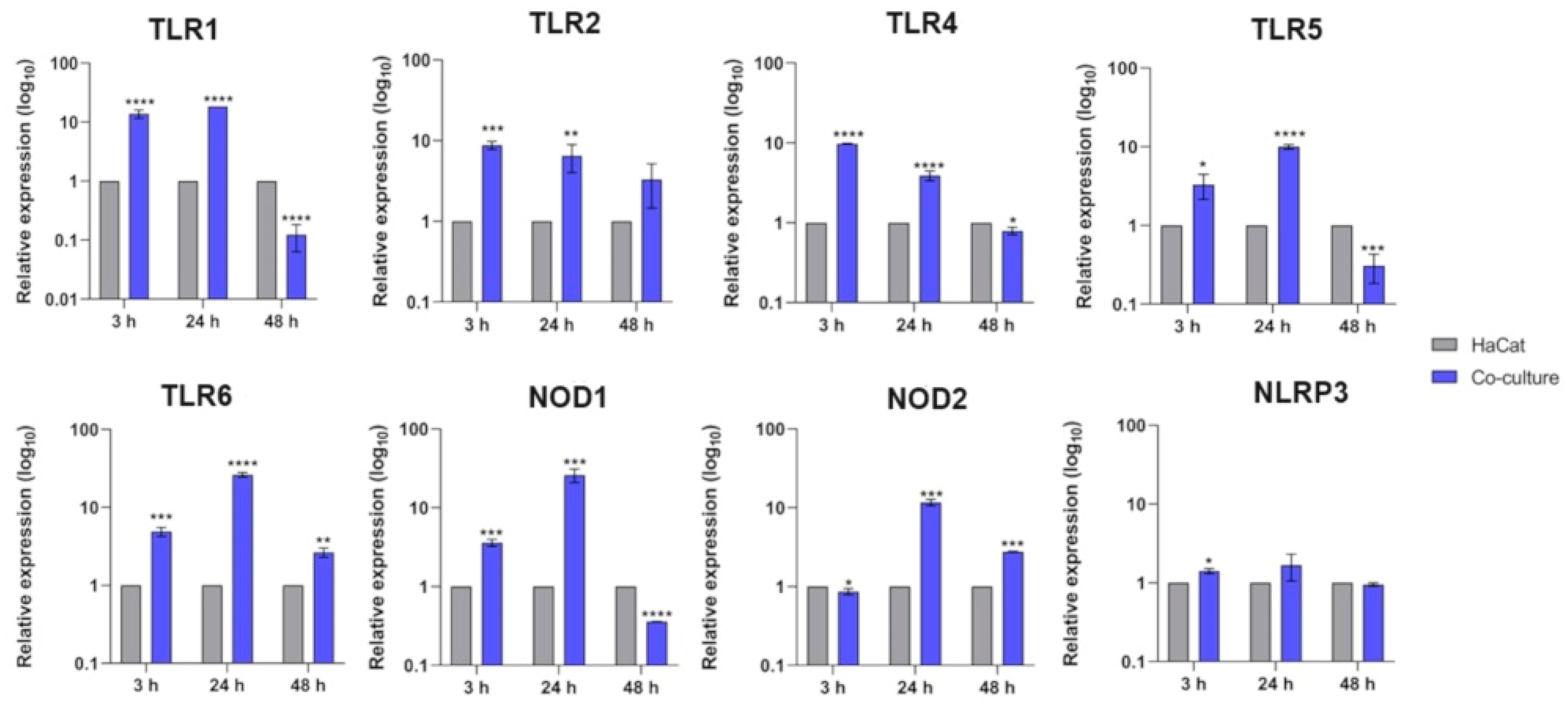

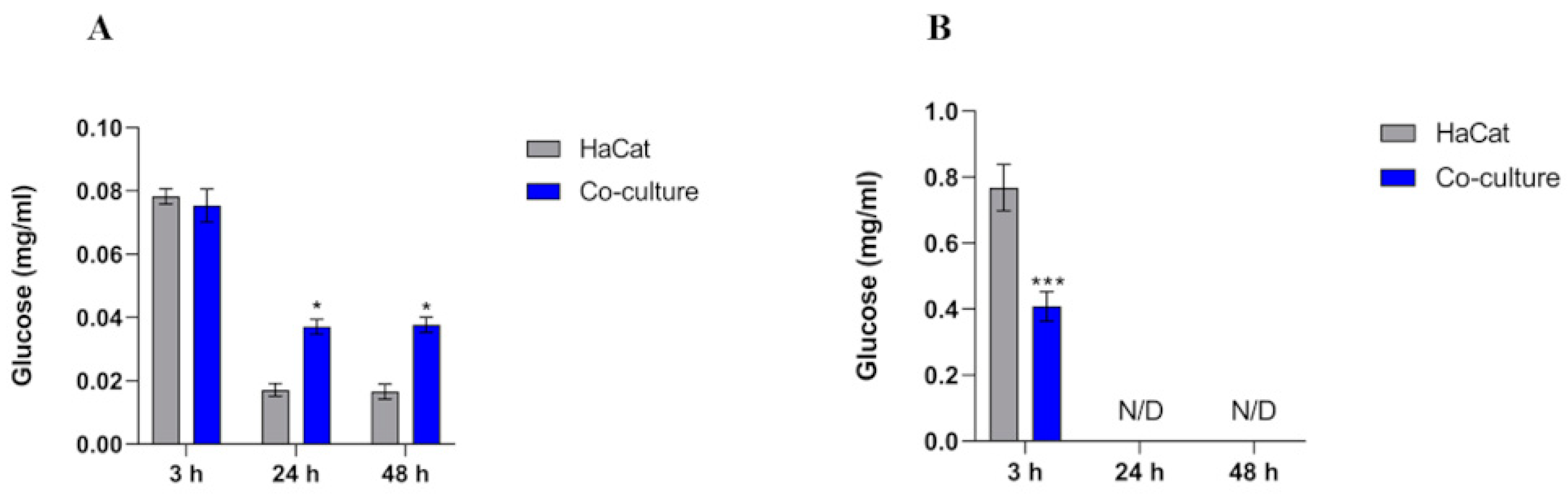

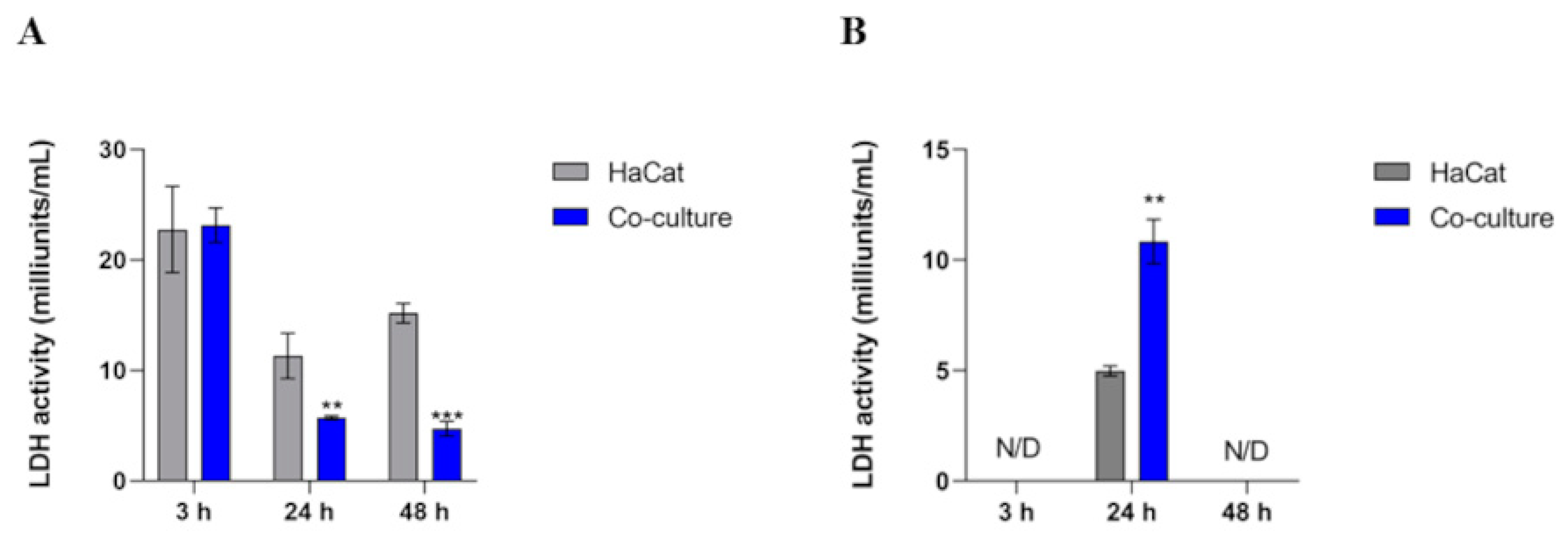
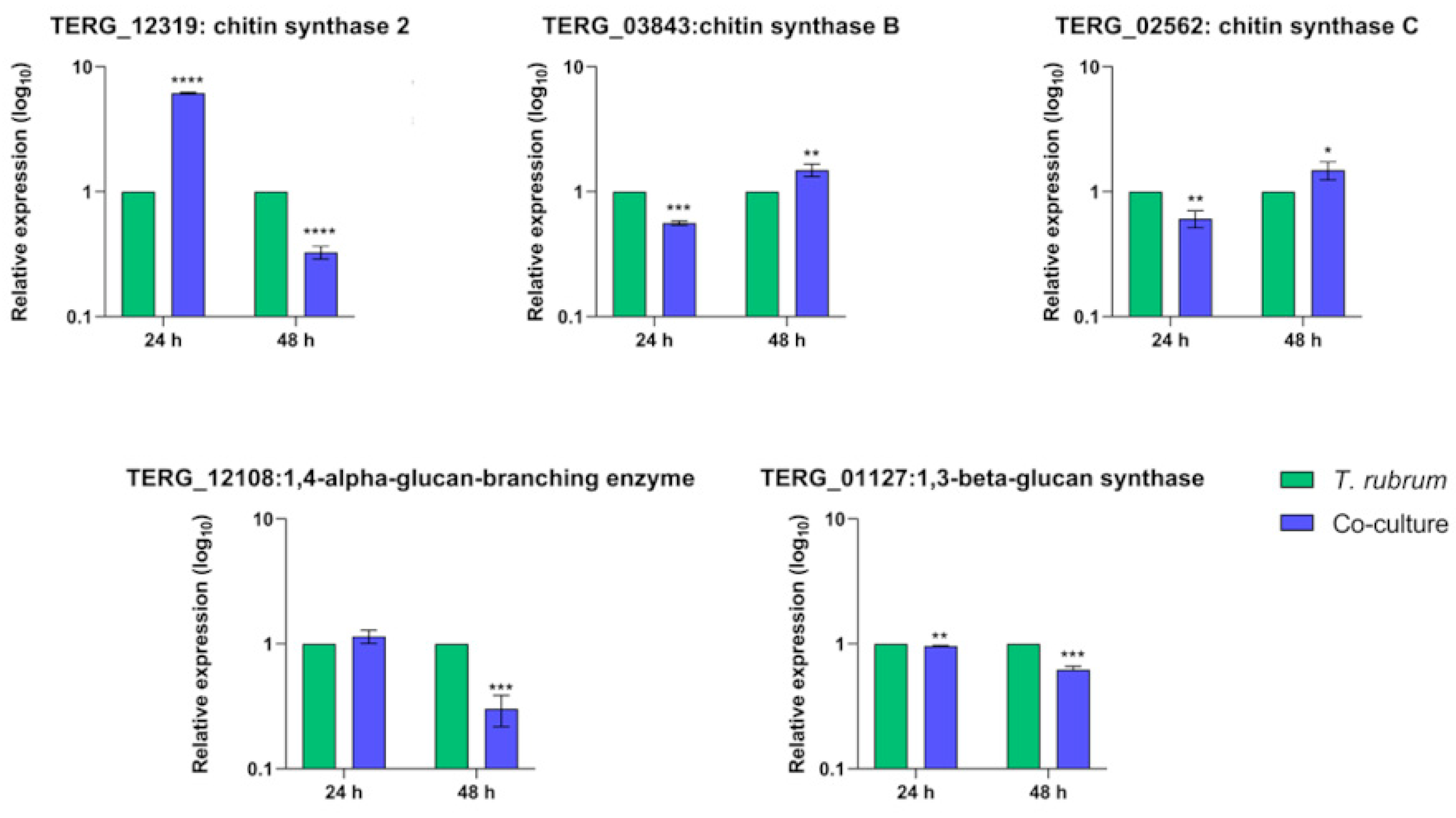
| Time | HaCat | Co-Culture |
|---|---|---|
| 3 h | 6.8 ± 0.15 | 7.2 ± 0.3 |
| 24 h | 7.2 ± 0.3 | 7.1 ± 0.5 |
| 48 h | 6.9 ± 0.5 | 6.5 ± 0.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrucelli, M.F.; Martins-Santana, L.; Rossi, A.; Martinez-Rossi, N.M. Molecular Signaling and Metabolic Responses during the Interaction between Human Keratinocytes (HaCaT) and the Dermatophyte Trichophyton rubrum. J. Fungi 2024, 10, 72. https://doi.org/10.3390/jof10010072
Petrucelli MF, Martins-Santana L, Rossi A, Martinez-Rossi NM. Molecular Signaling and Metabolic Responses during the Interaction between Human Keratinocytes (HaCaT) and the Dermatophyte Trichophyton rubrum. Journal of Fungi. 2024; 10(1):72. https://doi.org/10.3390/jof10010072
Chicago/Turabian StylePetrucelli, Monise Fazolin, Leonardo Martins-Santana, Antonio Rossi, and Nilce Maria Martinez-Rossi. 2024. "Molecular Signaling and Metabolic Responses during the Interaction between Human Keratinocytes (HaCaT) and the Dermatophyte Trichophyton rubrum" Journal of Fungi 10, no. 1: 72. https://doi.org/10.3390/jof10010072






