Effector-Mediated Suppression of Programmed Cell Death by Phytophthora palmivora in Oil Palm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Effector Selection
2.2. Alignment and Structure Prediction
2.3. Plasmid Construction
2.4. Plant Material
2.5. Particle Co-Bombardment of Oil Palm Leaflet Assays
3. Results
3.1. Effector-Mediated Suppression of PCD Induced in Oil Palm Leaflets
3.2. Effect of Effectors on PCD Induced in Oil Palm Leaflets: A Comparative Analysis of Genotype Responses
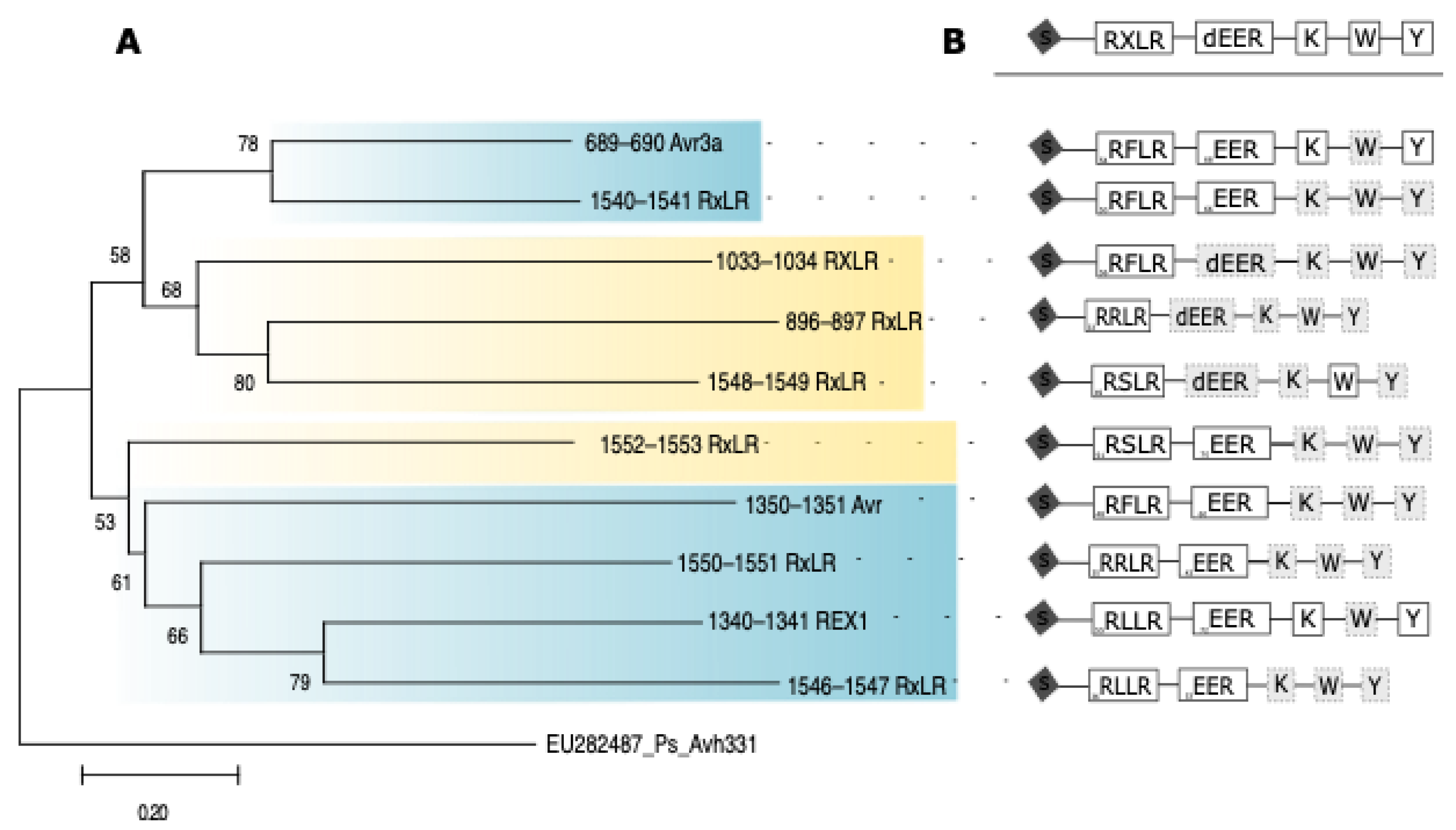
3.3. Suppression of PCD by Diverse Predicted Effector Containing Motifs
4. Discussion
4.1. Effector-Mediated PCD Suppression
4.2. Variable Anti-PCD Activity
4.3. Unique Effector Behavior
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barcelos, E.; De Almeida Rios, S.; Cunha, R.N.V.; Lopes, R.; Motoike, S.Y.; Babiychuk, E.; Skirycz, A.; Kushnir, S. Oil palm natural diversity and the potential for yield improvement. Front. Plant Sci. 2015, 6, 190. [Google Scholar] [CrossRef] [PubMed]
- Fedepalma. Statistical Yearbook 2023. In The Oil Palm Agroindustry in Colombia and the World 2018–2022; Fedepalma: Bogota, Colombia, 2023; p. 237. [Google Scholar]
- Torres, G.; Sarria, G.; Martinez, G.; Varon, F.; Drenth, A.; Guest, D. Bud Rot Caused by Phytophthora palmivora: A Destructive Emerging Disease of Oil Palm. Phytopathology 2016, 106, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Drenth, A.; Guest, D.I. Fungal and Oomycete Diseases of Tropical Tree Fruit Crops. Annu. Rev. Phytopathol. 2016, 54, 373–395. [Google Scholar] [CrossRef] [PubMed]
- Drenth, A.; Guest, D.I. Diversity and Management of Phytophthora in Southeast Asia; ACIAR: Canberra, Australia, 2004; Volume 114, pp. 7–9. Available online: https://www.aciar.gov.au/publication/books-and-manuals/diversity-and-management-phytophthora-southeast-asia (accessed on 1 October 2024).
- Mohamed Azni, I.N.A.; Sundram, S.; Ramachandran, V. Pathogenicity of Malaysian Phytophthora palmivora on cocoa, durian, rubber and oil palm determines the threat of bud rot disease. For. Pathol. 2019, 49, e12557. [Google Scholar] [CrossRef]
- Benitez, E.; Garcia, C. The history of research on oil palm bud rot (Elaeis guineensis Jacq.) in Colombia. Agron. Colombiana 2015, 32, 390–398. [Google Scholar] [CrossRef]
- Cock, J.H.; Ayala-Diaz, I.M.; Romero, H.M. A scheme for distribution of genetically improved oil palm plants well suited to local conditions. Agric. Syst. 2023, 212, 103756. [Google Scholar] [CrossRef]
- John Martin, J.J.; Yarra, R.; Wei, L.; Cao, H. Oil palm breeding in the modern era: Challenges and opportunities. Plants 2022, 11, 1395. [Google Scholar] [CrossRef]
- Wang, L.; Lee, M.; Yi Wan, Z.; Bai, B.; Ye, B.; Alfiko, Y.; Rahmadsyah, R.; Purwantomo, S.; Song, Z.; Suwanto, A.; et al. Chromosome-level reference genome of African oil palm provides insights into its divergence and stress adaptation. Genom. Proteom. Bioinform. 2023, 21, 440. [Google Scholar] [CrossRef]
- Vleeshouwers, V.G.A.A.; Oliver, R.P. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant-Microbe Interact. 2014, 27, 196–206. [Google Scholar] [CrossRef]
- Singh, R.; Ong-Abdullah, M.; Low, E.-T.L.; Manaf, M.A.A.; Rosli, R.; Nookiah, R.; Ooi, L.C.-L.; Ooi, S.E.; Chan, K.-L.; Halim, M.A. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 2013, 500, 335–339. [Google Scholar] [CrossRef]
- Gil, J.; Herrera, M.; Duitama, J.; Sarria, G.; Restrepo, S.; Romero, H.M. Genomic variability of Phytophthora palmivora isolates from different oil palm cultivation regions in Colombia. Phytopathology 2020, 110, 1553–1564. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Shao, J.; Lary, D.J.; Kronmiller, B.A.; Shen, D.; Strem, M.D.; Amoako-Attah, I.; Akrofi, A.Y.; Begoude, B.A.D.; ten Hoopen, G.M.; et al. Phytophthora megakarya and Phytophthora palmivora, closely related causal agents of cacao black pod rot, underwent increases in genome sizes and gene numbers by different mechanisms. Genome Biol. Evol. 2017, 9, 536–557. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Newman, M.; Yu, H.; Rashidzade, M.; Martínez-Soto, D.; Caicedo, A.; Allen, K.S.; Ma, L.-J. Fungal effectors: Past, present, and future. Curr. Opin. Microbiol. 2024, 81, 102526. [Google Scholar] [CrossRef] [PubMed]
- Uhse, S.; Djamei, A. Effectors of plant-colonizing fungi and beyond. PLoS Path. 2018, 14, e1006992. [Google Scholar] [CrossRef]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef]
- Whisson, S.C.; Boevink, P.C.; Moleleki, L.; Avrova, A.O.; Morales, J.G.; Gilroy, E.M.; Armstrong, M.R.; Grouffaud, S.; Van West, P.; Chapman, S.; et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 2007, 450, 115–118. [Google Scholar] [CrossRef]
- van Damme, M.; Cano, L.M.; Oliva, R.; Schornack, S.; Segretin, M.E.; Kamoun, S.; Raffaele, S. Evolutionary and Functional Dynamics of Oomycete Effector Genes. In Effectors in Plant–Microbe Interactions; Martin, F., Kamoun, S., Eds.; Wiley-Blackwell: Chichester, UK, 2011; pp. 101–120. [Google Scholar]
- Stam, R.; Jupe, J.; Howden, A.J.M.; Morris, J.A.; Boevink, P.C.; Hedley, P.E.; Huitema, E. Identification and Characterisation CRN Effectors in Phytophthora capsici Shows Modularity and Functional Diversity. PLoS ONE 2013, 8, e59517. [Google Scholar] [CrossRef]
- Tyler, B.M.; Tripathy, S.; Zhang, X.; Dehal, P.; Jiang, R.H.Y.; Aerts, A.; Arredondo, F.D.; Baxter, L.; Bensasson, D.; Beynon, J.L.; et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 2006, 313, 1261–1266. [Google Scholar] [CrossRef]
- Haas, B.J.; Kamoun, S.; Zody, M.C.; Jiang, R.H.Y.; Handsaker, R.E.; Cano, L.M.; Grabherr, M.; Kodira, C.D.; Raffaele, S.; Torto-Alalibo, T.; et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 2009, 461, 393–398. [Google Scholar] [CrossRef]
- Schornack, S.; Van Damme, M.; Bozkurt, T.O.; Cano, L.M.; Smoker, M.; Thines, M.; Gaulin, E.; Kamoun, S.; Huitema, E. Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA 2010, 107, 17421–17426. [Google Scholar] [CrossRef]
- Birch, P.R.J.; Rehmany, A.P.; Pritchard, L.; Kamoun, S.; Beynon, J.L. Trafficking arms: Oomycete effectors enter host plant cells. Trends Microbiol. 2006, 14, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Morgan, W.; Kamoun, S. RXLR effectors of plant pathogenic oomycetes. Curr. Opin. Microbiol. 2007, 10, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Deb, D.; Anderson, R.G.; How-Yew-Kin, T.; Tyler, B.M.; McDowell, J.M. Conserved RxLR effectors from oomycetes Hyaloperonospora arabidopsidis and Phytophthora sojae suppress PAMP- and effector-triggered immunity in diverse plants. Mol. Plant-Microbe Interact. 2018, 31, 374–385. [Google Scholar] [CrossRef]
- Evangelisti, E.; Gogleva, A.; Hainaux, T.; Doumane, M.; Tulin, F.; Quan, C.; Yunusov, T.; Floch, K.; Schornack, S. Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors. BMC Biol. 2017, 15, 39. [Google Scholar] [CrossRef]
- Masani, M.Y.A.; Noll, G.A.; Parveez, G.K.A.; Sambanthamurthi, R.; Prüfer, D. Efficient transformation of oil palm protoplasts by PEG-mediated transfection and DNA microinjection. PLoS ONE 2014, 9, e96831. [Google Scholar] [CrossRef]
- Mohan Jain, S. Date palm biotechnology: Current status and prospective-an overview. Emir. J. Food Agric. 2012, 24, 386–399. [Google Scholar]
- Dou, D.; Kale, S.D.; Wang, X.; Chen, Y.; Wang, Q.; Wang, X.; Jiang, R.H.Y.; Arredondo, F.D.; Anderson, R.G.; Thakur, P.B.; et al. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 2008, 20, 1118–1133. [Google Scholar] [CrossRef]
- Kale, S.D.; Tyler, B.M. Assaying effector function in planta using double-barreled particle bombardment. Methods Mol. Biol. 2011, 712, 153–172. [Google Scholar]
- Zubaidah, R.; Nurniwalis, A.W.; Chan, P.L.; Siti Masura, S.; Siti Nor Akmar, A.; Parveez, G.K.A. Tissue-specific promoters: The importance and potential application for genetic engineering in oil palm. J. Oil Palm Res. 2018, 30, 1–12. [Google Scholar]
- Ahmad Parveez, G.K.; Majid, N.i.A.; Zainal, A.; Rasid, O.A. Determination of minimal inhibitory concentration of selection agents for selecting transformed immature embryos of oil palm. Asia-Pac. J. Mol. Biol. Biotechnol. 2007, 15, 133–146. [Google Scholar]
- Omidvar, V.; Siti Nor Akmar, A.; Marziah, M.; Maheran, A.A. A transient assay to evaluate the expression of polyhydroxybutyrate genes regulated by oil palm mesocarp-specific promoter. Plant Cell Rep. 2008, 27, 1451–1459. [Google Scholar] [CrossRef]
- Parveez, G.K.A.; Majid, N.I.A. Factors affecting green fluorescence protein (GFP) gene expression in oil palm after microprojectile bombardment. J. Oil Palm Res. 2008, 20, 495–507. [Google Scholar]
- Baek, D.; Nam, J.; Koo, Y.D.; Kim, D.H.; Lee, J.; Jeong, J.C.; Kwak, S.S.; Chung, W.S.; Lim, C.O.; Bahk, J.D.; et al. Bax-induced cell death of Arabidopsis is meditated through reactive oxygen-dependent and -independent processes. Plant Mol. Biol. 2004, 56, 15–27. [Google Scholar] [CrossRef]
- Lacomme, C.; Santa Cruz, S. Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. USA 1999, 96, 7956–7961. [Google Scholar] [CrossRef]
- Li, Z.; Yin, Z.; Fan, Y.; Xu, M.; Kang, Z.; Huang, L. Candidate effector proteins of the necrotrophic apple canker pathogen Valsa mali can suppress BAX-induced PCD. Front. Plant Sci. 2015, 6, 579. [Google Scholar] [CrossRef]
- Avila-Mendez, K.; Rodrigo, Á.; Araque, L.; Romero, H.M. Simultaneous transcriptome analysis of oil palm clones and Phytophthora palmivora reveals oil palm defense strategies. PLoS ONE 2019, 14, e0222774. [Google Scholar] [CrossRef]
- Ávila-Méndez, K.; Avila-Diazgranados, R.; Pardo, A.; Herrera, M.; Sarria, G.; Romero, H.M. Response of in vitro obtained oil palm and interspecific OxG hybrids to inoculation with Phytophthora palmivora. For. Pathol. 2019, 49, e12486. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of Apoplastic and Cytoplasmic Effectors in Fungi and Oomycetes. Mol. Plant-Microbe Interact. MPMI 2022, 35, 146–156. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N.; Singh, K.B.; Taylor, J.M. ApoplastP: Prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 2017, 217, 1764–1778. [Google Scholar] [CrossRef]
- Gogleva, A.; Drost, H.G.; Schornack, S. SecretSanta: Flexible pipelines for functional secretome prediction. Bioinformatics 2018, 34, 2295–2296. [Google Scholar] [CrossRef]
- García-Gaona, M.; Botero-Rozo, D.; Araque, L.; Romero, H.M. The dynamic interaction between oil palm and Phytophthora palmivora in bud rot disease: Insights from transcriptomic analysis and network modelling. J. Fungi 2024, 10, 164. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Dou, D.; Kale, S.D.; Wang, X.; Jiang, R.H.; Bruce, N.A.; Arredondo, F.D.; Zhang, X.; Tyler, B.M. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 2008, 20, 1930–1947. [Google Scholar] [CrossRef]
- Parveez, G.K.A.; Masri, M.M.; Zainal, A.; Majid, N.i.A.; Masani, A.; Yunus, M.; Fadilah, H.H.; Rasid, O.; Cheah, S.-C. Transgenic oil palm: Production and projection. Biochem. Soc. Trans. 2000, 28, 969–972. [Google Scholar] [CrossRef]
- Prasad, L.; Katoch, S.; Shahid, S. Microbial interaction mediated programmed cell death in plants. 3 Biotech 2022, 12, 43. [Google Scholar] [CrossRef]
- Thanthrige, N.; Jain, S.; Bhowmik, S.D.; Ferguson, B.J.; Kabbage, M.; Mundree, S.; Williams, B. Centrality of BAGs in plant PCD, stress responses, and host defense. Trends Plant Sci. 2020, 25, 1131–1140. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Si, J.; Han, Z.; Chen, D. Action mechanisms of effectors in plant-pathogen interaction. Int. J. Mol. Sci. 2022, 23, 6758. [Google Scholar] [CrossRef]
- Li, S.; Hanlon, R.; Wise, H.; Pal, N.; Brar, H.; Liao, C.; Gao, H.; Perez, E.; Zhou, L.; Tyler, B.M.; et al. Interaction of Phytophthora sojae Effector Avr1b With E3 Ubiquitin Ligase GmPUB1 Is Required for Recognition by Soybeans Carrying Phytophthora Resistance Rps1-b and Rps1-k Genes. Front. Plant Sci. 2021, 12, 725571. [Google Scholar] [CrossRef]
- Du, Y.; Mpina, M.H.; Birch, P.R.J.; Bouwmeester, K.; Govers, F. Phytophthora infestans RXLR Effector AVR1 Interacts with Exocyst Component Sec5 to Manipulate Plant Immunity. Plant Physiol. 2015, 169, 1975–1990. [Google Scholar] [CrossRef]
- Mindrinos, M.; Katagiri, F.; Yu, G.L.; Ausubel, F.M. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 1994, 78, 1089–1099. [Google Scholar] [CrossRef]
- Bos, J.I.B.; Armstrong, M.R.; Gilroy, E.M.; Boevink, P.C.; Hein, I.; Taylor, R.M.; Zhendong, T.; Engelhardt, S.; Vetukuri, R.R.; Harrower, B.; et al. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 2010, 107, 9909–9914. [Google Scholar] [CrossRef]
- Boutemy, L.S.; King, S.R.F.; Win, J.; Hughes, R.K.; Clarke, T.A.; Blumenschein, T.M.A.; Kamoun, S.; Banfield, M.J. Structures of Phytophthora RXLR effector proteins: A conserved but adaptable fold underpins functional diversity. J. Biol. Chem. 2011, 286, 35834–35842. [Google Scholar] [CrossRef]
- Combier, M.; Evangelisti, E.; Piron, M.C.; Schornack, S.; Mestre, P. Candidate effector proteins from the oomycetes Plasmopara viticola and Phytophthora parasitica share similar predicted structures and induce cell death in Nicotiana species. PLoS ONE 2022, 17, e0278778. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018, 219, 17–24. [Google Scholar] [CrossRef]

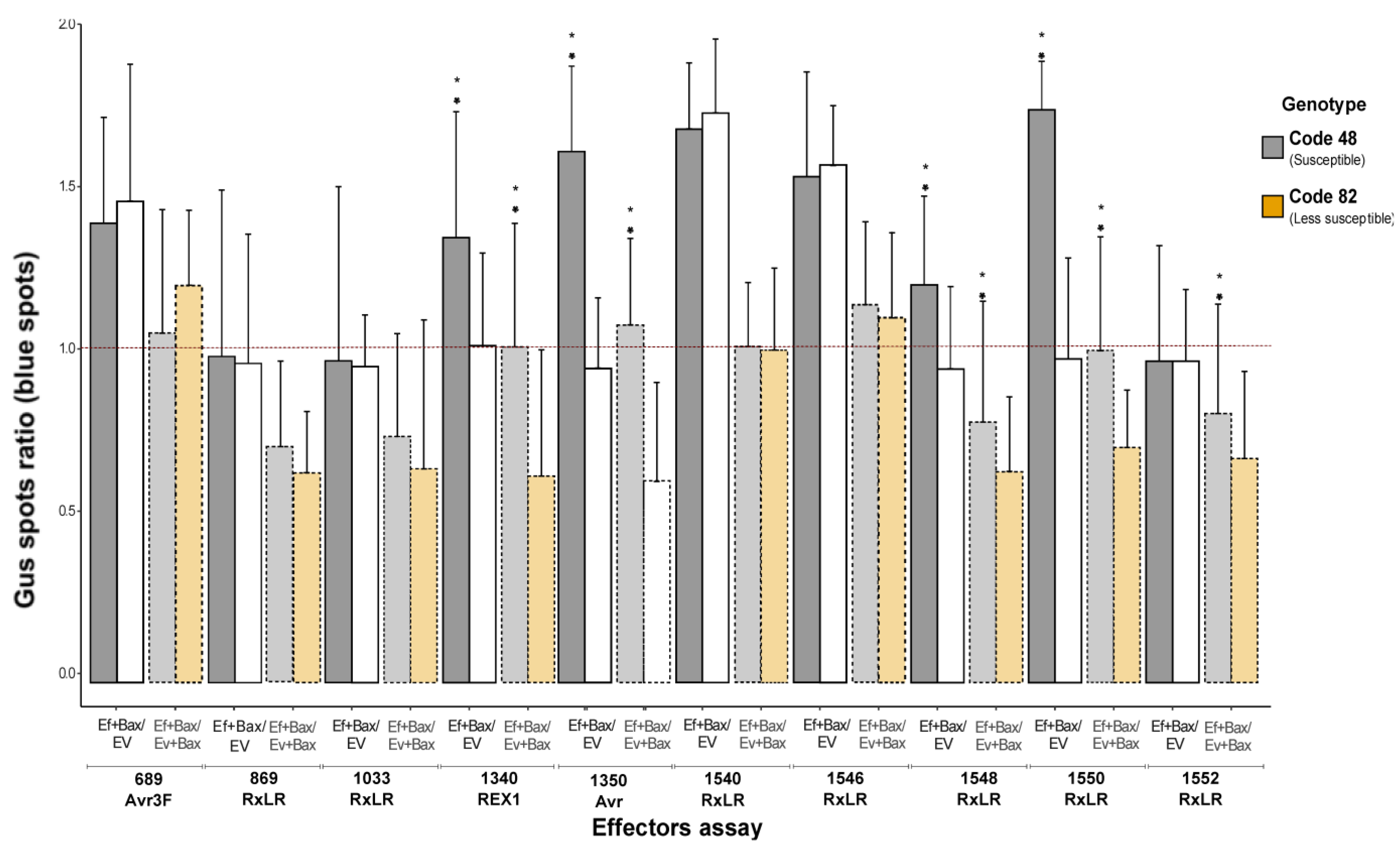
| Indirect Assay | Direct Assay | |||||||
|---|---|---|---|---|---|---|---|---|
| Genotype * | Effector | (BAX + EV)/Ev (a) | (Effector)/EV (a) | Ratio (b) | p Value (c1) | (BAX + effector + EV)/(BAX + EV) (a) | p Value (c2) | Anti-PCD activity (d) |
| 48 | 689-690-Avr3F | 0.82 ± 0.08 | 1.14 ± 0.88 | 1.39 | p < 0.05 | 1.07 ± 0.41 | p < 0.01 | Yes |
| 896-897-RxLR | 0.82 ± 0.08 | 0.80 ± 0.45 | 0.98 | p > 0.01 | 0.71 ± 0.26 | p < 0.01 | No | |
| 1033-1034-RxLR | 0.82 ± 0.08 | 0.80 ± 0.38 | 0.97 | p > 0.01 | 0.73 ± 0.14 | p > 0.01 | No | |
| 1340-1341-REX1 | 0.82 ± 0.08 | 1.12 ± 0.42 | 1.36 | p < 0.01 | 1.04 ± 0.38 | p < 0.01 | Yes | |
| 1350-1351-Avr | 0.82 ± 0.08 | 1.32 ± 0.42 | 1.61 | p < 0.05 | 1.10 ± 0.27 | p < 0.01 | Yes | |
| 1540-1541-RxLR | 0.82 ± 0.08 | 1.38 ± 0.29 | 1.68 | p < 0.01 | 1.06 ± 0.39 | p < 0.05 | Yes | |
| 1546-147-RxLR | 0.82 ± 0.08 | 1.25 ± 0.32 | 1.53 | p < 0.01 | 1.15 ± 0.27 | p > 0.01 | Yes | |
| 1548-1549-RxLR | 0.82 ± 0.08 | 1.03 ± 0.40 | 1.25 | p > 0.1 | 0.77 ± 0.37 | p > 0.1 | No | |
| 1550-1551-RxLR | 0.82 ± 0.08 | 1.44 ± 0.25 | 1.76 | p < 0.01 | 1.01 ± 0.41 | p < 0.01 | Yes | |
| 1552-1553-RxLR | 0.82 ± 0.08 | 0.81 ± 0.35 | 0.98 | p < 0.05 | 0.79 ± 0.23 | p < 0.01 | No | |
| 82 | 689-690-Avr3F | 0.73 ± 0.22 | 1.06 ± 0.51 | 1.45 | p < 0.01 | 1.20 ± 0.45 | p < 0.01 | Yes |
| 896-897-RxLR | 0.73 ± 0.22 | 0.70 ± 0.15 | 0.96 | p < 0.01 | 0.63 ± 0.20 | p < 0.01 | No | |
| 1033-1034-RxLR | 0.73 ± 0.22 | 0.70 ± 0.18 | 0.96 | p < 0.01 | 0.63 ± 0.28 | p < 0.01 | No | |
| 1340-1341-REX1 | 0.73 ± 0.22 | 0.78 ± 0.29 | 1.07 | p < 0.05 | 0.62 ± 0.39 | p < 0.01 | No | |
| 1350-1351-Avr | 0.73 ± 0.22 | 0.69 ± 0.22 | 0.95 | p < 0.1 | 0.59 ± 0.30 | p < 0.01 | No | |
| 1540-1541-RxLR | 0.73 ± 0.22 | 1.28 ± 0.42 | 1.76 | p < 0.05 | 1.01 ± 0.25 | p < 0.01 | Yes | |
| 1546-147-RxLR | 0.73 ± 0.22 | 1.14 ± 0.20 | 1.57 | p < 0.05 | 1.10 ± 0.32 | p < 0.01 | Yes | |
| 1548-1549-RxLR | 0.73 ± 0.22 | 0.69 ± 0.20 | 0.95 | p < 0.01 | 0.63 ± 0.23 | p < 0.01 | No | |
| 1550-1551-RxLR | 0.73 ± 0.22 | 0.71 ± 0.30 | 0.98 | p > 0.1 | 0.70 ± 0.17 | p < 0.01 | No | |
| 1552-1553-RxLR | 0.73 ± 0.22 | 0.71 ± 0.18 | 0.98 | p > 0.1 | 0.68 ± 0.27 | p > 0.1 | No | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Cruz, M.C.; Montoya, C.; Ayala-Diaz, I.; Araque, L.; Romero, H.M. Effector-Mediated Suppression of Programmed Cell Death by Phytophthora palmivora in Oil Palm. J. Fungi 2024, 10, 750. https://doi.org/10.3390/jof10110750
Rodríguez-Cruz MC, Montoya C, Ayala-Diaz I, Araque L, Romero HM. Effector-Mediated Suppression of Programmed Cell Death by Phytophthora palmivora in Oil Palm. Journal of Fungi. 2024; 10(11):750. https://doi.org/10.3390/jof10110750
Chicago/Turabian StyleRodríguez-Cruz, María Camila, Carmenza Montoya, Iván Ayala-Diaz, Leonardo Araque, and Hernán Mauricio Romero. 2024. "Effector-Mediated Suppression of Programmed Cell Death by Phytophthora palmivora in Oil Palm" Journal of Fungi 10, no. 11: 750. https://doi.org/10.3390/jof10110750
APA StyleRodríguez-Cruz, M. C., Montoya, C., Ayala-Diaz, I., Araque, L., & Romero, H. M. (2024). Effector-Mediated Suppression of Programmed Cell Death by Phytophthora palmivora in Oil Palm. Journal of Fungi, 10(11), 750. https://doi.org/10.3390/jof10110750






