Regulation of Catalase Expression and Activity by DhHog1 in the Halotolerant Yeast Debaryomyces hansenii Under Saline and Oxidative Conditions
Abstract
1. Introduction
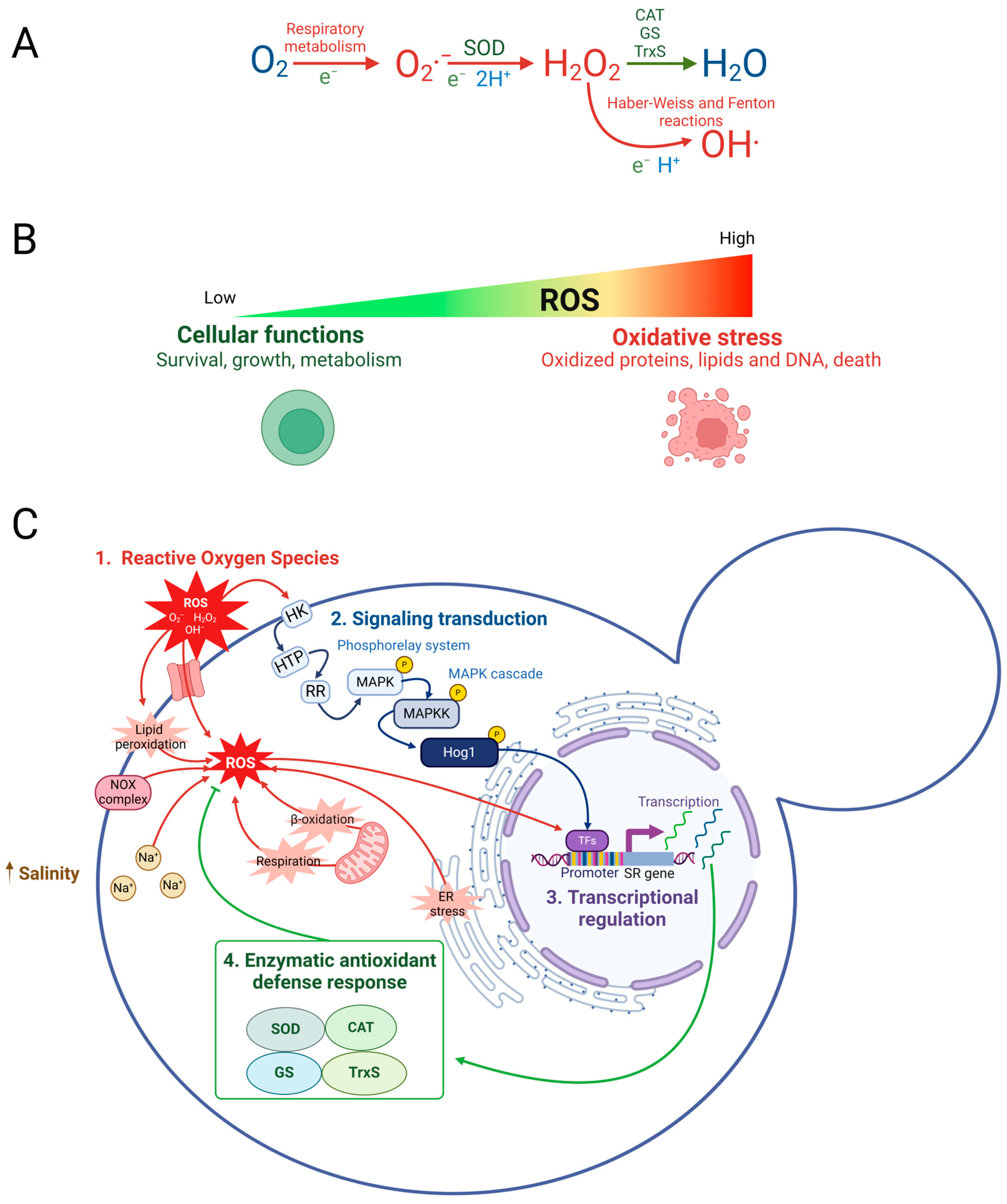
2. Material and Methods
2.1. Strains and Growth Conditions
2.2. Growth Curves
2.3. Cell Viability After H2O2 Gradient Shock Assay
2.4. Specific Catalase Activity Determination
2.5. RNA Extraction
2.6. Analysis of Gene Expression
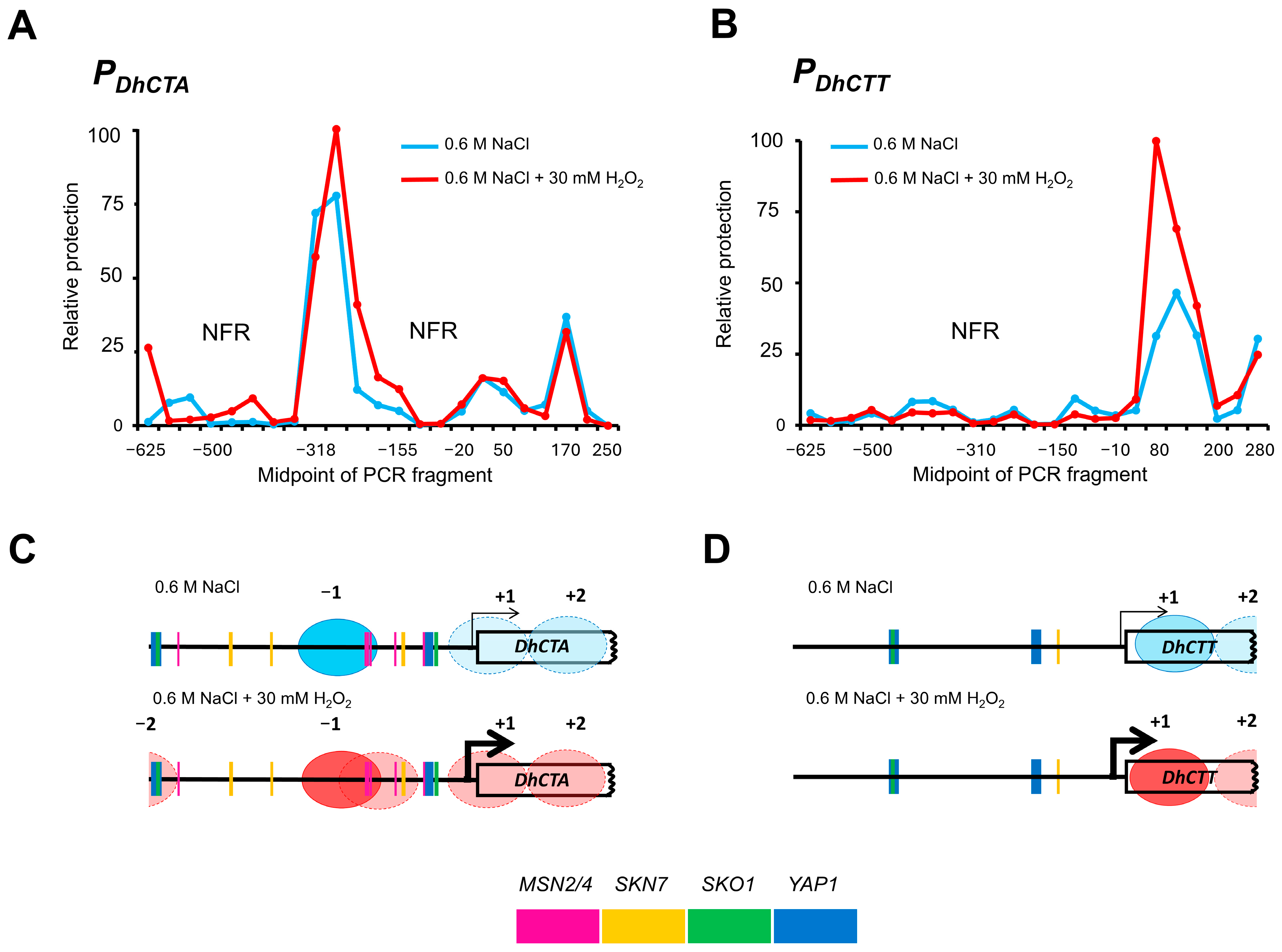
2.7. Nucleosome Scanning Assay
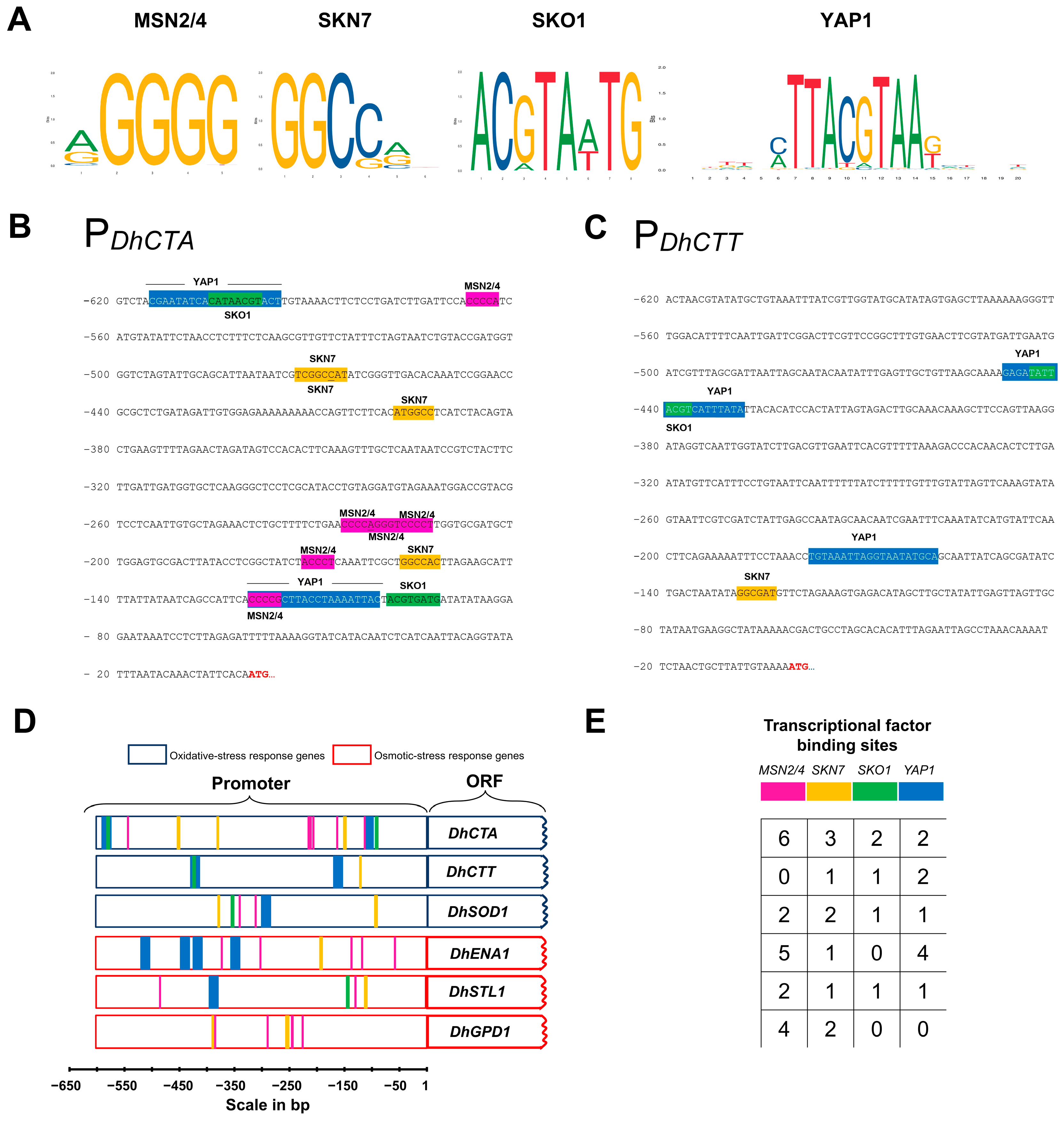
2.8. Promoter Analysis
2.9. Identification of Putative Orthologs of Transcription Factors
2.10. Statistical Analysis
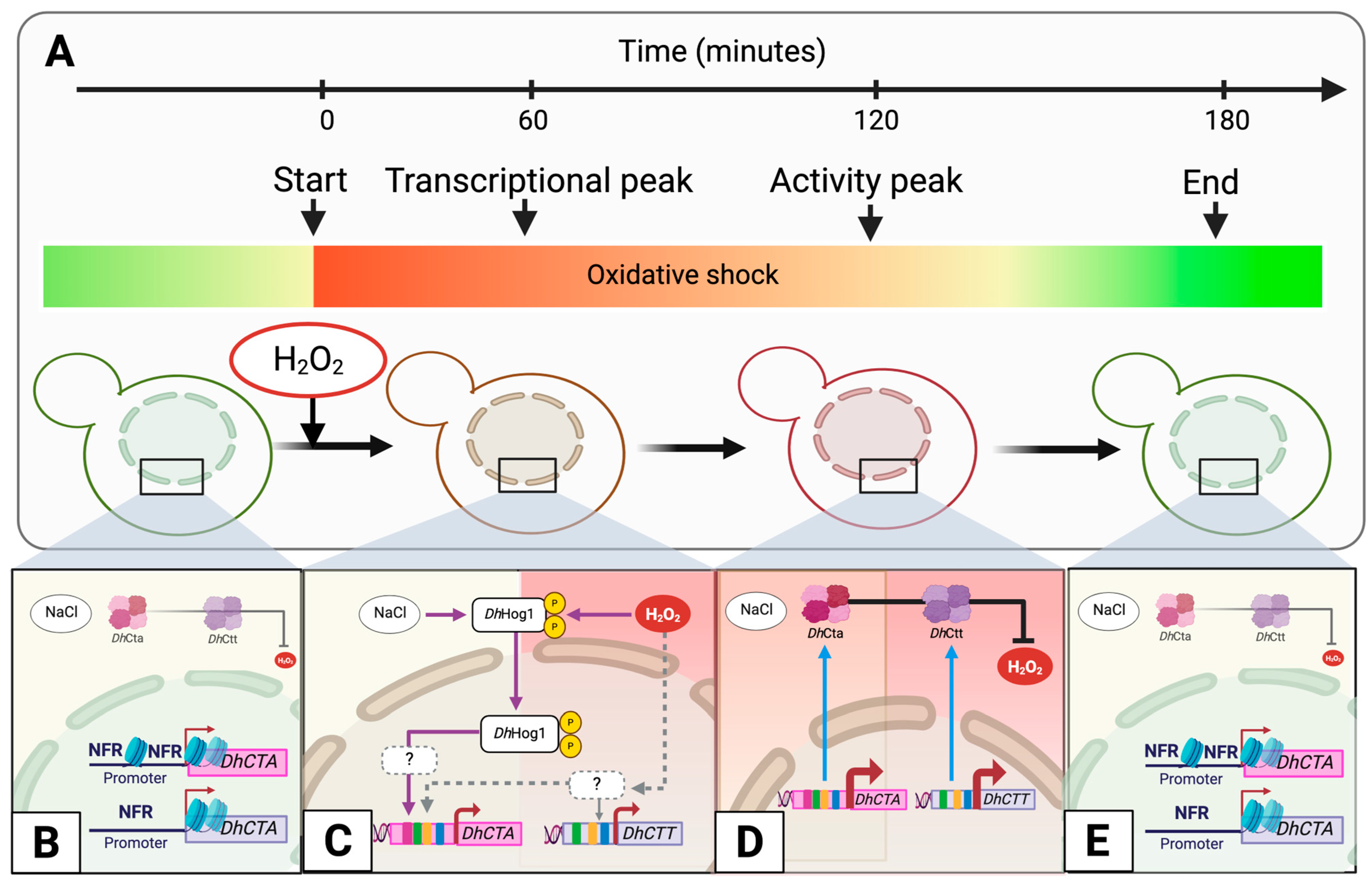
3. Results
3.1. H2O2-Induced Oxidative Stress Triggers Catalase Expression and Activity in D. hansenii
3.2. Nucleosome Occupancy and Putative TF Binding Motifs Distribution in DhCTA and DhCTT Promoters
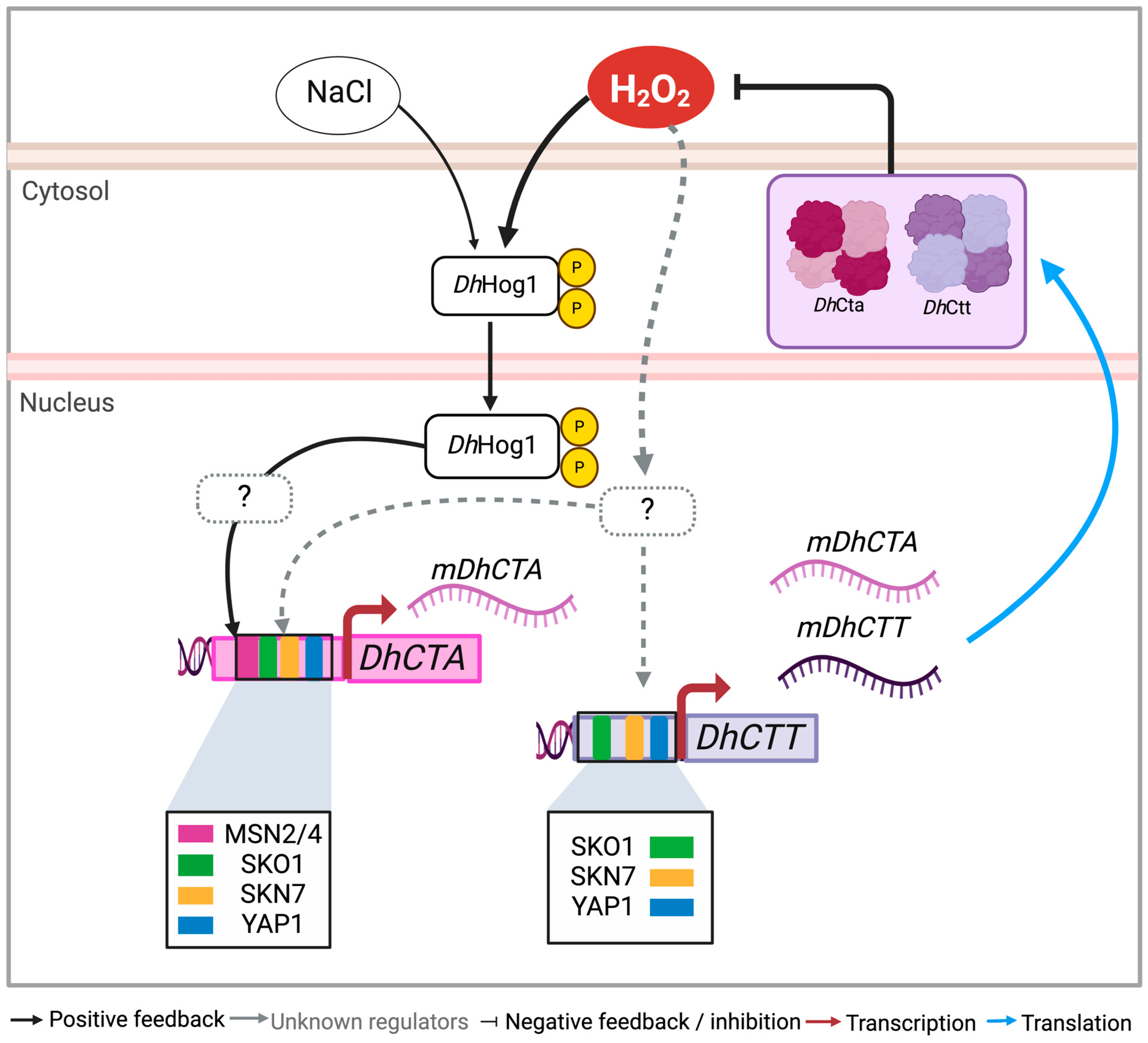
3.3. DhHog1 Regulates the Activity and Expression of Catalase A Under Saline and Oxidative Conditions
4. Discussion
4.1. The Expression of DhCTA and DhCTT Genes Increases After Exposure to H2O2 Under NaCl Condition
4.2. Nucleosome Arrangement and Putative TF-Binding Motifs Configurations Between DhCTA and DhCTT Promoters Reveal Divergent Transcriptional Regulation
4.3. DhHog1 Regulates the Induction of Catalase A to Adapt Under Saline and Oxidative Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dolz-Edo, L.; Rienzo, A.; Poveda-Huertes, D.; Pascual-Ahuir, A.; Proft, M. Deciphering dynamic dose responses of natural promoters and single cis elements upon osmotic and oxidative stress in yeast. Mol. Cell. Biol. 2013, 33, 2228–2240. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.M.S.; Lowry, C.V.; Davies, K.J.A. Transient adaptation to oxidative stress in yeast. Arch. Biochem. Biophys. 1995, 317, 1–6. [Google Scholar] [CrossRef]
- Izawa, S.; Inoue, Y.; Kimura, A. Importance of catalase in the adaptive response to hydrogen peroxide: Analysis of acatalasaemic Saccharomyces cerevisiae. Biochem. J. 1996, 320 Pt 1, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Yale, J.; Bohnert, H.J. Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 2001, 276, 15996–16007. [Google Scholar] [CrossRef] [PubMed]
- Parrou, J.L.; Teste, M.-A.; François, J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: Genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiol. 1997, 143, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- Taymaz-Nikerel, H.; Cankorur-Cetinkaya, A.; Kirdar, B. Genome-wide transcriptional response of Saccharomyces cerevisiae to stress-induced perturbations. Front. Bioeng. Biotechnol. 2016, 4, 17. [Google Scholar] [CrossRef]
- Jamieson, D.J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 1998, 14, 1511–1527. [Google Scholar] [CrossRef]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef]
- Farrugia, G.; Balzan, R. Oxidative stress and programmed cell death in yeast. Front. Oncol. 2012, 2, 64. [Google Scholar] [CrossRef]
- Perrone, G.G.; Tan, S.-X.; Dawes, I.W. Reactive oxygen species and yeast apoptosis. BBA Mol. Cell Res. 2008, 1783, 1354–1368. [Google Scholar] [CrossRef]
- González, J.; Castillo, R.; García-Campos, M.A.; Noriega-Samaniego, D.; Escobar-Sánchez, V.; Romero-Aguilar, L.; Alba-Lois, L.; Segal-Kischinevzky, C. Tolerance to oxidative stress in budding yeast by heterologous expression of catalases A and T from Debaryomyces hansenii. Curr. Microbiol. 2020, 77, 4000–4015. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Moreno, L.; Ramos, J.; Michán, C. Overlapping responses between salt and oxidative stress in Debaryomyces hansenii. World J. Microbiol. Biotechnol. 2019, 35, 170. [Google Scholar] [CrossRef] [PubMed]
- Sadeh, A.; Movshovich, N.; Volokh, M.; Gheber, L.; Aharoni, A. Fine-tuning of the Msn2/4–mediated yeast stress responses as revealed by systematic deletion of Msn2/4 partners. MBoC 2011, 22, 3127–3138. [Google Scholar] [CrossRef] [PubMed]
- Ikner, A.; Shiozaki, K. Yeast signaling pathways in the oxidative stress response. MR/Fundam. Mol. Mech. Mutagen. 2005, 569, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Auesukaree, C. Molecular mechanisms of the yeast adaptive response and tolerance to stresses encountered during ethanol fermentation. J. Bioscien. Bioengin. 2017, 124, 133–142. [Google Scholar] [CrossRef]
- Vázquez, J.; González, B.; Sempere, V.; Mas, A.; Torija, M.J.; Beltran, G. Melatonin reduces oxidative stress damage induced by hydrogen peroxide in Saccharomyces cerevisiae. Front. Microbiol. 2017, 8, 66. [Google Scholar] [CrossRef]
- Gościńska, K.; Shahmoradi Ghahe, S.; Domogała, S.; Topf, U. Eukaryotic elongation factor 3 protects Saccharomyces cerevisiae yeast from oxidative stress. Genes. 2020, 11, 1432. [Google Scholar] [CrossRef]
- Martins, D.; English, A.M. Catalase activity is stimulated by H2O2 in rich culture medium and is required for H2O2 resistance and adaptation in yeast. Redox Biol. 2014, 2, 308–313. [Google Scholar] [CrossRef]
- Qi, Y.; Qin, Q.; Liao, G.; Tong, L.; Jin, C.; Wang, B.; Fang, W. Unveiling the super tolerance of Candida nivariensis to oxidative stress: Insights into the involvement of a catalase. Microbiol. Spectr. 2024, 12, e03169-23. [Google Scholar] [CrossRef]
- Yaakoub, H.; Mina, S.; Calenda, A.; Bouchara, J.-P.; Papon, N. Oxidative stress response pathways in fungi. Cell. Mol. Life Sci. 2022, 79, 333. [Google Scholar] [CrossRef]
- Godon, C.; Lagniel, G.; Lee, J.; Buhler, J.M.; Kieffer, S.; Perrot, M.; Boucherie, H.; Toledano, M.B.; Labarre, J. The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 22480–22489. [Google Scholar] [CrossRef] [PubMed]
- Klopf, E.; Schmidt, H.A.; Clauder-Münster, S.; Steinmetz, L.M.; Schüller, C. INO80 represses osmostress induced gene expression by resetting promoter proximal nucleosomes. Nucl. Acids Res. 2017, 45, 3752–3766. [Google Scholar] [CrossRef] [PubMed]
- Salas-Delgado, G.; Ongay-Larios, L.; Kawasaki-Watanabe, L.; López-Villaseñor, I.; Coria, R. The yeasts phosphorelay Ssystems: A comparative view. World J. Microbiol. Biotechnol. 2017, 33, 111. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Gao, Z.; Shi, T.-Q.; Song, P.; Ren, L.-J.; Huang, H.; Ji, X.-J. Reactive oxygen species-mediated cellular stress response and lipid accumulation in oleaginous microorganisms: The state of the art and future perspectives. Front. Microbiol. 2017, 8, 793. [Google Scholar] [CrossRef]
- de Nadal, E.; Alepuz, P.M.; Posas, F. Dealing with osmostress through MAP kinase activation. EMBO Rep. 2002, 3, 735–740. [Google Scholar] [CrossRef]
- Saito, H.; Posas, F. Response to hyperosmotic stress. Genetics 2012, 192, 289–318. [Google Scholar] [CrossRef]
- Hohmann, S. An integrated view on a eukaryotic osmoregulation system. Curr. Genet. 2015, 61, 373–382. [Google Scholar] [CrossRef]
- Gonzalez, R.; Morales, P.; Tronchoni, J.; Cordero-Bueso, G.; Vaudano, E.; Quirós, M.; Novo, M.; Torres-Pérez, R.; Valero, E. New genes involved in osmotic stress tolerance in Saccharomyces cerevisiae. Front. Microbiol. 2016, 7, 1545. [Google Scholar] [CrossRef]
- Ruiz-Roig, C.; Noriega, N.; Duch, A.; Posas, F.; de Nadal, E. The Hog1 SAPK controls the Rtg1/Rtg3 transcriptional complex activity by multiple regulatory mechanisms. MBoC 2012, 23, 4286–4296. [Google Scholar] [CrossRef]
- Pascual-Ahuir, A.; Proft, M. The Sch9 kinase is a chromatin-associated transcriptional activator of osmostress-responsive genes. EMBO J. 2007, 26, 3098–3108. [Google Scholar] [CrossRef]
- Prista, C.; Loureiro-Dias, M.C.; Montiel, V.; García, R.; Ramos, J. Mechanisms underlying the halotolerant way of Debaryomyces hansenii. FEMS Yeast Res. 2005, 5, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Gunde-Cimerman, N.; Ramos, J.; Plemenitaš, A. Halotolerant and halophilic fungi. Mycol. Res. 2009, 113, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Michán, C.; Martínez, J.L.; Alvarez, M.C.; Turk, M.; Sychrova, H.; Ramos, J. Salt and oxidative stress tolerance in Debaryomyces hansenii and Debaryomyces fabryi. FEMS Yeast Res. 2013, 13, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Meena, N.; Aggarwal, M.; Mondal, A.K. Debaryomyces hansenii, a highly osmo-tolerant and halo-tolerant yeast, maintains activated Dhog1p in the cytoplasm during its growth under severe osmotic stress. Curr. Genet. 2005, 48, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, N.S.; Calahorra, M.; González, J.; Defosse, T.; Papon, N.; Peña, A.; Coria, R. Contribution of the mitogen-activated protein kinase Hog1 to the halotolerance of the marine yeast Debaryomyces hansenii. Curr. Genet. 2020, 66, 1135–1153. [Google Scholar] [CrossRef]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef]
- Segal-Kischinevzky, C.; Rodarte-Murguía, B.; Valdés-López, V.; Mendoza-Hernández, G.; González, A.; Alba-Lois, L. The euryhaline yeast Debaryomyces hansenii has two catalase genes encoding enzymes with differential activity profile. Curr. Microbiol. 2011, 62, 933–943. [Google Scholar] [CrossRef]
- Gostinčar, C.; Gunde-Cimerman, N. Overview of oxidative stress response genes in selected halophilic fungi. Genes 2018, 9, 143. [Google Scholar] [CrossRef]
- Komalapriya, C.; Kaloriti, D.; Tillmann, A.T.; Yin, Z.; Herrero-de-Dios, C.; Jacobsen, M.D.; Belmonte, R.C.; Cameron, G.; Haynes, K.; Grebogi, C.; et al. Integrative model of oxidative stress adaptation in the fungal pathogen Candida albicans. PLoS ONE 2015, 10, e0137750. [Google Scholar] [CrossRef]
- Cuéllar-Cruz, M.; Briones-Martin-del-Campo, M.; Cañas-Villamar, I.; Montalvo-Arredondo, J.; Riego-Ruiz, L.; Castaño, I.; De Las Peñas, A. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot. Cell 2008, 7, 814–825. [Google Scholar] [CrossRef]
- Aebi, H. [13] Catalase in vitro. In Methods in Enzymology; Oxygen Radicals in Biological Systems; Academic Press: Cambridge, MA, USA, 1984; Volume 105, pp. 121–126. [Google Scholar] [CrossRef]
- Schmitt, M.E.; Brown, T.A.; Trumpower, B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucl. Acids Res. 1990, 18, 3091–3092. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Gutiérrez, D.; Reyes-Torres, A.M.; de la Fuente-Colmenares, I.; Escobar-Sánchez, V.; González, J.; Ortiz-Hernández, R.; Torres-Ramírez, N.; Segal-Kischinevzky, C. Alternative CUG codon usage in the halotolerant yeast Debaryomyces hansenii: Gene expression profiles provide new insights into ambiguous translation. JoF 2022, 8, 970. [Google Scholar] [CrossRef] [PubMed]
- González, J.; López, G.; Argueta, S.; Escalera-Fanjul, X.; el Hafidi, M.; Campero-Basaldua, C.; Strauss, J.; Riego-Ruiz, L.; González, A. Diversification of transcriptional regulation determines subfunctionalization of paralogous branched chain aminotransferases in the yeast Saccharomyces cerevisiae. Genetics 2017, 207, 975–991. [Google Scholar] [CrossRef] [PubMed][Green Version]
- González, J.; Quezada, H.; Campero-Basaldua, J.C.; Ramirez-González, É.; Riego-Ruiz, L.; González, A. Transcriptional regulation of the genes encoding branched-chain aminotransferases in Kluyveromyces lactis and Lachancea kluyveri is independent of chromatin remodeling. Microbiol. Res. 2024, 15, 1225–1238. [Google Scholar] [CrossRef]
- Infante, J.J.; Law, G.L.; Young, E.T. Analysis of nucleosome positioning using a nucleosome-scanning assay. In Chromatin Remodeling: Methods and Protocols; Morse, R.H., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 63–87. [Google Scholar] [CrossRef]
- Santana-Garcia, W.; Castro-Mondragon, J.A.; Padilla-Gálvez, M.; Nguyen, N.T.T.; Elizondo-Salas, A.; Ksouri, N.; Gerbes, F.; Thieffry, D.; Vincens, P.; Contreras-Moreira, B.; et al. RSAT 2022: Regulatory sequence analysis tools. Nucl. Acids Res. 2022, 50, W670–W676. [Google Scholar] [CrossRef]
- Maguire, S.L.; ÓhÉigeartaigh, S.S.; Byrne, K.P.; Schröder, M.S.; O’Gaora, P.; Wolfe, K.H.; Butler, G. Comparative genome analysis and gene finding in Candida species using CGOB. Mol. Biol. Evol. 2013, 30, 1281–1291. [Google Scholar] [CrossRef]
- Nikolaou, E.; Agrafioti, I.; Stumpf, M.; Quinn, J.; Stansfield, I.; Brown, A.J. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol. Biol. 2009, 9, 44. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI job dispatcher sequence analysis tools framework in 2024. Nucl. Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- González-Hernández, J.C.; Peña, A. [Adaptation strategies of halophilic microorganisms and Debaryomyces hansenii (halophilic yeast)]. Rev. Latinoam. Microbiol. 2002, 44, 137–156. [Google Scholar]
- Pascual-Ahuir, A.; González-Cantó, E.; Juyoux, P.; Pable, J.; Poveda-Huertes, D.; Saiz-Balbastre, S.; Squeo, S.; Ureña-Marco, A.; Vanacloig-Pedros, E.; Zaragoza-Infante, L.; et al. Dose dependent gene expression is dynamically modulated by the history, physiology and age of yeast cells. BBA Gene Regul. Mech. 2019, 1862, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, C.; Sánchez, B.J.; Savickas, S.; Martínez, J.L. DebaryOmics: An integrative –omics study to understand the halophilic behaviour of Debaryomyces hansenii. Microb. Biotechnol. 2022, 15, 1133–1151. [Google Scholar] [CrossRef] [PubMed]
- Capaldi, A.P.; Kaplan, T.; Liu, Y.; Habib, N.; Regev, A.; Friedman, N.; O’Shea, E.K. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat. Genet. 2008, 40, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Proft, M.; Pascual-Ahuir, A.; de Nadal, E.; Ariño, J.; Serrano, R.; Posas, F. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 2001, 20, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Proft, M.; Remize, F.; Tamás, M.; Serrano, R.; Thevelein, J.M.; Hohmann, S. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 2001, 40, 1067–1083. [Google Scholar] [CrossRef]
- Buzzini, P.; Turchetti, B.; Yurkov, A. Extremophilic yeasts: The toughest yeast around? Yeast 2018, 35, 487–497. [Google Scholar] [CrossRef]
- Segal-Kischinevzky, C.; Romero-Aguilar, L.; Alcaraz, L.D.; López-Ortiz, G.; Martínez-Castillo, B.; Torres-Ramírez, N.; Sandoval, G.; González, J. Yeasts inhabiting extreme environments and their biotechnological applications. Microorganisms 2022, 10, 794. [Google Scholar] [CrossRef]
- Lin, N.-X.; He, R.-Z.; Xu, Y.; Yu, X.-W. Augmented peroxisomal ROS buffering capacity renders oxidative and thermal stress cross-tolerance in yeast. Microb. Cell Fact. 2021, 20, 131. [Google Scholar] [CrossRef]
- Thines, L.; Morsomme, P. Manganese superoxide dismutase activity assay in the yeast Saccharomyces cerevisiae. Bio Protoc. 2020, 10, e3542. [Google Scholar] [CrossRef]
- Toledano, M.B.; Delaunay-Moisan, A.; Outten, C.E.; Igbaria, A. Functions and cellular compartmentation of the thioredoxin and glutathione pathways in yeast. Antioxid. Redox Sign. 2013, 18, 1699–1711. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Ramakrishna, S.; Vikineswary, S.; Das, D.; Bahkali, A.H.; Guo, S.-Y.; Pang, K.-L. How do fungi survive in the sea and respond to climate change? JoF 2022, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Gunde-Cimerman, N.; Butinar, L.; Sonjak, S.; Turk, M.; Uršič, V.; Zalar, P.; Plemenitaš, A. Halotolerant and halophilic fungi from coastal environments in the Arctics. In Adaptation to Life at High Salt Concentrations in Archaea, Bacteria, and Eukarya (Cellular Origin, Life in Extreme Habitats and Astrobiology); Gunde-Cimerman, N., Oren, A., Plemenitaš, A., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 397–423. [Google Scholar]
- Butinar, L.; Zalar, P.; Frisvad, J.C.; Gunde-Cimerman, N. The genus Eurotium– members of indigenous fungal community in hypersaline waters of salterns. FEMS Microbiol. Ecol. 2005, 51, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Plemenitaš, A.; Lenassi, M.; Konte, T.; Kejžar, A.; Zajc, J.; Gostinčar, C.; Gunde-Cimerman, N. Adaptation to high salt concentrations in halotolerant/halophilic fungi: A molecular perspective. Front. Microbiol. 2014, 5, 199. [Google Scholar] [CrossRef] [PubMed]
- Prista, C.; Michán, C.; Miranda, I.M.; Ramos, J. The halotolerant Debaryomyces hansenii, the cinderella of non-conventional yeasts. Yeast 2016, 33, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, C.; Siles, A.; Martínez, J.L.; Calero, F.; Ramos, J. Oxidative stress sensitivity in Debaryomyces hansenii. FEMS Yeast Res. 2009, 9, 582–590. [Google Scholar] [CrossRef]
- Marchler, G.; Schüller, C.; Adam, G.; Ruis, H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993, 12, 1997–2003. [Google Scholar] [CrossRef]
- Rep, M.; Reiser, V.; Gartner, U.; Thevelein, J.M.; Hohmann, S.; Ammerer, G.; Ruis, H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol. Cell. Biol. 1999, 19, 5474–5485. [Google Scholar] [CrossRef]
- Kathiresan, M.; Martins, D.; English, A.M. Respiration triggers heme transfer from cytochrome c peroxidase to catalase in yeast mitochondria. Proc. Natl. Acad. Sci. USA 2014, 111, 17468–17473. [Google Scholar] [CrossRef]
- Strucko, T.; Andersen, N.L.; Mahler, M.R.; Martínez, J.L.; Mortensen, U.H. A CRISPR/Cas9 method facilitates efficient oligo-mediated gene editing in Debaryomyces hansenii. Synth. Biol. 2021, 6, ysab031. [Google Scholar] [CrossRef]
- Spasskaya, D.S.; Kotlov, M.I.; Lekanov, D.S.; Tutyaeva, V.V.; Snezhkina, A.V.; Kudryavtseva, A.V.; Karpov, V.L.; Karpov, D.S. CRISPR/Cas9-mediated genome engineering reveals the contribution of the 26S proteasome to the extremophilic nature of the yeast Debaryomyces hansenii. ACS Synth. Biol. 2021, 10, 297–308. [Google Scholar] [CrossRef]
- Mendoza-Téllez, B.; Zamora-Bello, A.; Rosas-Paz, M.; Villarreal-Huerta, D.; de la Fuente, I.; Segal-Kischinevzky, C.; González, J. Introducción a los sistemas CRISPR y sus aplicaciones en levaduras. TIP Rev. Espec. Cienc. Quím-Biol. 2022, 25, 1–21. [Google Scholar] [CrossRef]
- Scacchetti, A.; Becker, P.B. Variation on a theme: Evolutionary strategies for H2A.Z exchange by SWR1-type remodelers. Curr. Opin. Cell Biol. 2021, 70, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Tremethick, D.J. Multifunctional histone variants in genome function. Nat. Rev. Genet. 2024, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Jansen, A.; Verstrepen, K.J. Nucleosome positioning in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2011, 75, 301–320. [Google Scholar] [CrossRef] [PubMed]
- Campero-Basaldua, C.; Quezada, H.; Riego-Ruíz, L.; Márquez, D.; Rojas, E.; González, J.; El-Hafidi, M.; González, A. Diversification of the kinetic properties of yeast NADP-glutamate-dehydrogenase isozymes proceeds independently of their evolutionary origin. Microbiol. Open 2017, 6, e00419. [Google Scholar] [CrossRef]
- Escalera-Fanjul, X.; Campero-Basaldua, C.; Colón, M.; González, J.; Márquez, D.; González, A. Evolutionary diversification of alanine transaminases in yeast: Catabolic specialization and biosynthetic redundancy. Front. Microbiol. 2017, 8, 1150. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Liu, C.L.; Humphrey, E.L.; Perlstein, E.O.; Schreiber, S.L. Global nucleosome occupancy in yeast. Genome Biol. 2004, 5, R62. [Google Scholar] [CrossRef]
- Boeger, H.; Griesenbeck, J.; Strattan, J.S.; Kornberg, R.D. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 2003, 11, 1587–1598. [Google Scholar] [CrossRef]
- Reinke, H.; Hörz, W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 2003, 11, 1599–1607. [Google Scholar] [CrossRef]
- Proft, M.; Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 2002, 9, 1307–1317. [Google Scholar] [CrossRef]
- Hasan, R.; Leroy, C.; Isnard, A.-D.; Labarre, J.; Boy-Marcotte, E.; Toledano, M.B. The control of the yeast H2O2 response by the Msn2/4 transcription factors. Mol. Microbiol. 2002, 45, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Mat Nanyan, N.S.; Takagi, H. Proline homeostasis in Saccharomyces cerevisiae: How does the stress-responsive transcription factor Msn2 play a role? Front. Genet. 2020, 11, 534672. [Google Scholar] [CrossRef] [PubMed]
- Alepuz, P.M.; Jovanovic, A.; Reiser, V.; Ammerer, G. Stress-induced MAP kinase Hog1 is part of transcription activation complexes. Mol. Cell 2001, 7, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Schüller, C.; Brewster, J.L.; Alexander, M.R.; Gustin, M.C.; Ruis, H. The HOG pathway controls osmotic regulation of transcription via the Stress Response Element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994, 13, 4382–4389. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Monge, R.; Navarro-García, F.; Román, E.; Negredo, A.I.; Eisman, B.; Nombela, C.; Pla, J. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2003, 2, 351–361. [Google Scholar] [CrossRef]
- Smith, D.A.; Nicholls, S.; Morgan, B.A.; Brown, A.J.P.; Quinn, J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. MBoC 2004, 15, 4179–4190. [Google Scholar] [CrossRef]
- Sadeh, A.; Volokh, M.; Aharoni, A. Conserved motifs in the Msn2-activating domain are important for Msn2-mediated yeast stress response. J. Cell Sci. 2012, 125, 3333–3342. [Google Scholar] [CrossRef]
- Brown, J.L.; Bussey, H.; Stewart, R.C. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994, 13, 5186–5194. [Google Scholar] [CrossRef]
- Ketela, T.; Brown, J.L.; Stewart, R.C.; Bussey, H. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol. Gen. Genet. 1998, 259, 372–378. [Google Scholar] [CrossRef]
- Fassler, J.S.; West, A.H. Fungal Skn7 stress responses and their relationship to virulence. Eukaryot. Cell 2011, 10, 156–167. [Google Scholar] [CrossRef]
- Alberts, A.S.; Bouquin, N.; Johnston, L.H. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein β subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 1998, 273, 8616–8622. [Google Scholar] [CrossRef] [PubMed]
- Lettow, J.; Kliewe, F.; Aref, R.; Schüller, H.-J. Functional characterization and comparative analysis of gene repression-mediating domains interacting with yeast pleiotropic corepressors Sin3, Cyc8 and Tup1. Curr. Genet. 2023, 69, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Moye-Rowley, W.S.; Harshman, K.D.; Parker, C.S. Yeast YAP1 encodes a novel form of the Jun family of transcriptional activator proteins. Genes Dev. 1989, 3, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, L.; Rodrigues-Pousada, C.; Struhl, K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell Biol. 1997, 17, 6982–6993. [Google Scholar] [CrossRef]
- Rodrigues-Pousada, C.; Devaux, F.; Caetano, S.M.; Pimentel, C.; da Silva, S.; Cordeiro, A.C.; Amaral, C. Yeast AP-1 like transcription factors (Yap) and stress response: A current overview. Microb. Cell 2019, 6, 267–285. [Google Scholar] [CrossRef]
- Harshman, K.D.; Moye-Rowley, W.S.; Parker, C.S. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell 1988, 53, 321–330. [Google Scholar] [CrossRef]
- Kuge, S.; Jones, N.; Nomoto, A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997, 16, 1710–1720. [Google Scholar] [CrossRef]
- Yan, L.; Lee, L.H.; Davis, L.I. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998, 17, 7416–7429. [Google Scholar] [CrossRef]
- Azevedo, D.; Tacnet, F.; Delaunay, A.; Rodrigues-Pousada, C.; Toledano, M.B. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Rad. Biol. Med. 2003, 35, 889–900. [Google Scholar] [CrossRef]
- Mendoza-Martínez, A.E.; Cano-Domínguez, N.; Aguirre, J. Yap1 homologs mediate more than the redox regulation of the antioxidant response in filamentous fungi. Fungal Biol. 2020, 124, 253–262. [Google Scholar] [CrossRef]




| Gene | Primer | Sequence (5′–3′) | Amplicon Size (bp) |
|---|---|---|---|
| DhCTA | DhCTAFw | AAGGTCTTGCCACATAAGG | 145 |
| DhCTARv | GATCAGCAGAAGCTTCCATG | ||
| DhCTT | DhCTTFw | ATGAAAGAATTGCTGCTGG | 150 |
| DhCTTRv | GGCCAAACTTTCTTAATGGG | ||
| DhSTL1 | DhSTL1Fw | TGGGAATGGCTGACACTTATG | 118 |
| DhSTL1Rv | GCTCTTCTACCCAACCTATCAATC | ||
| DhRPS3 | DhS3Fw | AAGGACCCAGCAACCAACAA | 148 |
| DhS3Rv | AAGCGGAAGCTTCAACTGGT | ||
| DhACT1 | DhACT1Fw | CCCAGAAGAACACCCAGTTT | 125 |
| DhACT1Rv | CGGCTTGGATAGAAACGTAGAA |
| Protein in D. hansenii | Position | Domain | % Identity | ||
|---|---|---|---|---|---|
| S. cerevisiae | C. albicans | ||||
| DhMsn2/4 | 368–419 | C2H2 Zinc Finger | 75 | 79 | 89 |
| DhSkn7 | 26–131 | HSF-type | 61 | 85 | |
| 397–513 | Response Regulator | 73 | 95 | ||
| DhSko1 | 74–97 | Hog1-interaction domain | 52 | 58 | |
| 226–372 | Repression domain | 14 | 25 | ||
| 233–243 | Hydrophobic patch | 63 | 82 | ||
| 413–430 | PKA interaction region | 33 | 24 | ||
| 466–487 | bZIP | 82 | 91 | ||
| 491–514 | bZIP2 | 33 | 75 | ||
| DhYap1 | 39–99 | bZIP | 62 | 80 | |
| 41–48 | NLS | 100 | 100 | ||
| 243–281 | nCRD | 44 | 69 | ||
| 417–462 | cCRD | 66 | 91 | ||
| 439–457 | NES | 63 | 100 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de la Fuente-Colmenares, I.; González, J.; Sánchez, N.S.; Ochoa-Gutiérrez, D.; Escobar-Sánchez, V.; Segal-Kischinevzky, C. Regulation of Catalase Expression and Activity by DhHog1 in the Halotolerant Yeast Debaryomyces hansenii Under Saline and Oxidative Conditions. J. Fungi 2024, 10, 740. https://doi.org/10.3390/jof10110740
de la Fuente-Colmenares I, González J, Sánchez NS, Ochoa-Gutiérrez D, Escobar-Sánchez V, Segal-Kischinevzky C. Regulation of Catalase Expression and Activity by DhHog1 in the Halotolerant Yeast Debaryomyces hansenii Under Saline and Oxidative Conditions. Journal of Fungi. 2024; 10(11):740. https://doi.org/10.3390/jof10110740
Chicago/Turabian Stylede la Fuente-Colmenares, Ileana, James González, Norma Silvia Sánchez, Daniel Ochoa-Gutiérrez, Viviana Escobar-Sánchez, and Claudia Segal-Kischinevzky. 2024. "Regulation of Catalase Expression and Activity by DhHog1 in the Halotolerant Yeast Debaryomyces hansenii Under Saline and Oxidative Conditions" Journal of Fungi 10, no. 11: 740. https://doi.org/10.3390/jof10110740
APA Stylede la Fuente-Colmenares, I., González, J., Sánchez, N. S., Ochoa-Gutiérrez, D., Escobar-Sánchez, V., & Segal-Kischinevzky, C. (2024). Regulation of Catalase Expression and Activity by DhHog1 in the Halotolerant Yeast Debaryomyces hansenii Under Saline and Oxidative Conditions. Journal of Fungi, 10(11), 740. https://doi.org/10.3390/jof10110740








