Comparative Genomics of Different Lifestyle Fungi in Helotiales (Leotiomycetes) Reveals Temperature and Ecosystem Adaptations
Abstract
:1. Introduction
2. Materials and Methods
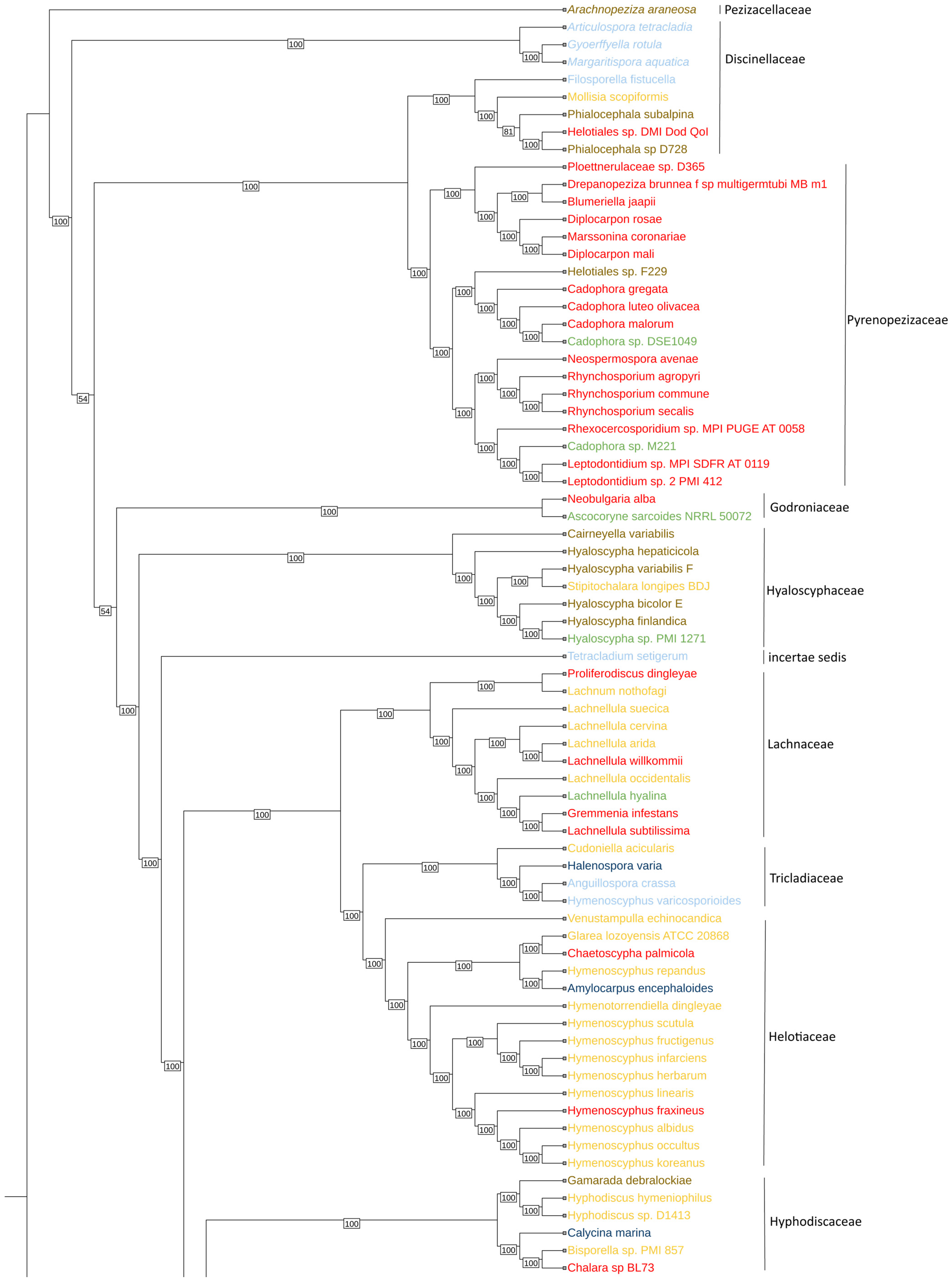
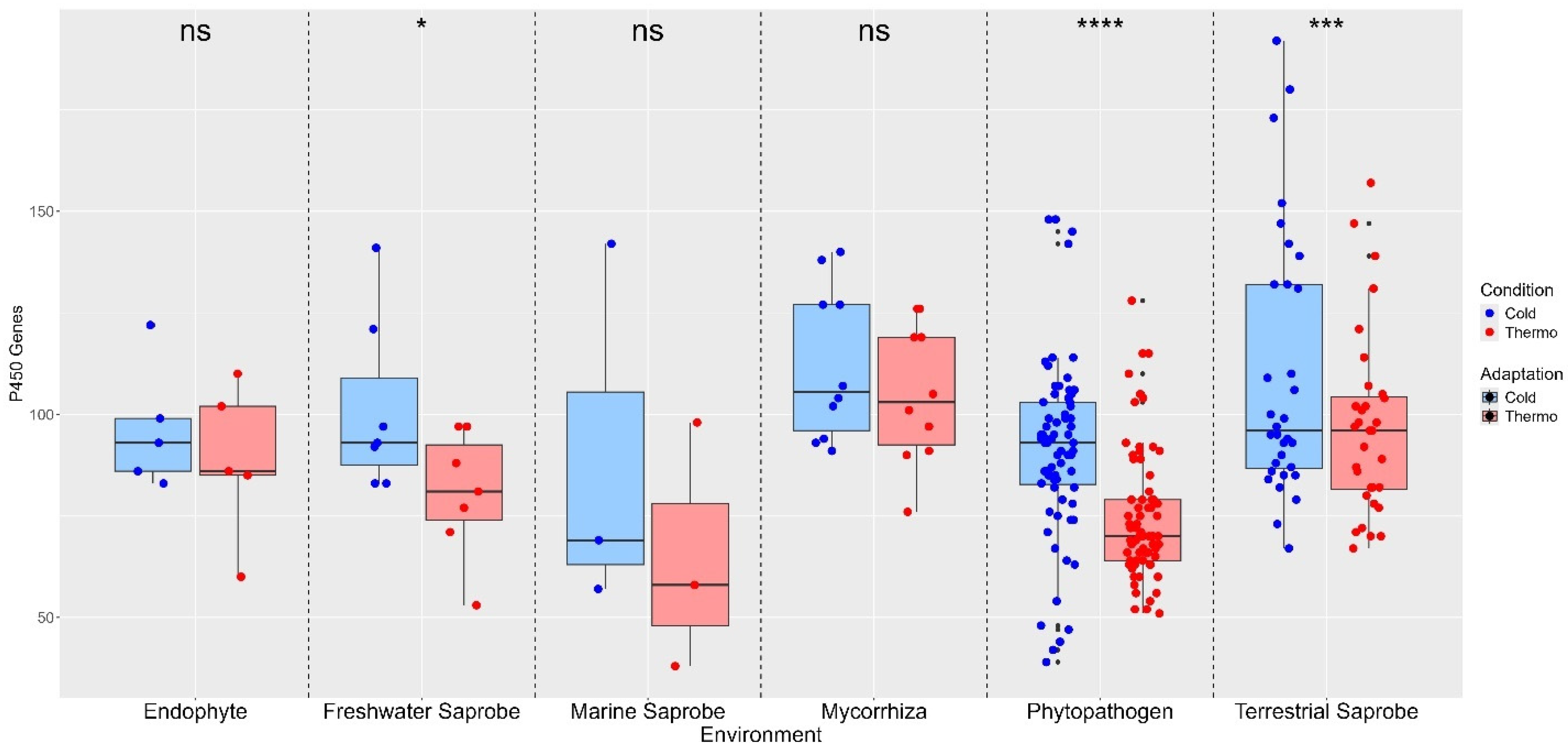
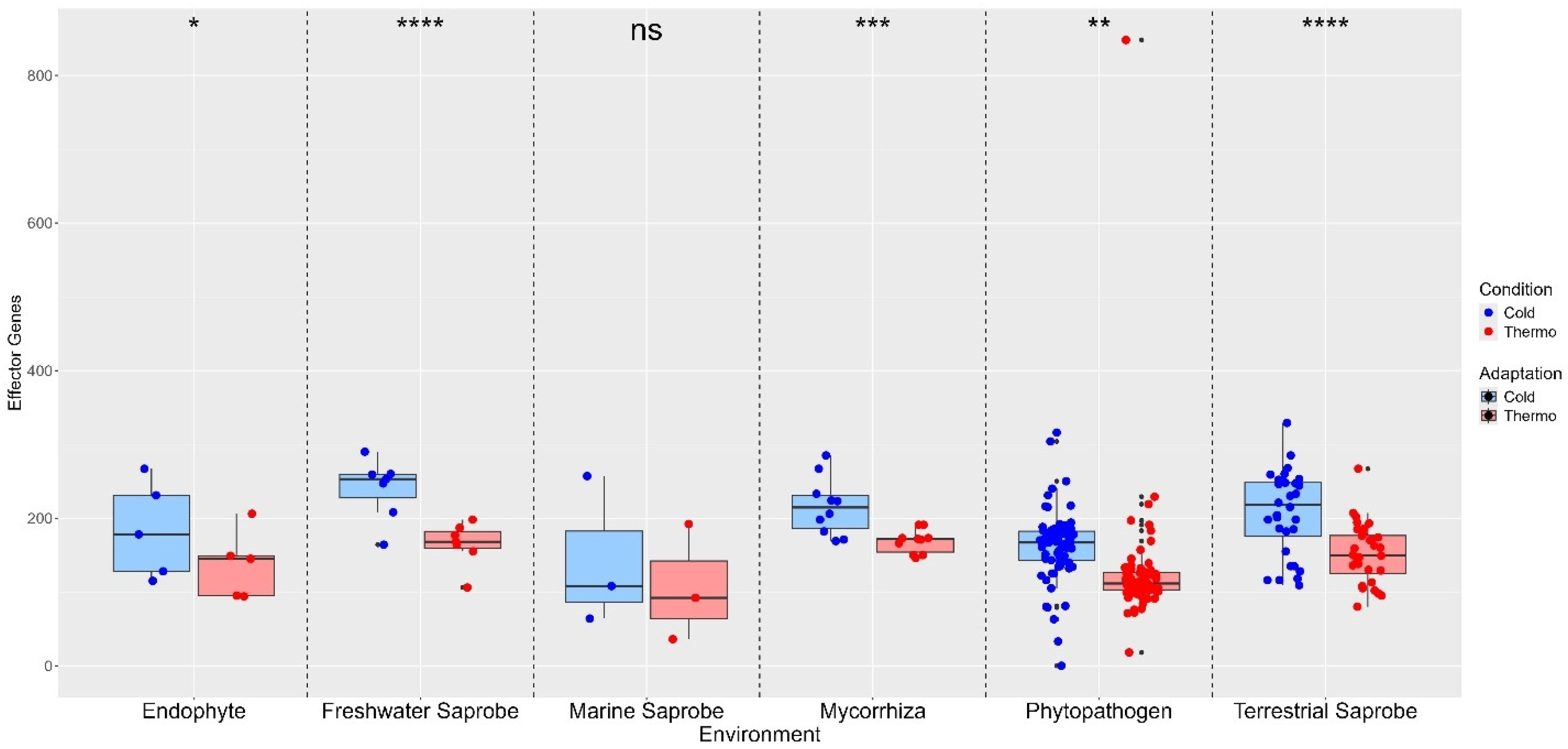
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Z.; Johnston, P.R.; Takamatsu, S.; Spatafora, J.W.; Hibbett, D.S. Toward a Phylogenetic Classification of the Leotiomycetes Based on rDNA Data. Mycologia 2006, 98, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Johnston, P.R.; Quijada, L.; Smith, C.A.; Baral, H.O.; Hosoya, T.; Baschien, C.; Pärtel, K.; Zhuang, W.Y.; Haelewaters, D.; Park, D.; et al. A Multigene Phylogeny toward a New Phylogenetic Classification of Leotiomycetes. IMA Fungus 2019, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, H.T.; Liu, J.K.; Maharachchikumbura, S.S.N.; Ekanayaka, A.H.; Tian, Q.; Phookamsak, R. Outline of Ascomycota: 2017. Fungal Divers. 2018, 88, 167–263. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Rajeshkumar, K.C.; Hawksworth, D.L.; Madrid, H.; Kirk, P.M.; Braun, U.; Singh, R.V.; Crous, P.W.; Kukwa, M.; et al. Notes for Genera: Ascomycota. Fungal Divers. 2017, 86, 1–594. [Google Scholar]
- Dai, D.Q.; Wijayawardene, N.N.; Zhang, G.Q.; Gao, Y.; Wijayawardene, N.N.; Hyde, K.D.; Pem, D.; Thiyagaraja, V.; Dong, W.; Sánchez-García, M.; et al. Outline of Fungi and Fungus-like Taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Quandt, C.A.; Haelewaters, D. Phylogenetic Advances in Leotiomycetes, an Understudied Clade of Taxonomically and Ecologically Diverse Fungi. Encycl. Mycol. 2021, 1, 284–294. [Google Scholar] [CrossRef]
- Baral, H.O.; Marson, G.; Bogale, M.; Untereiner, W.A. Xerombrophila crystallifera, a New Genus and Species in the Helotiales. Mycol. Prog. 2013, 12, 475–488. [Google Scholar] [CrossRef]
- Wang, Z.; Binder, M.; Schoch, C.L.; Johnston, P.R.; Spatafora, J.W.; Hibbett, D.S. Evolution of Helotialean Fungi (Leotiomycetes, Pezizomycotina): A Nuclear rDNA Phylogeny. Mol. Phylogenet Evol. 2006, 41, 295–312. [Google Scholar] [CrossRef]
- Hosoya, T. Systematics, Ecology, and Application of Helotiales: Recent Progress and Future Perspectives for Research with Special Emphasis on Activities within Japan. Mycoscience 2021, 62, 1–9. [Google Scholar] [CrossRef]
- Liu, S.L.; Wang, X.W.; Li, G.J.; Deng, C.Y.; Rossi, W.; Leonardi, M.; Liimatainen, K.; Kekki, T.; Niskanen, T.; Smith, M.E.; et al. Fungal Diversity Notes 1717–1817: Taxonomic and Phylogenetic Contributions on Genera and Species of Fungal Taxa. Fungal Divers. 2024, 124, 1–216. [Google Scholar] [CrossRef]
- Vrålstad, T.; Myhre, E.; Schumacher, T. Molecular Diversity and Phylogenetic Affinities of Symbiotic Root-Associated Ascomycetes of the Helotiales in Burnt and Metal Polluted Habitats. New Phytol. 2002, 155, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An Open Annotation Tool for Parsing Fungal Community Datasets by Ecological Guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Bruyant, P.; Moënne-Loccoz, Y.; Almario, J. Root-Associated Helotiales Fungi: Overlooked Players in Plant Nutrition. Soil. Biol. Biochem. 2024, 191, 109363. [Google Scholar] [CrossRef]
- Newsham, K.K. A Meta-Analysis of Plant Responses to Dark Septate Root Endophytes. New Phytol. 2011, 190, 783–793. [Google Scholar] [CrossRef]
- Deslippe, J.R.; Hartmann, M.; Simard, S.W.; Mohn, W.W. Long-Term Warming Alters the Composition of Arctic Soil Microbial Communities. FEMS Microbiol. Ecol. 2012, 82, 303–315. [Google Scholar] [CrossRef]
- Geml, J.; Morgado, L.N.; Semenova, T.A.; Welker, J.M.; Walker, M.D.; Smets, E. Long-Term Warming Alters Richness and Composition of Taxonomic and Functional Groups of Arctic Fungi. FEMS Microbiol. Ecol. 2015, 91, 95. [Google Scholar] [CrossRef]
- Newsham, K.K.; Garnett, M.H.; Robinson, C.H.; Cox, F. Discrete Taxa of Saprotrophic Fungi Respire Different Ages of Carbon from Antarctic Soils. Sci. Rep. 2018, 8, 7866. [Google Scholar] [CrossRef]
- Gajdošová, Z.; Caboň, M.; Kolaříková, Z.; Sudová, R.; Rydlová, J.; Turisová, I.; Turis, P.; Kučera, J.; Slovák, M. Environmental Heterogeneity Structures Root-Associated Fungal Communities in Daphne arbuscula (Thymelaeaceae), a Shrub Adapted to Extreme Rocky Habitats. Mol. Ecol. 2024, 33, e17441. [Google Scholar] [CrossRef]
- Kluge, M.; Wauthy, M.; Clemmensen, K.E.; Wurzbacher, C.; Hawkes, J.A.; Einarsdottir, K.; Rautio, M.; Stenlid, J.; Peura, S. Declining Fungal Diversity in Arctic Freshwaters along a Permafrost Thaw Gradient. Glob. Change Biol. 2021, 27, 5889–5906. [Google Scholar] [CrossRef]
- de Souza, L.M.D.; Ogaki, M.B.; Teixeira, E.A.A.; de Menezes, G.C.A.; Convey, P.; Rosa, C.A.; Rosa, L.H. Communities of Culturable Freshwater Fungi Present in Antarctic Lakes and Detection of Their Low-Temperature-Active Enzymes. Braz. J. Microbiol. 2022, 54, 1923–1933. [Google Scholar] [CrossRef]
- Rissi, D.V.; Ijaz, M.; Baschien, C. Comparative Genome Analysis of the Freshwater Fungus Filosporella fistucella Indicates Potential for Plant-Litter Degradation at Cold Temperatures. G3 Genes|Genomes|Genetics 2023, 13, jkad190. [Google Scholar] [CrossRef] [PubMed]
- Conway, K.E. The Aquatic Hyphomycetes of Central New York. Mycologia 1970, 62, 516–530. [Google Scholar] [CrossRef]
- Suberkropp, K.F.; Klug, M.J. Decomposition of Deciduous Leaf Litter in a Woodland Stream—I. A. Scanning Electron Microscopic Study. Microb. Ecol. 1974, 1, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Wood-Eggenschwiler, S.; Barlocher, F. Aquatic Hyphomycetes in Sixteen Streams in France, Germany and Switzerland. Trans. Br. Mycol. Soc. 1983, 81, 371–379. [Google Scholar] [CrossRef]
- Siles, J.A.; Vera, A.; Díaz-López, M.; García, C.; van den Hoogen, J.; Crowther, T.W.; Eisenhauer, N.; Guerra, C.; Jones, A.; Orgiazzi, A.; et al. Land-Use- and Climate-Mediated Variations in Soil Bacterial and Fungal Biomass across Europe and Their Driving Factors. Geoderma 2023, 434, 116474. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, Y.; Maestre, F.T.; Berdugo, M.; Wang, J.; Coleine, C.; Sáez-Sandino, T.; García-Velázquez, L.; Singh, B.K.; Delgado-Baquerizo, M. Water Availability Creates Global Thresholds in Multidimensional Soil Biodiversity and Functions. Nat. Ecol. Evol. 2023, 7, 1002–1011. [Google Scholar] [CrossRef]
- Peltoniemi, K.; Velmala, S.; Lloret, E.; Ollio, I.; Hyvönen, J.; Liski, E.; Brandt, K.K.; Campillo-Cora, C.; Fritze, H.; Iivonen, S.; et al. Soil and Climatic Characteristics and Farming System Shape Fungal Communities in European Wheat Fields. Agric. Ecosyst. Environ. 2024, 370, 109035. [Google Scholar] [CrossRef]
- Dunleavy, H.R.; Mack, M.C. Long-Term Experimental Warming and Fertilization Have Opposing Effects on Ectomycorrhizal Root Enzyme Activity and Fungal Community Composition in Arctic Tundra. Soil. Biol. Biochem. 2021, 154, 108151. [Google Scholar] [CrossRef]
- Bärlocher, F. Water-Borne Conidia of Aquatic Hyphomycetes: Seasonal and Yearly Patterns in Catamaran Brook, New Brunswick, Canada. Can. J. Bot. 2000, 78, 157–167. [Google Scholar] [CrossRef]
- Nikolcheva, L.G.; Bärlocher, F. Taxon-Specific Fungal Primers Reveal Unexpectedly High Diversity during Leaf Decomposition in a Stream. Mycol. Progress. 2004, 3, 41–49. [Google Scholar] [CrossRef]
- Yang, L.N.; Ren, M.; Zhan, J. Modeling Plant Diseases under Climate Change: Evolutionary Perspectives. Trends Plant Sci. 2023, 28, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Frantzeskakis, L.; Di Pietro, A.; Rep, M.; Schirawski, J.; Wu, C.H.; Panstruga, R. Rapid Evolution in Plant–Microbe Interactions—A Molecular Genomics Perspective. New Phytol. 2020, 225, 1134–1142. [Google Scholar] [CrossRef] [PubMed]
- Campos-Avelar, I.; Montoya-Martínez, A.C.; Villa-Rodríguez, E.D.; Valenzuela-Ruiz, V.; Ayala Zepeda, M.; Parra-Cota, F.I.; de los Santos Villalobos, S. The Mitigation of Phytopathogens in Wheat under Current and Future Climate Change Scenarios: Next-Generation Microbial Inoculants. Sustainability 2023, 15, 15250. [Google Scholar] [CrossRef]
- Zerouki, C.; Chakraborty, K.; Kuittinen, S.; Pappinen, A.; Turunen, O. Whole-Genome Sequence and Mass Spectrometry Study of the Snow Blight Fungus Phacidium infestans (Karsten) DSM 5139 Growing at Freezing Temperatures. Mol. Genet. Genom. 2023, 298, 1449–1466. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS Primers with Enhanced Specificity for Basidiomycetes—Application to the Identification of Mycorrhizae and Rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid Genetic Identification and Mapping of Enzymatically Amplified Ribosomal DNA from Several Cryptococcus Species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Freire, B.; Ladra, S.; Parama, J.R. Memory-Efficient Assembly Using Flye. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 19, 3564–3577. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Author, C.; Zhao, Q.; Ehc, M.; Circolo Micologico, A.; Carini, G. Preliminary Classification of Leotiomycetes. Mycosphere 2019, 10, 310–489. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. TRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Gabriel, L.; Brůna, T.; Hoff, K.J.; Ebel, M.; Lomsadze, A.; Borodovsky, M.; Stanke, M. BRAKER3: Fully Automated Genome Annotation Using RNA-Seq and Protein Evidence with GeneMark-ETP, AUGUSTUS, and TSEBRA. Genome Res. 2024, 34, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Ter-Hovhannisyan, V.; Lomsadze, A.; Chernoff, Y.O.; Borodovsky, M. Gene Prediction in Novel Fungal Genomes Using an Ab Initio Algorithm with Unsupervised Training. Genome Res. 2008, 18, 1979–1990. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab Initio Prediction of Alternative Transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; Von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting Sequence Signals in Targeting Peptides Using Deep Learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of Apoplastic and Cytoplasmic Effectors in Fungi and Oomycetes. Mol. Plant-Microbe Interact. 2022, 35, 146–156. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. DbCAN3: Automated Carbohydrate-Active Enzyme and Substrate Annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- Lu, T.; Yao, B.; Zhang, C. DFVF: Database of Fungal Virulence Factors. Database 2012, 2012, bas032. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, S.; Choi, J.; Ahn, K.; Park, B.; Park, J.; Kang, S.; Lee, Y.H. Fungal Cytochrome P450 Database. BMC Genom. 2008, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.T.; Cheng, J.T.; Cao, F.; Cao, F.; Chen, X.A.; Chen, X.A.; Li, Y.Q.; Li, Y.Q.; Mao, X.M.; Mao, X.M. Genomic and Transcriptomic Survey of an Endophytic Fungus Calcarisporium arbuscula NRRL 3705 and Potential Overview of Its Secondary Metabolites. BMC Genom. 2020, 21, 424. [Google Scholar] [CrossRef] [PubMed]
- Erickson, E.; Gado, J.E.; Avilán, L.; Bratti, F.; Brizendine, R.K.; Cox, P.A.; Gill, R.; Graham, R.; Kim, D.J.; König, G.; et al. Sourcing Thermotolerant Poly(Ethylene Terephthalate) Hydrolase Scaffolds from Natural Diversity. Nat. Commun. 2022, 13, 7850. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Johnston, P.R.; Baschien, C. Tricladiaceae Fam. Nov. (Helotiales, Leotiomycetes). Fungal Syst. Evol. 2020, 6, 233–242. [Google Scholar] [CrossRef]
- Baschien, C.; Tsui, C.K.M.; Gulis, V.; Szewzyk, U.; Marvanová, L. The Molecular Phylogeny of Aquatic Hyphomycetes with Affinity to the Leotiomycetes. Fungal Biol. 2013, 117, 660–672. [Google Scholar] [CrossRef]
- Carl, S.; Mohr, S.; Sahm, R.; Baschien, C. Laboratory Conditions Can Change the Complexity and Composition of the Natural Aquatic Mycobiome on Alnus glutinosa Leaf Litter. Fungal Ecol. 2022, 57–58, 101142. [Google Scholar] [CrossRef]
- Kebert, M.; Kostić, S.; Zlatković, M.; Stojnic, S.; Čapelja, E.; Zorić, M.; Kiprovski, B.; Budakov, D.; Orlović, S. Ectomycorrhizal Fungi Modulate Biochemical Response against Powdery Mildew Disease in Quercus robur L. Forests 2022, 13, 1491. [Google Scholar] [CrossRef]
- Zhan, J.; McDonald, B.A. Thermal Adaptation in the Fungal Pathogen Mycosphaerella graminicola. Mol. Ecol. 2011, 20, 1689–1701. [Google Scholar] [CrossRef]
- Marquez, N.; Giachero, M.L.; Declerck, S.; Ducasse, D.A. Macrophomina phaseolina: General Characteristics of Pathogenicity and Methods of Control. Front. Plant Sci. 2021, 12, 634397. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A. Global Warming Could Drive the Emergence of New Fungal Pathogens. Nat. Microbiol. 2023, 8, 2217–2219. [Google Scholar] [CrossRef]
- Matsushita, K.; Azuma, Y.; Kosaka, T.; Yakushi, T.; Hoshida, H.; Akada, R.; Yamada, M. Genomic Analyses of Thermotolerant Microorganisms Used for High-Temperature Fermentations. Biosci. Biotechnol. Biochem. 2016, 80, 655–668. [Google Scholar] [CrossRef]
- Bárcenas-Moreno, G.; Brandón, M.G.; Rousk, J.; Bååth, E. Adaptation of Soil Microbial Communities to Temperature: Comparison of Fungi and Bacteria in a Laboratory Experiment. Glob. Change Biol. 2009, 15, 2950–2957. [Google Scholar] [CrossRef]
- Zhao, S.; Shi, R.; Liang, X.; Li, P.; Bai, X.; Wang, Y.; Zhang, Y. Sulfadiazine Degradation by Bjerkandera adusta DH0817 at Low Temperatures and Its Cold-Adaptation Mechanisms. Bioresour. Technol. 2024, 407, 131108. [Google Scholar] [CrossRef]
- Kutyła, M.; Fiedurek, J.; Gromada, A.; Jedrzejewski, K.; Trytek, M. Mutagenesis and Adaptation of the Psychrotrophic Fungus Chrysosporium pannorum A-1 as a Method for Improving β-Pinene Bioconversion. Molecules 2020, 25, 2589. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. Low-Temperature Extremophiles and Their Applications. Curr. Opin. Biotechnol. 2002, 13, 253–261. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Charlton, T.; Ertan, H.; Omar, S.M.; Siddiqui, K.S.; Williams, T.J. Biotechnological Uses of Enzymes from Psychrophiles. Microb. Biotechnol. 2011, 4, 449–460. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Cavicchioli, R. Cold-Adapted Enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Williams, T.J.; Wilkins, D.; Yau, S.; Allen, M.A.; Brown, M.V.; Lauro, F.M.; Cavicchioli, R. Psychrophiles. Annu. Rev. Earth Planet. Sci. 2013, 41, 87–115. [Google Scholar] [CrossRef]
- Siddiqui, K.S. Some like It Hot, Some like It Cold: Temperature Dependent Biotechnological Applications and Improvements in Extremophilic Enzymes. Biotechnol. Adv. 2015, 33, 1912–1922. [Google Scholar] [CrossRef] [PubMed]
- Fields, P.A. Review: Protein Function at Thermal Extremes: Balancing Stability and Flexibility. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2001, 129, 417–431. [Google Scholar] [CrossRef] [PubMed]
- Duarte, S.; Fernandes, I.; Nogueira, M.J.; Cássio, F.; Pascoal, C. Temperature Alters Interspecific Relationships among Aquatic Fungi. Fungal Ecol. 2013, 6, 187–191. [Google Scholar] [CrossRef]
- Hughes, R.M.; Paulsen, S.G.; Stoddard, J.L. EMAP-Surface Waters: A Multiassemblage, Probability Survey of Ecological Integrity in the U.S.A. Hydrobiologia 2000, 422–423, 429–443. [Google Scholar] [CrossRef]
- Bärlocher, F.; Seena, S.; Wilson, K.P.; Dudley Williams, D. Raised Water Temperature Lowers Diversity of Hyporheic Aquatic Hyphomycetes. Freshw. Biol. 2008, 53, 368–379. [Google Scholar] [CrossRef]
- Barlocher, F.; Kendrick, B. Dynamics of the Fungal Population on Leaves in a Stream. J. Ecol. 1974, 62, 761. [Google Scholar] [CrossRef]
- Bärlocher, F. Research on Aquatic Hyphomycetes: Historical Background and Overview; Springer: Berlin/Heidelberg, Germany, 1992; pp. 1–15. [Google Scholar] [CrossRef]
- Sridhar, K.R.; Bärlocher, F. Effect of Temperature on Growth and Survival of Five Aquatic Hyphomycetes. Sydowia 1993, 45, 377–387. [Google Scholar]
- Lin, S.; Wei, J.; Yang, B.; Zhang, M.; Zhuo, R. Bioremediation of Organic Pollutants by White Rot Fungal Cytochrome P450: The Role and Mechanism of CYP450 in Biodegradation. Chemosphere 2022, 301, 134776. [Google Scholar] [CrossRef]
- Zhang, L.J.; Zhang, Z.R.; Zheng, Y.Q.; Zhang, L.J.; Wang, M.Y.; Wang, X.T.; Yuan, M.L. Genome-Wide Gene Expression Profiles of the Pea Aphid (Acyrthosiphon pisum) under Cold Temperatures Provide Insights into Body Color Variation. Arch. Insect Biochem. Physiol. 2021, 108, e21797. [Google Scholar] [CrossRef]
- Nandanwar, S.K.; Borkar, S.B.; Lee, J.H.; Kim, H.J. Taking Advantage of Promiscuity of Cold-Active Enzymes. Appl. Sci. 2020, 10, 8128. [Google Scholar] [CrossRef]
- Shin, J.; Kim, J.E.; Lee, Y.W.; Son, H. Fungal Cytochrome P450s and the P450 Complement (CYPome) of Fusarium graminearum. Toxins 2018, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.; Ahmed, M.; Khan, T.A.; Akhter, Y. Fungal P450 Monooxygenases—The Diversity in Catalysis and Their Promising Roles in Biocontrol Activity. Appl. Microbiol. Biotechnol. 2020, 104, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, G.; Schachner, M.; Gabbiani, G.; Minerdi, D.; Savoi, S.; Sabbatini, P. Role of Cytochrome P450 Enzyme in Plant Microorganisms’ Communication: A Focus on Grapevine. Int. J. Mol. Sci. 2023, 24, 4695. [Google Scholar] [CrossRef] [PubMed]
- Nsele, N.N.; Padayachee, T.; Nelson, D.R.; Syed, K. Pezizomycetes Genomes Reveal Diverse P450 Complements Characteristic of Saprotrophic and Ectomycorrhizal Lifestyles. J. Fungi 2023, 9, 830. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zheng, C.; Shi, F.; Xu, Y.; Zong, S.; Tao, J. Expression Analysis of Genes Related to Cold Tolerance in Dendroctonus valens. PeerJ 2021, 9, e10864. [Google Scholar] [CrossRef]
- Shin, J.Y.; Bui, D.C.; Lee, Y.; Nam, H.; Jung, S.; Fang, M.; Kim, J.C.; Lee, T.; Kim, H.; Choi, G.J.; et al. Functional Characterization of Cytochrome P450 Monooxygenases in the Cereal Head Blight Fungus Fusarium graminearum. Environ. Microbiol. 2017, 19, 2053–2067. [Google Scholar] [CrossRef]
- Dauda, W.P.; Glen, E.; Abraham, P.; Adetunji, C.O.; Morumda, D.; Abraham, S.E.; Wabba, G.P.; Ogwuche, I.O.; Azameti, M.K. Comparative Phylogenomic Analysis of Cytochrome P450 Monooxygenases From Fusarium Species. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Zhang, J.; Jin, X.; Wang, Y.; Zhang, B.; Liu, T. A Cytochrome P450 Monooxygenase in Nondefoliating Strain of Verticillium dahliae Manipulates Virulence via Scavenging Reactive Oxygen Species. Phytopathology 2022, 112, 1723–1729. [Google Scholar] [CrossRef]
- Oberhofer, M.; Malfent, F.; Zehl, M.; Urban, E.; Wackerlig, J.; Reznicek, G.; Vignolle, G.A.; Rückert, C.; Busche, T.; Wibberg, D.; et al. Biosynthetic Potential of the Endophytic Fungus Helotiales Sp. BL73 Revealed via Compound Identification and Genome Mining. Appl. Environ. Microbiol. 2022, 88, e0251021. [Google Scholar] [CrossRef]
- Dauda, W.P.; Morumda, D.; Abraham, P.; Adetunji, C.O.; Ghazanfar, S.; Glen, E.; Abraham, S.E.; Peter, G.W.; Ogra, I.O.; Ifeanyi, U.J.; et al. Genome-Wide Analysis of Cytochrome P450s of Alternaria Species: Evolutionary Origin, Family Expansion and Putative Functions. J. Fungi 2022, 8, 324. [Google Scholar] [CrossRef]
- Bowman, J.S.; Deming, J.W. Alkane Hydroxylase Genes in Psychrophile Genomes and the Potential for Cold Active Catalysis. BMC Genom. 2014, 15, 1120. [Google Scholar] [CrossRef] [PubMed]
- Grossart, H.P.; Hassan, E.A.; Masigol, H.; Arias-Andres, M.; Rojas-Jimenez, K. Inland Water Fungi in the Anthropocene: Current and Future Perspectives. In Encyclopedia of Inland Waters, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2022; Volume 4, pp. 667–684. [Google Scholar] [CrossRef]
- Baroncelli, R.; Sreenivasaprasad, S.; Sukno, S.A.; Thon, M.R.; Holub, E. Draft Genome Sequence of Colletotrichum acutatum Sensu Lato (Colletotrichum fioriniae). Genome Announc. 2014, 2, e00112-14. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Salvatore, M.M.; Andolfi, A. Phytopathogenic Fungi and Toxicity. Toxins 2021, 13, 689. [Google Scholar] [CrossRef] [PubMed]
- Fragner, D.; Zomorrodi, M.; Kües, U.; Majcherczyk, A. Optimized Protocol for the 2-DE of Extracellular Proteins from Higher Basidiomycetes Inhabiting Lignocellulose. Electrophoresis 2009, 30, 2431–2441. [Google Scholar] [CrossRef]
- Yike, I. Fungal Proteases and Their Pathophysiological Effects. Mycopathologia 2011, 171, 299–323. [Google Scholar] [CrossRef]
- Guegan, H.; Poirier, W.; Ravenel, K.; Dion, S.; Delabarre, A.; Desvillechabrol, D.; Pinson, X.; Sergent, O.; Gallais, I.; Gangneux, J.P.; et al. Deciphering the Role of PIG1 and DHN-Melanin in Scedosporium apiospermum Conidia. J. Fungi 2023, 9, 134. [Google Scholar] [CrossRef]
- Paulson, O.B.; Waldemar, G. Role of the Local Renin-Angiotensin System in the Autoregulation of the Cerebral Circulation. J. Vasc. Res. 1991, 28, 231–235. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Tyler, B.M.; Wang, Y.Y. Defense and Counterdefense during Plant-Pathogenic Oomycete Infection. Annu. Rev. Microbiol. 2019, 73, 667–696. [Google Scholar] [CrossRef]
- Rodríguez-Sifuentes, L.; Marszalek, J.E.; Chuck-Hernández, C.; Serna-Saldívar, S.O. Legumes Protease Inhibitors as Biopesticides and Their Defense Mechanisms against Biotic Factors. Int. J. Mol. Sci. 2020, 21, 3322. [Google Scholar] [CrossRef]
- Redkar, A.; Sabale, M.; Zuccaro, A.; Di Pietro, A. Determinants of Endophytic and Pathogenic Lifestyle in Root Colonizing Fungi. Curr. Opin. Plant Biol. 2022, 67, 102226. [Google Scholar] [CrossRef] [PubMed]
- Al-Qaissi, A.R.M.; Hamoudi, A.H.M.; Hassoun, K.W.S. Morphological and Molecular Identification of Aquatic Fungi in Tigris River for Some Areas in Salah Al-Din Province and Evaluating Their Enzymatic Activity. Int. J. Drug Deliv. Technol. 2022, 12, 463–471. [Google Scholar]
- Subapriya, S.; Senthil, N.; Vairamuthu, S.; Nagarajan, B.; Thirunavukkarasu, C. Opportunistic Fungi as Etiologic Agents of Dermatitis—A Case of Alternaria Fungal Infestation in Canines. Int. J. Livest. Res. 2015, 5, 24. [Google Scholar] [CrossRef]
- Read, S.J.; Moss, S.T.; Jones, E.B.G. Attachment Studies of Aquatic Hyphomycetes. Philos. Trans.—R. Soc. Lond. B 1991, 334, 449–457. [Google Scholar] [CrossRef]
- Kearns, S.G.; Bärlocher, F. Leaf Surface Roughness Influences Colonization Success of Aquatic Hyphomycete Conidia. Fungal Ecol. 2008, 1, 13–18. [Google Scholar] [CrossRef]
- Au, D.W.T.; Jones, E.B.G.; Moss, S.T. Spore Attachment and Extracellular Mucilage of Aquatic Hyphomycetes. Biofouling 1996, 10, 123–140. [Google Scholar] [CrossRef]
- Harrison, S.J.; Moss, S.T.; Jones, E.B.G. Fungal Adhesion in Aquatic Hyphomycetes. Int. Biodeterior. 1988, 24, 271–276. [Google Scholar] [CrossRef]
- Shah, P.A.; Pell, J.K. Entomopathogenic Fungi as Biological Control Agents. Appl. Microbiol. Biotechnol. 2003, 61, 413–423. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; Zhang, X.; Yin, Y.; Li, R.; Lin, Y.; Deng, C.; Yang, K.; Liu, X.; Wang, Z. Pathogenicity of Metarhizium rileyi against Spodoptera litura Larvae: Appressorium Differentiation, Proliferation in Hemolymph, Immune Interaction, and Reemergence of Mycelium. Fungal Genet. Biol. 2021, 150, 103508. [Google Scholar] [CrossRef]
- Lopez, R.O.; Chiocchio, V.M.; Ruscitti, M.F.; Taborda, C.P.; Saparrat, M.C.N. Towards a Better Understanding of Melanins from Dark Septate Endophytes (DSEs): Their Variability, Synthesis Pathways and Biological Roles. J. Soil. Sci. Plant Nutr. 2024, 24, 1650–1664. [Google Scholar] [CrossRef]
- Gil, S.S.; Cappellari, L.d.R.; Giordano, W.; Banchio, E. Antifungal Activity and Alleviation of Salt Stress by Volatile Organic Compounds of Native Pseudomonas Obtained from Mentha piperita. Plants 2023, 12, 1488. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J.G.M.; Mehrabi, R.; Van Den Burg, H.A.; Stergiopoulos, I. Fungal Effector Proteins: Past, Present and Future. Mol. Plant Pathol. 2009, 10, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Garcia, E.; Yan, X.; Oses-Ruiz, M.; de Paula, S.; Talbot, N.J. Effector-Triggered Susceptibility by the Rice Blast Fungus Magnaporthe oryzae. New Phytol. 2024, 241, 1007–1020. [Google Scholar] [CrossRef]
- Duxbury, Z.; Ma, Y.; Furzer, O.J.; Huh, S.U.; Cevik, V.; Jones, J.D.G.; Sarris, P.F. Pathogen Perception by NLRs in Plants and Animals: Parallel Worlds. BioEssays 2016, 38, 769–781. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Katagiri, F.; Tsuda, K. Understanding the Plant Immune System. Mol. Plant. Microbe. Interact. 2010, 23, 1531–1536. [Google Scholar] [CrossRef]
- Gay, E.J.; Jacques, N.; Lapalu, N.; Cruaud, C.; Laval, V.; Balesdent, M.H.; Rouxel, T. Location and Timing Govern Tripartite Interactions of Fungal Phytopathogens and Host in the Stem Canker Species Complex. BMC Biol. 2023, 21, 247. [Google Scholar] [CrossRef]
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. Comparative Genomic and Transcriptional Analyses of the Carbohydrate-Active Enzymes and Secretomes of Phytopathogenic Fungi Reveal Their Significant Roles during Infection and Development. Sci. Rep. 2015, 5, 15565. [Google Scholar] [CrossRef]
- Rissi, D.V.; Ijaz, M.; Baschien, C. Comparative Genomics of Fungi in Nectriaceae Reveals Their Environmental Adaptation and Conservation Strategies. J. Fungi 2024, 10, 632. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.R. Comparative Analysis of Fungal Genomes Reveals Different Plant Cell Wall Degrading Capacity in Fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef]
- Czislowski, E.; Zeil-rolfe, I.; Aitken, E.A.B. Effector Profiles of Endophytic Fusarium Associated with Asymptomatic Banana (Musa Sp.) Hosts. Int. J. Mol. Sci. 2021, 22, 2508. [Google Scholar] [CrossRef] [PubMed]
- Promputtha, I.; Hyde, K.D.; McKenzie, E.H.C.; Peberdy, J.F.; Lumyong, S. Can Leaf Degrading Enzymes Provide Evidence That Endophytic Fungi Becoming Saprobes? Fungal Divers. 2010, 41, 89–99. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N.; Sofyin, A.V.; Revina, T.A.; Gvozdeva, E.L.; Ievleva, E.V.; Valueva, T.A. Secretion of Proteolytic Enzymes by Three Phytopathogenic Microorganisms. Appl. Biochem. Microbiol. 2013, 49, 514–520. [Google Scholar] [CrossRef]
- Yarullina, L.G.; Akhatova, A.R.; Kasimova, R.I. Hydrolytic Enzymes and Their Proteinaceous Inhibitors in Regulation of Plant–Pathogen Interactions. Russ. J. Plant Physiol. 2016, 63, 193–203. [Google Scholar] [CrossRef]
- Li, X.; Zhen, L.X.; Cheng, D.G.; Jian, C. Use of Enzymatic Biodegradation for Protection of Plant against Microbial Disease. Curr. Top. Biotechnol. 2008, 4, 1–12. [Google Scholar]
- Wang, B.; Liang, X.; Gleason, M.L.; Zhang, R.; Sun, G. Comparative Genomics of Botryosphaeria dothidea and B. kuwatsukai, Causal Agents of Apple Ring Rot, Reveals Both Species Expansion of Pathogenicityrelated Genes and Variations in Virulence Gene Content during Speciation. IMA Fungus 2018, 9, 243–257. [Google Scholar] [CrossRef]
- Yu, C.; Diao, Y.; Lu, Q.; Zhao, J.; Cui, S.; Xiong, X.; Lu, A.; Zhang, X.; Liu, H. Comparative Genomics Reveals Evolutionary Traits, Mating Strategies, and Pathogenicity-Related Genes Variation of Botryosphaeriaceae. Front. Microbiol. 2022, 13, 800981. [Google Scholar] [CrossRef]
- Hage, H.; Rosso, M.-N.; Faulds, C.; Chomnunti, P. Evolution of Fungal Carbohydrate-Active Enzyme Portfolios and Adaptation to Plant Cell-Wall Polymers. J. Fungi 2021, 7, 185. [Google Scholar] [CrossRef]
- Ospina-Giraldo, M.D.; Griffith, J.G.; Laird, E.W.; Mingora, C. The CAZyome of Phytophthora Spp.: A Comprehensive Analysis of the Gene Complement Coding for Carbohydrate-Active Enzymes in Species of the Genus Phytophthora. BMC Genom. 2010, 11, 525. [Google Scholar] [CrossRef]
- Haridas, S.; Albert, R.; Binder, M.; Bloem, J.; LaButti, K.; Salamov, A.; Andreopoulos, B.; Baker, S.E.; Barry, K.; Bills, G.; et al. 101 Dothideomycetes Genomes: A Test Case for Predicting Lifestyles and Emergence of Pathogens. Stud. Mycol. 2020, 96, 141–153. [Google Scholar] [CrossRef]
- Frey, S.D.; Drijber, R.; Smith, H.; Melillo, J. Microbial Biomass, Functional Capacity, and Community Structure after 12 Years of Soil Warming. Soil. Biol. Biochem. 2008, 40, 2904–2907. [Google Scholar] [CrossRef]
- D’Alò, F.; Tosadori, G.; Zucconi, L.; Onofri, S.; Canini, F.; Roos, R.E.; Klanderud, K.; Voříšková, J. Soil Microbial Community Responses to Long-Term Experimental Warming in an Alpine Dryas Octopetala Heath in Norway. Appl. Soil. Ecol. 2024, 200, 105430. [Google Scholar] [CrossRef]
- Newsham, K.K.; Misiak, M.; Goodall-Copestake, W.P.; Dahl, M.S.; Boddy, L.; Hopkins, D.W.; Davey, M.L. Experimental Warming Increases Fungal Alpha Diversity in an Oligotrophic Maritime Antarctic Soil. Front. Microbiol. 2022, 13, 1050372. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.; Larrañaga, A.; Pérez, J.; Descals, E.; Pozo, J. Temperature Affects Leaf Litter Decomposition in Low-Order Forest Streams: Field and Microcosm Approaches. FEMS Microbiol. Ecol. 2014, 87, 257–267. [Google Scholar] [CrossRef]
- Chauvet, E.; Suberkropp, K. Temperature and Sporulation of Aquatic Hyphomycetes. Appl. Environ. Microbiol. 1998, 64, 1522–1525. [Google Scholar] [CrossRef]
- Sridhar, K.R. Dimensions, Diversity and Ecology of Aquatic Mycobiome. Kavaka 2020, 54, 10. [Google Scholar] [CrossRef]
- Sati, S.C.; Belwal, M. Aquatic Hyphomycetes as Endophytes of Riparian Plant Roots. Mycologia 2005, 97, 45–49. [Google Scholar] [CrossRef]
- Marvanova, L.; Fisher, P.J. A New Endophytic Hyphomycete from Alder Roots. Nova Hedwig. 1991, 52, 33–37. [Google Scholar]
- Jia, M.; Gong, X.; Fan, M.; Liu, H.; Zhou, H.; Gu, S.; Liu, Y.; Dong, J. Identification and Analysis of the Secretome of Plant Pathogenic Fungi Reveals Lifestyle Adaptation. Front. Microbiol. 2023, 14, 1171618. [Google Scholar] [CrossRef]
- Edwards, A.; Pachebat, J.A.; Swain, M.; Hegarty, M.; Hodson, A.J.; Irvine-Fynn, T.D.L.; Rassner, S.M.E.; Sattler, B. A Metagenomic Snapshot of Taxonomic and Functional Diversity in an Alpine Glacier Cryoconite Ecosystem. Environ. Res. Lett. 2013, 8, 035003. [Google Scholar] [CrossRef]
- Sajjad, W.; Rafiq, M.; Din, G.; Hasan, F.; Iqbal, A.; Zada, S.; Ali, B.; Hayat, M.; Irfan, M.; Kang, S. Resurrection of Inactive Microbes and Resistome Present in the Natural Frozen World: Reality or Myth? Sci. Total Environ. 2020, 735, 139275. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rissi, D.V.; Ijaz, M.; Baschien, C. Comparative Genomics of Different Lifestyle Fungi in Helotiales (Leotiomycetes) Reveals Temperature and Ecosystem Adaptations. J. Fungi 2024, 10, 869. https://doi.org/10.3390/jof10120869
Rissi DV, Ijaz M, Baschien C. Comparative Genomics of Different Lifestyle Fungi in Helotiales (Leotiomycetes) Reveals Temperature and Ecosystem Adaptations. Journal of Fungi. 2024; 10(12):869. https://doi.org/10.3390/jof10120869
Chicago/Turabian StyleRissi, Daniel Vasconcelos, Maham Ijaz, and Christiane Baschien. 2024. "Comparative Genomics of Different Lifestyle Fungi in Helotiales (Leotiomycetes) Reveals Temperature and Ecosystem Adaptations" Journal of Fungi 10, no. 12: 869. https://doi.org/10.3390/jof10120869
APA StyleRissi, D. V., Ijaz, M., & Baschien, C. (2024). Comparative Genomics of Different Lifestyle Fungi in Helotiales (Leotiomycetes) Reveals Temperature and Ecosystem Adaptations. Journal of Fungi, 10(12), 869. https://doi.org/10.3390/jof10120869






