Fungal Endophytes: Discovering What Lies within Some of Canada’s Oldest and Most Resilient Grapevines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Grape Samples and Varietal Testing
2.2. Leaf Sterilization and Isolation of Foliar Fungal Endophytes
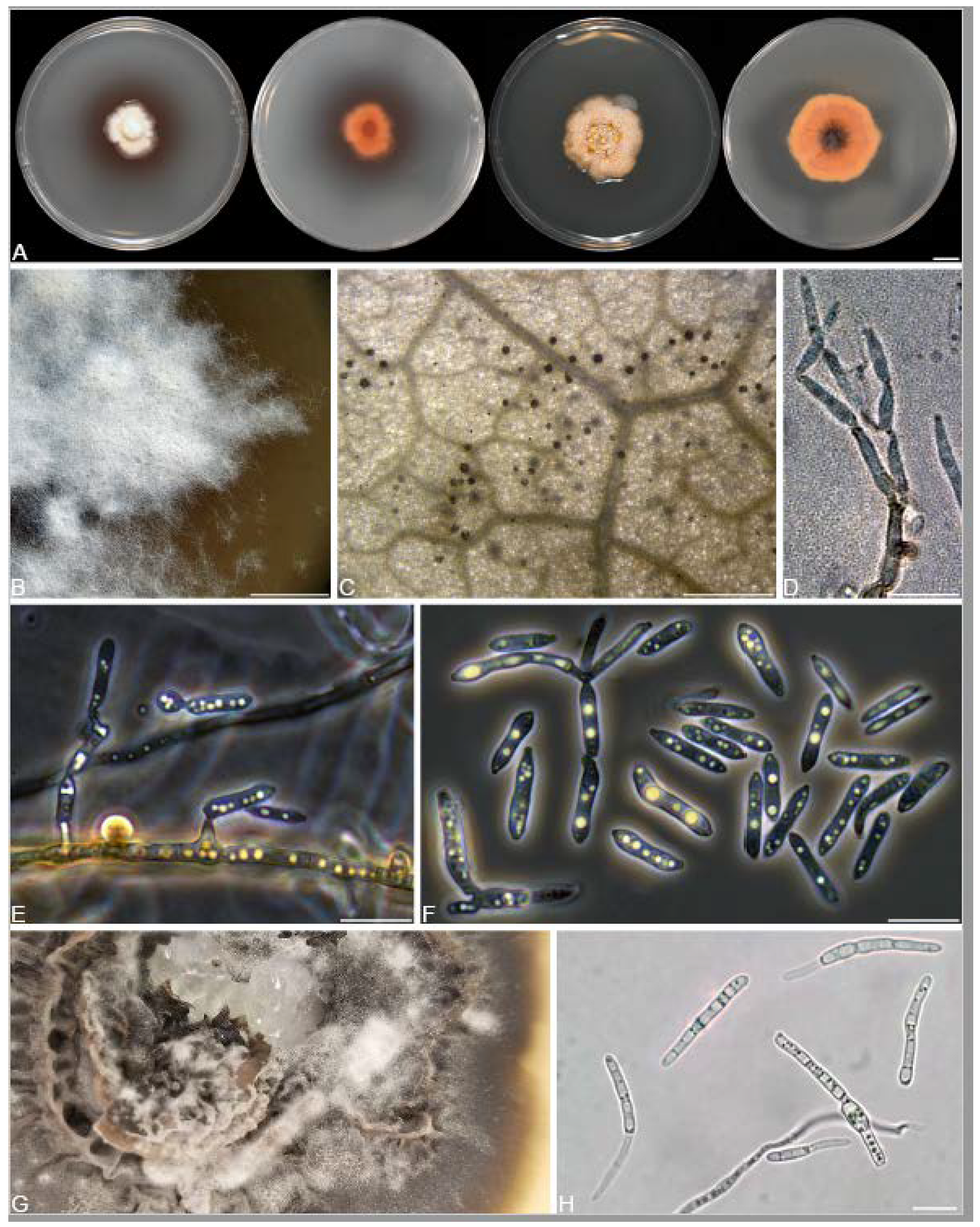
2.3. Culturing and Morphological Observations
2.4. DNA Extraction, Amplification, Sequencing, and Analysis
2.5. Screening of Endophyte Bioactivity against Botrytis
2.6. Metabolite Screening by High-Resolution HPLC-MS
3. Results
3.1. Grape Site/DNA Barcoding Results
3.2. Isolation and Identification of Endophyte Strains
3.3. Screening of Antimicrobial Activity of Endophytic Fungi against Botrytis
3.4. Extrolite Screening of Bioactive strains
4. Discussion
4.1. Feral Endophyte Host Identification
4.2. Botrytis Suppression
4.3. Endophyte Diversity and Ecology
5. Significance Statement
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lawson, G. On the northern limit of wild grape vines. Proc. Nova Scotian Inst. Sci. 1884, 6, 1883–1886. [Google Scholar]
- Wallace, B. L’Anse Aux Meadows, Leif Eriksson’s Home in Vinland. J. North Atl. 2009, 2, 114–125. [Google Scholar] [CrossRef]
- Speck, F.G.; Dexter, R.W. Utilization of animals and plants by the Malecite Indians of New Brunswick. J. Wash. Acad. Sci. 1952, 42, 1–7. [Google Scholar]
- Murray, R.A.; Wright, B. The Tangled Vine: Winegrowing in Nova Scotia; Blue Frog: Bridgewater, NS, Canada, 2004. [Google Scholar]
- Frank, R.; Eyler, R. The Economic Impact of the Wine and Grape Industry in Canada 2015; Canadian Vintners Association, Winery and Grower Alliance of Ontario British Columbia, Wine Institute Winery Association of Nova Scotia: Halifax, NS, Canada, 2017. [Google Scholar]
- Bishop, R.; Craig, D.; MacEachern, C. Observations on the Performance of Grape Cultivars in Nova Scotia1. HortScience 1970, 5, 154–156. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Courty, P.E.; Oger, P. Plant symbionts are engineers of the plant-associated microbiome. Trends Plant Sci. 2019, 24, 905–916. [Google Scholar] [CrossRef]
- Yan, L.; Zhu, J.; Zhao, X.; Shi, J.; Jiang, C.; Shao, D. Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 2019, 103, 3327–3340. [Google Scholar] [CrossRef]
- De Vries, S.; von Dahlen, J.K.; Schnake, A.; Ginschel, S.; Schulz, B.; Rose, L.E. Broad-spectrum inhibition of Phytophthora infestans by fungal endophytes. FEMS Microbiol. Ecol. 2018, 94, fiy037. [Google Scholar] [CrossRef] [PubMed]
- Glare, T.; Caradus, J.; Gelernter, W.; Jackson, T.; Keyhani, N.; Kohl, J.; Marrone, P.; Morin, L.; Stewart, A. Have biopesticides come of age? Trends Biotechnol. 2012, 30, 250–258. [Google Scholar] [CrossRef]
- Alabouvette, C.; Olivain, C.; Steinberg, C. Biological Control of Plant Diseases: The European Situation. Eur. J. Plant Pathol. 2006, 114, 329–341. [Google Scholar] [CrossRef]
- Grabka, R.; d’Entremont, T.W.; Adams, S.J.; Walker, A.K.; Tanney, J.B.; Abbasi, P.A.; Ali, S. Fungal endophytes and their role in agricultural plant protection against pests and pathogens. Plants 2022, 11, 384. [Google Scholar] [CrossRef] [PubMed]
- Bamisile, B.S.; Dash, C.K.; Akutse, K.S.; Keppanan, R.; Afolabi, O.G.; Hussain, M.; Qasim, M.; Wang, L. Prospects of endophytic fungal entomopathogens as biocontrol and plant growth promoting agents: An insight on how artificial inoculation methods affect endophytic colonization of host plants. Microbiol. Res. 2018, 217, 34–50. [Google Scholar] [CrossRef]
- De Silva, N.I.; Brooks, S.; Lumyong, S.; Hyde, K.D. Use of endophytes as biocontrol agents. Fungal Biol. Rev. 2019, 33, 133–148. [Google Scholar] [CrossRef]
- Latz, M.A.; Jensen, B.; Collinge, D.B.; Jørgensen, H.J. Endophytic fungi as biocontrol agents: Elucidating mechanisms in disease suppression. Plant Ecol. Divers. 2018, 11, 555–567. [Google Scholar] [CrossRef]
- Omomowo, O.I.; Babalola, O.O. Bacterial and fungal endophytes: Tiny giants with immense beneficial potential for plant growth and sustainable agricultural productivity. Microorganisms 2019, 7, 481. [Google Scholar] [CrossRef]
- Segaran, G.; Sathiavelu, M. Fungal endophytes: A potent biocontrol agent and a bioactive metabolites reservoir. Biocatal. Agric. Biotechnol. 2019, 21, 101284. [Google Scholar] [CrossRef]
- Brum, M.; Araújo, W.L.; Maki, C.S.; Azevedo, J.L.D. Endophytic fungi from Vitis labrusca L.(‘Niagara Rosada’) and its potential for the biological control of Fusarium oxysporum. Genet. Mol. Res. GMR 2012, 11, 4187–4197. [Google Scholar] [CrossRef] [PubMed]
- Kernaghan, G.; Mayerhofer, M.; Griffin, A. Fungal endophytes of wild and hybrid Vitis leaves and their potential for vineyard biocontrol. Can. J. Microbiol. 2017, 63, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Ginn, F.M. Endophytic Fungi in Vaccinium macrocarpon (Cranberry) and Vaccinium angustifolium (Blueberry); The University of New Brunswick: Fredericton, NS, Canada, 2001. [Google Scholar]
- Tanney, J.B.; Di Stefano, J.; Miller, J.D.; McMullin, D.R. Natural products from the Picea foliar endophytes Niesslia endophytica sp. nov. and Strasseria geniculata. Mycol. Prog. 2023, 22, 17. [Google Scholar] [CrossRef]
- Abbasi, P.A.; Ali, S.; Renderos, W.; Naeem, H.A.; Papadopoulos, Y. First report of Alternaria alternata causing leaf spot and blight symptoms on alfalfa in Canada. Can. J. Plant Pathol. 2018, 40, 451–455. [Google Scholar] [CrossRef]
- Ali, S.; Renderos, W.; Bevis, E.; Hebb, J.; Abbasi, P.A. Diaporthe eres causes stem cankers and death of young apple rootstocks in Canada. Can. J. Plant Pathol. 2020, 42, 218–227. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Crous, P.W.; Summerell, B.; Shivas, R.; Burgess, T.; Decock, C.; Dreyer, L.; Granke, L.; Guest, D.; Hardy, G.S.; Hausbeck, M. Fungal Planet description sheets: 107–127. Persoonia-Mol. Phylogeny Evol. Fungi 2012, 28, 138–182. [Google Scholar] [CrossRef]
- Damm, U.; Cannon, P.; Woudenberg, J.; Crous, P. The Colletotrichum acutatum species complex. Stud. Mycol. 2012, 73, 37–113. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Groenewald, J.Z.; Polizzi, G.; Crous, P.W. High species diversity in Colletotrichum associated with citrus diseases in Europe. Persoonia-Mol. Phylogeny Evol. Fungi 2017, 39, 32–50. [Google Scholar] [CrossRef]
- Kraus, C.; Damm, U.; Bien, S.; Voegele, R.; Fischer, M. New species of Phaeomoniellales from a German vineyard and their potential threat to grapevine (Vitis vinifera) health. Fungal Syst. Evol. 2020, 6, 139–155. [Google Scholar] [CrossRef]
- Damm, U.; Woudenberg, J.; Cannon, P.; Crous, P. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Divers. 2009, 39, 45. [Google Scholar]
- Gomes, R.; Glienke, C.; Videira, S.; Lombard, L.; Groenewald, J.; Crous, P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia-Mol. Phylogeny Evol. Fungi 2013, 31, 1–41. [Google Scholar] [CrossRef]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Chukeatirote, E.; Hyde, K.D. The Diaporthe sojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biol. 2015, 119, 383–407. [Google Scholar] [CrossRef]
- Du, Z.; Fan, X.-L.; Hyde, K.D.; Yang, Q.; Liang, Y.-M.; Tian, C.-M. Phylogeny and morphology reveal two new species of Diaporthe from Betula spp. in China. Phytotaxa 2016, 269, 90–102. [Google Scholar] [CrossRef]
- Gao, Y.; Su, Y.; Sun, W.; Cai, L. Diaporthe species occurring on Lithocarpus glabra in China, with descriptions of five new species. Fungal Biol. 2015, 119, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Guarnaccia, V.; Groenewald, J.Z.; Woodhall, J.; Armengol, J.; Cinelli, T.; Eichmeier, A.; Ezra, D.; Fontaine, F.; Gramaje, D.; Gutierrez-Aguirregabiria, A. Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia-Mol. Phylogeny Evol. Fungi 2018, 40, 135–153. [Google Scholar] [CrossRef]
- Thompson, S.; Tan, Y.P.; Shivas, R.G.; Neate, S.M.; Morin, L.; Bissett, A.; Aitken, E. Green and brown bridges between weeds and crops reveal novel Diaporthe species in Australia. Persoonia-Mol. Phylogeny Evol. Fungi 2015, 35, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, N.; Tian, C.-M. Three new Diaporthe species from Shaanxi province, China. MycoKeys 2020, 67, 1. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.M.; Correia, V.G.; Phillips, A.J. Primers for mating-type diagnosis in Diaporthe and Phomopsis: Their use in teleomorph induction in vitro and biological species definition. Fungal Biol. 2010, 114, 255–270. [Google Scholar] [CrossRef]
- Udayanga, D.; Liu, X.; Crous, P.W.; McKenzie, E.H.; Chukeatirote, E.; Hyde, K.D. A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Divers. 2012, 56, 157–171. [Google Scholar] [CrossRef]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Chukeatirote, E.; Hyde, K.D. Insights into the genus Diaporthe: Phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 2014, 67, 203–229. [Google Scholar] [CrossRef]
- Manawasinghe, I.S.; Dissanayake, A.J.; Li, X.; Liu, M.; Wanasinghe, D.N.; Xu, J.; Zhao, W.; Zhang, W.; Zhou, Y.; Hyde, K.D. High genetic diversity and species complexity of Diaporthe associated with grapevine dieback in China. Front. Microbiol. 2019, 10, 1936. [Google Scholar] [CrossRef]
- Schoch, C.L.; Robbertse, B.; Robert, V.; Vu, D.; Cardinali, G.; Irinyi, L.; Meyer, W.; Nilsson, R.H.; Hughes, K.; Miller, A.N. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for Fungi. Database 2014, 2014, bau061. [Google Scholar] [CrossRef]
- Thompson, S.; Tan, Y.; Young, A.; Neate, S.; Aitken, E.; Shivas, R. Stem cankers on sunflower (Helianthus annuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia-Mol. Phylogeny Evol. Fungi 2011, 27, 80–89. [Google Scholar] [CrossRef]
- Tanney, J.B.; McMullin, D.R.; Green, B.D.; Miller, J.D.; Seifert, K.A. Production of antifungal and antiinsectan metabolites by the Picea endophyte Diaporthe maritima sp. nov. Fungal Biol. 2016, 120, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.; Phillips, A.; Hyde, K.; Yan, J.; Li, X. The current status of species in Diaporthe. Mycosphere 2017, 8, 1106–1156. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Phukhamsakda, C.; Hyde, K.D.; Jeewon, R.; Lee, H.B.; Gareth Jones, E.; Tibpromma, S.; Tennakoon, D.S.; Dissanayake, A.J.; Jayasiri, S.C. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Zhou, H.; Hou, C.-L. Three new species of Diaporthe from China based on morphological characters and DNA sequence data analyses. Phytotaxa 2019, 422, 157–174. [Google Scholar] [CrossRef]
- Feng, X.; Chen, J.; Wang, G.; Cao, T.; Zheng, Y.; Zhang, C. Diaporthe sinensis, a new fungus from Amaranthus sp. in China. Phytotaxa 2019, 425, 259–268. [Google Scholar] [CrossRef]
- Hilário, S.; Amaral, I.A.; Gonçalves, M.F.; Lopes, A.; Santos, L.; Alves, A. Diaporthe species associated with twig blight and dieback of Vaccinium corymbosum in Portugal, with description of four new species. Mycologia 2020, 112, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Luangsa-Ard, J.; Wingfield, M.; Carnegie, A.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.; Baseia, I.; Cano-Lira, J. Fungal Planet description sheets: 785–867. Persoonia-Mol. Phylogeny Evol. Fungi 2018, 41, 238–417. [Google Scholar] [CrossRef] [PubMed]
- Sogonov, M.V.; Castlebury, L.; Rossman, A.; Mejía, L.C.; White, J. Leaf-inhabiting genera of the Gnomoniaceae, Diaporthales. Stud. Mycol. 2008, 62, 1–77. [Google Scholar] [CrossRef] [PubMed]
- Mejía, L.C.; Castlebury, L.A.; Rossman, A.Y.; Sogonov, M.V.; White, J.F. Phylogenetic placement and taxonomic review of the genus Cryptosporella and its synonyms Ophiovalsa and Winterella (Gnomoniaceae, Diaporthales). Mycol. Res. 2008, 112, 23–35. [Google Scholar] [CrossRef]
- Sánchez-Ballesteros, J.; González, V.; Salazar, O.; Acero, J.; Portal, M.A.; Julián, M.; Rubio, V.; Bills, G.F.; Polishook, J.D.; Platas, G. Phylogenetic study of Hypoxylon and related genera based on ribosomal ITS sequences. Mycologia 2000, 92, 964–977. [Google Scholar] [CrossRef]
- Newcombe, G.; Shipunov, A.; Eigenbrode, S.; Raghavendra, A.K.; Ding, H.; Anderson, C.L.; Menjivar, R.; Crawford, M.; Schwarzländer, M. Endophytes influence protection and growth of an invasive plant. Commun. Integr. Biol. 2009, 2, 29–31. [Google Scholar] [CrossRef]
- Joan, T.; Chapela, I.H.; Bruns, T.; Taylor, J. The Phyloecology of Hypaxylan sensu lata. Master’s Thesis, University of California, Berkeley, CA, USA, 2001. [Google Scholar]
- Fournier, J.; Lechat, C.; Ribes Ripoll, M. Record of Nemania aureolutea (Xylariaceae) from the southernmost region of Spain. Ascomycete.org 2020, 12, 221–226. [Google Scholar]
- U’Ren, J.M.; Arnold, A.E. Diversity, taxonomic composition, and functional aspects of fungal communities in living, senesced, and fallen leaves at five sites across North America. PeerJ 2016, 4, e2768. [Google Scholar] [CrossRef]
- Stadler, M.; Fournier, J.; Læssøe, T.; Chlebicki, A.; Lechat, C.; Flessa, F.; Rambold, G.; Peršoh, D. Chemotaxonomic and phylogenetic studies of Thamnomyces (Xylariaceae). Mycoscience 2010, 51, 189–207. [Google Scholar] [CrossRef]
- Lagarde, A.; Millot, M.; Pinon, A.; Liagre, B.; Girardot, M.; Imbert, C.; Ouk, T.-S.; Jargeat, P.; Mambu, L. Antiproliferative and antibiofilm potentials of endolichenic fungi associated with the lichen Nephroma laevigatum. J. Appl. Microbiol. 2019, 126, 1044–1058. [Google Scholar] [CrossRef]
- Ding, W.; Chen, H.-W.; Wang, M.; Wan, M.; Hu, J.-F.; Li, J. Three new eremophilane sesquiterpenes and one new related derivative from Nemania sp. HDF-Br-5, an endophytic fungus of the endangered conifer Pseudotsuga gaussenii Flous. Phytochem. Lett. 2022, 49, 5–11. [Google Scholar] [CrossRef]
- Crous, P.; Summerell, B.; Shivas, R.; Romberg, M.; Mel’nik, V.; Verkley, G.; Groenewald, J. Fungal Planet description sheets: 92–106. Persoonia Mol. Phylogeny Evol. Fungi 2011, 27, 130–162. [Google Scholar] [CrossRef]
- Spies, C.; Mostert, L.; Carlucci, A.; Moyo, P.; Van Jaarsveld, W.; du Plessis, I.; Van Dyk, M.; Halleen, F. Dieback and decline pathogens of olive trees in South Africa. Persoonia-Mol. Phylogeny Evol. Fungi 2020, 45, 196–220. [Google Scholar] [CrossRef]
- Damm, U.; Fourie, P.; Crous, P.W. Coniochaeta (Lecythophora), Collophora gen. nov. and Phaeomoniella species associated with wood necroses of Prunus trees. Persoonia-Mol. Phylogeny Evol. Fungi 2010, 24, 60–80. [Google Scholar] [CrossRef]
- Ferreira, A.B.M.; Leite, L.G.; Hernandes, J.L.; Harakava, R.; Padovani, C.R.; Bueno, C.J. Colonization of vines by Petri disease fungi, susceptibility of rootstocks to Phaeomoniella chlamydospora and their disinfection. Arq. Inst. Biológico 2018, 85, e0882017. [Google Scholar] [CrossRef]
- Videira, S.; Groenewald, J.Z.; Braun, U.; Shin, H.; Crous, P.W. All that glitters is not Ramularia. Stud. Mycol. 2016, 83, 49–163. [Google Scholar] [CrossRef]
- Videira, S.; Groenewald, J.Z.; Kolecka, A.; van Haren, L.; Boekhout, T.; Crous, P.W. Elucidating the Ramularia eucalypti species complex. Persoonia-Mol. Phylogeny Evol. Fungi 2015, 34, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Verkley, G.; Quaedvlieg, W.; Shin, H.-D.; Crous, P. A new approach to species delimitation in Septoria. Stud. Mycol. 2013, 75, 213–305. [Google Scholar] [CrossRef] [PubMed]
- Quaedvlieg, W.; Binder, M.; Groenewald, J.; Summerell, B.; Carnegie, A.; Burgess, T.; Crous, P.W. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia-Mol. Phylogeny Evol. Fungi 2014, 33, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.I.; Batzer, J.C.; Harrington, T.C.; Crous, P.W.; Lavrov, D.V.; Li, H.; Gleason, M.L. Ancestral state reconstruction infers phytopathogenic origins of sooty blotch and flyspeck fungi on apple. Mycologia 2016, 108, 292–302. [Google Scholar] [CrossRef]
- Crous, P.; Braun, U.; Groenewald, J. Mycosphaerella is polyphyletic. Stud. Mycol. 2007, 58, 1–32. [Google Scholar] [CrossRef]
- Ghildiyal, A.; Pandey, A. Isolation of cold tolerant antifungal strains of Trichoderma sp. from glacial sites of Indian Himalayan region. Res. J. Microbiol. 2008, 3, 559–564. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Laatsch, H. AntiBase: The Natural Compound Identifier; Wiley-Vch Weinheim: Weinheim, Germany, 2017. [Google Scholar]
- Badalyan, S.M.; Garibyan, N.; Innocenti, G. Antagonistic activity of xylotrophic mushrooms against pathogenic fungi of cereals in dual culture. Phytopathol. Mediterr. 2002, 41, 220–225. [Google Scholar]
- Flores, A.C.; Pamphile, J.A.; Sarragiotto, M.H.; Clemente, E. Production of 3-nitropropionic acid by endophytic fungus Phomopsis longicolla isolated from Trichilia elegans A. JUSS ssp. elegans and evaluation of biological activity. World J. Microbiol. Biotechnol. 2013, 29, 923–932. [Google Scholar] [CrossRef]
- Specian, V.; Sarragiotto, M.H.; Pamphile, J.A.; Clemente, E. Chemical characterization of bioactive compounds from the endophytic fungus Diaporthe helianthi isolated from Luehea divaricata. Braz. J. Microbiol. 2012, 43, 1174–1182. [Google Scholar] [CrossRef]
- Carvalho, C.d.; Ferreira-D’Silva, A.; Wedge, D.; Cantrell, C.; Rosa, L. Antifungal activities of cytochalasins produced by Diaporthe miriciae, an endophytic fungus associated with tropical medicinal plants. Can. J. Microbiol. 2018, 64, 835–843. [Google Scholar] [CrossRef]
- Hedrick, U.P.; Booth, N.O.; Maxwell, J.D.; Taylor, O.M.; Wellington, R. The Grapes of New York; State of New York Department of Agriculture: Albany, NY, USA, 1908. [Google Scholar]
- Sessa, L.; Abreo, E.; Lupo, S. Diversity of fungal latent pathogens and true endophytes associated with fruit trees in Uruguay. J. Phytopathol. 2018, 166, 633–647. [Google Scholar] [CrossRef]
- Udayanga, D.; Liu, X.; McKenzie, E.H.; Chukeatirote, E.; Bahkali, A.H.; Hyde, K.D. The genus Phomopsis: Biology, applications, species concepts and names of common phytopathogens. Fungal Divers. 2011, 50, 189–225. [Google Scholar] [CrossRef]
- Abramczyk, B.; Marzec-Grządziel, A.; Grządziel, J.; Król, E.; Gałązka, A.; Oleszek, W. Biocontrol Potential and Catabolic Profile of Endophytic Diaporthe eres Strain 1420S from Prunus domestica L. in Poland—A Preliminary Study. Agronomy 2022, 12, 165. [Google Scholar] [CrossRef]
- Polonio, J.; Almeida, T.; Garcia, A.; Mariucci, G.; Azevedo, J.; Rhoden, S.; Pamphile, J. Biotechnological prospecting of foliar endophytic fungi of guaco (Mikania glomerata Spreng.) with antibacterial and antagonistic activity against phytopathogens. Genet. Mol. Res. 2015, 14, 7297–7309. [Google Scholar] [CrossRef]
- Da Silva Ribeiro, A.; Polonio, J.C.; Costa, A.T.; Dos Santos, C.M.; Rhoden, S.A.; Azevedo, J.L.; Pamphile, J.A. Bioprospection of culturable endophytic fungi associated with the ornamental plant Pachystachys lutea. Curr. Microbiol. 2018, 75, 588–596. [Google Scholar] [CrossRef]
- Verma, P.; Hiremani, N.S.; Gawande, S.P.; Sain, S.K.; Nagrale, D.T.; Narkhedkar, N.G.; Prasad, Y. Modulation of plant growth and antioxidative defense system through endophyte biopriming in cotton (Gossypium spp.) and non-host crops. Heliyon 2022, 8, e09487. [Google Scholar] [CrossRef]
- Webber, J.; Gibbs, J. Colonization of elm bark by Phomopsis oblonga. Trans. Br. Mycol. Soc. 1984, 82, 348–352. [Google Scholar] [CrossRef]
- Uecker, F. A World List of Phomopsis Names with Notes on Nomenclature,-Morphology and Biology; Cramer Publishers: Berlin, Germany, 1988. [Google Scholar]
- Hilário, S.; Gonçalves, M.F.; Alves, A. Using genealogical concordance and coalescent-based species delimitation to assess species boundaries in the Diaporthe eres complex. J. Fungi 2021, 7, 507. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.J.; Liu, M.; Zhang, W.; Chen, Z.; Udayanga, D.; Chukeatirote, E.; Li, X.; Yan, J.; Hyde, K.D. Morphological and molecular characterisation of Diaporthe species associated with grapevine trunk disease in China. Fungal Biol. 2015, 119, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Reveglia, P.; Pacetti, A.; Masi, M.; Cimmino, A.; Carella, G.; Marchi, G.; Mugnai, L.; Evidente, A. Phytotoxic metabolites produced by Diaporthe eres involved in cane blight of grapevine in Italy. Nat. Prod. Res. 2021, 35, 2872–2880. [Google Scholar] [CrossRef] [PubMed]
- Makris, G.; Solonos, S.; Christodoulou, M.; Kanetis, L.I. First report of Diaporthe foeniculina associated with grapevine trunk diseases on Vitis vinifera in Cyprus. Plant Dis. 2022, 106, 1294. [Google Scholar] [CrossRef] [PubMed]
- Pine, T. Etiology of the dead-arm disease of Grapevines. Phytopathology 1958, 48, 192–197. [Google Scholar]
- Nita, M.; Ellis, M.; Madden, L. Variation in disease incidence of Phomopsis cane and leaf spot of grape in commercial vineyards in Ohio. Plant Dis. 2008, 92, 1053–1061. [Google Scholar] [CrossRef]
- Cinelli, T.; Mondello, V.; Marchi, G.; Burruano, S.; Alves, A.; Mugnai, L. First report of Diaporthe eres associated with cane blight of grapevine (Vitis vinifera) in Italy. Plant Dis. 2016, 100, 532. [Google Scholar] [CrossRef]
- Lesuthu, P.; Mostert, L.; Spies, C.F.; Moyo, P.; Regnier, T.; Halleen, F. Diaporthe nebulae sp. nov. and first report of D. cynaroidis, D. novem, and D. serafiniae on grapevines in South Africa. Plant Dis. 2019, 103, 808–817. [Google Scholar] [CrossRef]
- Reveglia, P.; Savocchia, S.; Billones-Baaijens, R.; Masi, M.; Cimmino, A.; Evidente, A. Phytotoxic metabolites by nine species of Botryosphaeriaceae involved in grapevine dieback in Australia and identification of those produced by Diplodia mutila, Diplodia seriata, Neofusicoccum australe and Neofusicoccum luteum. Nat. Prod. Res. 2019, 33, 2223–2229. [Google Scholar] [CrossRef]
- Barba, P.; Lillis, J.; Luce, R.S.; Travadon, R.; Osier, M.; Baumgartner, K.; Wilcox, W.F.; Reisch, B.I.; Cadle-Davidson, L. Two dominant loci determine resistance to Phomopsis cane lesions in F 1 families of hybrid grapevines. Theor. Appl. Genet. 2018, 131, 1173–1189. [Google Scholar] [CrossRef] [PubMed]
- Pinna, C.; Linaldeddu, B.T.; Deiana, V.; Maddau, L.; Montecchio, L.; Lentini, A. Plant pathogenic fungi associated with Coraebus florentinus (Coleoptera: Buprestidae) attacks in declining oak forests. Forests 2019, 10, 488. [Google Scholar] [CrossRef]
- Fort, T.; Pauvert, C.; Zanne, A.E.; Ovaskainen, O.; Caignard, T.; Barret, M.; Compant, S.; Hampe, A.; Delzon, S.; Vacher, C. Maternal effects shape the seed mycobiome in Quercus petraea. New Phytol. 2021, 230, 1594–1608. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.L.; Peter, K.A. Quantification of Colletotrichum fioriniae in orchards and deciduous forests indicates it is primarily a leaf endophyte. Phytopathology® 2021, 111, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Jankowiak, R.; Stępniewska, H.; Bilański, P.; Taerum, S.J. Fungi as potential factors limiting natural regeneration of pedunculate oak (Quercus robur) in mixed-species forest stands in Poland. Plant Pathol. 2022, 71, 805–817. [Google Scholar] [CrossRef]
- Costa, D.; Tavares, R.M.; Baptista, P.; Lino-Neto, T. Cork oak endophytic fungi as potential biocontrol agents against Biscogniauxia mediterranea and Diplodia corticola. J. Fungi 2020, 6, 287. [Google Scholar] [CrossRef] [PubMed]
- Tosi, L.; Beccari, G.; Rondoni, G.; Covarelli, L.; Ricci, C. Natural occurrence of Fusarium proliferatum on chestnut in Italy and its potential entomopathogenicity against the Asian chestnut gall wasp Dryocosmus kuriphilus. J. Pest Sci. 2015, 88, 369–381. [Google Scholar] [CrossRef]
- Patanita, M.; Albuquerque, A.; Campos, M.D.; Materatski, P.; Varanda, C.M.; Ribeiro, J.A.; Félix, M.d.R. Metagenomic assessment unravels fungal microbiota associated to grapevine trunk diseases. Horticulturae 2022, 8, 288. [Google Scholar] [CrossRef]
- Van Dyk, M.; Spies, C.F.; Mostert, L.; van der Rijst, M.; du Plessis, I.L.; Moyo, P.; van Jaarsveld, W.J.; Halleen, F. Pathogenicity testing of fungal isolates associated with olive trunk diseases in South Africa. Plant Dis. 2021, 105, 4060–4073. [Google Scholar] [CrossRef] [PubMed]
- Bushman, T.J.; Cunneely, Q.; Ciesla, L. Extraction, isolation, and Biological activity of natural cyclic dipeptides. Stud. Nat. Prod. Chem. 2023, 78, 75–99. [Google Scholar]
- Ivic, D.; Voncina, D.; Sever, Z.; Simon, S.; Pejic, I. Identification of Colletotrichum species causing bitter rot of apple and pear in Croatia. J. Phytopathol. 2013, 161, 284–286. [Google Scholar] [CrossRef]
- Munir, M.; Amsden, B.; Dixon, E.; Vaillancourt, L.; Gauthier, N.W. Characterization of Colletotrichum species causing bitter rot of apple in Kentucky orchards. Plant Dis. 2016, 100, 2194–2203. [Google Scholar] [CrossRef]
- Kim, C.H.; Hassan, O.; Chang, T. Diversity, pathogenicity, and fungicide sensitivity of Colletotrichum species associated with apple anthracnose in South Korea. Plant Dis. 2020, 104, 2866–2874. [Google Scholar] [CrossRef]
- Schoeneberg, A.; Hu, M.-J. First report of Anthracnose fruit rot caused by Colletotrichum fioriniae on red raspberry (Rubus idaeus) in the mid-atlantic region of the United States. Plant Dis. 2020, 104, 1855. [Google Scholar] [CrossRef]
- Luongo, L.; Galli, M.; Garaguso, I.; Petrucci, M.; Vitale, S. First Report of Colletotrichum fioriniae and C. nymphaeae as Causal Agents of Anthracnose on Walnut in Italy. Plant Dis. 2022, 106, 327. [Google Scholar] [CrossRef]
- Xu, S.-J.; Aktaruzzaman, M.; Kim, B.S.; Kim, J.; Shin, H.-D. First report of anthracnose caused by Colletotrichum fioriniae on eggplant fruits in Korea. Plant Dis. 2018, 102, 2642. [Google Scholar] [CrossRef]
- Kasson, M.; Pollok, J.; Benhase, E.; Jelesko, J. First report of seedling blight of eastern poison ivy (Toxicodendron radicans) by Colletotrichum fioriniae in Virginia. Plant Dis. 2014, 98, 995. [Google Scholar] [CrossRef]
- Garibaldi, A.; Bertetti, D.; Matić, S.; Luongo, I.; Guarnaccia, V.; Gullino, M. First report of leaf blight caused by Colletotrichum fioriniae on Mahonia aquifolium in Italy. Plant Dis. 2020, 104, 983. [Google Scholar] [CrossRef]
- Redman, R.S.; Dunigan, D.D.; Rodriguez, R.J. Fungal symbiosis from mutualism to parasitism: Who controls the outcome, host or invader? New Phytol. 2001, 151, 705–716. [Google Scholar] [CrossRef]
- Gonzaga, L.; Costa, L.; Santos, T.; Araújo, E.; Queiroz, M. Endophytic fungi from the genus Colletotrichum are abundant in the Phaseolus vulgaris and have high genetic diversity. J. Appl. Microbiol. 2015, 118, 485–496. [Google Scholar] [CrossRef]
- Nigar, Q.; Cadle-Davidson, L.; Gadoury, D.M.; Hassan, M.u. First report of Colletotrichum fioriniae causing grapevine anthracnose in New York. Plant Dis. 2023, 107, 223. [Google Scholar] [CrossRef]
- Cosseboom, S.D.; Hu, M. Predicting ripe rot of grape, caused by Colletotrichum fioriniae, with leaf wetness, temperature, and the crop growth stage. PhytoFrontiers™ 2023, 3, 303–313. [Google Scholar] [CrossRef]
- Marcelino, J.; Giordano, R.; Gouli, S.; Gouli, V.; Parker, B.L.; Skinner, M.; TeBeest, D.; Cesnik, R. Colletotrichum acutatum var. fioriniae (teleomorph: Glomerella acutata var. fioriniae var. nov.) infection of a scale insect. Mycologia 2008, 100, 353–374. [Google Scholar] [CrossRef]
- Marcelino, J.A.; Gouli, S.; Parker, B.L.; Skinner, M.; Schwarzberg, L.; Giordano, R. Host plant associations of an entomopathogenic variety of the fungus, Colletotrichum acutatum, recovered from the elongate hemlock scale, Fiorinia externa. J. Insect Sci. 2009, 9, 25. [Google Scholar] [CrossRef]
- González, J.B.; Lambert, C.A.; Foley, A.M.; Hajek, A.E. First report of Colletotrichum fioriniae infections in brown marmorated stink bugs, Halyomorpha halys. J. Invertebr. Pathol. 2023, 199, 107939. [Google Scholar] [CrossRef]
- Ibrahim, A.; Tanney, J.B.; Fei, F.; Seifert, K.A.; Cutler, G.C.; Capretta, A.; Miller, J.D.; Sumarah, M.W. Metabolomic-guided discovery of cyclic nonribosomal peptides from Xylaria ellisii sp. nov., a leaf and stem endophyte of Vaccinium angustifolium. Sci. Rep. 2020, 10, 4599. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Stadler, M. Recent progress in biodiversity research on the Xylariales and their secondary metabolism. J. Antibiot. 2021, 74, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Sørensen, D.; Jenkins, H.A.; Ejim, L.; Capretta, A.; Sumarah, M.W. Epoxynemanione A, nemanifuranones A–F, and nemanilactones A–C, from Nemania serpens, an endophytic fungus isolated from Riesling grapevines. Phytochemistry 2017, 140, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.P.; Araujo, A.R.; Batista, J.M., Jr.; Cardoso, C.L.; Seidl, C.; Vilela, A.F.; Domingos, H.V.; Costa-Lotufo, L.V.; Andersen, R.J.; Silva, D.H. Botryane terpenoids produced by Nemania bipapillata, an endophytic fungus isolated from red alga Asparagopsis taxiformis-Falkenbergia stage. Sci. Rep. 2019, 9, 12318. [Google Scholar] [CrossRef]
- Tibpromma, S.; Zhang, L.; Karunarathna, S.C.; Du, T.-Y.; Phukhamsakda, C.; Rachakunta, M.; Suwannarach, N.; Xu, J.; Mortimer, P.E.; Wang, Y.-H. Volatile constituents of endophytic fungi isolated from Aquilaria sinensis with descriptions of two new species of Nemania. Life 2021, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Bilański, P.; Kowalski, T. Fungal endophytes in Fraxinus excelsior petioles and their in vitro antagonistic potential against the ash dieback pathogen Hymenoscyphus fraxineus. Microbiol. Res. 2022, 257, 126961. [Google Scholar] [CrossRef] [PubMed]
- Aleynova, O.A.; Nityagovsky, N.N.; Kiselev, K.V. Biodiversity of endophytic bacteria and fungi of wild grapes Vitis amurensis Rupr. In Proceedings of the BIO Web of Conferences, Virtual, 23–24 November 2021; p. 05001. [Google Scholar]
- Vasilyeva, L.N.; Stephenson, S.L. Pyrenomycetes of the Great Smoky Mountains National Park. III. Cryptosphaeria, Eutypa and Eutypella (Diatrypaceae). Fungal Divers. 2006, 22, 243–254. [Google Scholar]
- Zíbarová, L.; Kout, J. Xylariaceous pyrenomycetes from Bohemia: Species of Biscogniauxia and Hypoxylon new to the Czech Republic, and notes on other rare species. Czech Mycol. 2017, 69, 77–108. [Google Scholar] [CrossRef]
- Herath, P.; Beauseigle, S.; Dhillon, B.; Ojeda, D.I.; Bilodeau, G.; Isabel, N.; Gros-Louis, M.-C.; Kope, H.; Zeglen, S.; Hamelin, R.C. Anthropogenic signature in the incidence and distribution of an emerging pathogen of poplars. Biol. Invasions 2016, 18, 1147–1161. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Verkley, G.; Shin, H.-D.; Barreto, R.; Alfenas, A.; Swart, W.; Groenewald, J.; Crous, P.W. Sizing up septoria. Stud. Mycol. 2013, 75, 307–390. [Google Scholar] [CrossRef]
- Ali, S.; Hildebrand, P.D.; Renderos, W.E.; Abbasi, P.A. Identification and Characterization of Sphaerulina vaccinii sp. nov. as the Cause of Leaf Spot and Stem Canker in Lowbush Blueberry and Its Epidemiology. Phytopathology 2021, 111, 1560–1570. [Google Scholar] [CrossRef]
- Gumbrewicz, R.; Calderwood, L. Comparison of wood mulch particle sizes for wild blueberry management in a changing climate. Int. J. Fruit Sci. 2022, 22, 551–567. [Google Scholar] [CrossRef]
- Abbasi, P.A.; Hildebrand, P.D.; Ali, S.; Moreau, D.L.; Renderos, W.E. Effect of RH, Temperature, Light, and Plant Age on Infection of Lowbush Blueberry by Sphaerulina vaccinii. Plant Dis. 2022, 106, 297–303. [Google Scholar] [CrossRef]
- Thach, T.; Munk, L.; Hansen, A.L.; Jørgensen, L.N. Disease variation and chemical control of Ramularia leaf spot in sugar beet. Crop Prot. 2013, 51, 68–76. [Google Scholar] [CrossRef]
- McGrann, G.R.; Havis, N.D. Ramularia leaf spot: A newly important threat to barley production. Outlooks Pest Manag. 2017, 28, 65–69. [Google Scholar] [CrossRef]
- Ding, C.-H.; Wang, Q.-B.; Guo, S.; Wang, Z.-Y. The improvement of bioactive secondary metabolites accumulation in Rumex gmelini Turcz through co-culture with endophytic fungi. Braz. J. Microbiol. 2018, 49, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Gasparoto, M.; Lourenço, S.; Tanaka, F.A.O.; Spósito, M.B.; Marchini, L.C.; Silva Junior, G.; Amorim, L. Honeybees can spread Colletotrichum acutatum and C. gloeosporioides among citrus plants. Plant Pathol. 2017, 66, 777–782. [Google Scholar] [CrossRef]
- González-Domínguez, E.; Caffi, T.; Languasco, L.; Latinovic, N.; Latinovic, J.; Rossi, V. Dynamics of Diaporthe ampelina conidia released from grape canes that overwintered in the vineyard. Plant Dis. 2021, 105, 3092–3100. [Google Scholar] [CrossRef]
- Nelson, A.; Vandegrift, R.; Carroll, G.C.; Roy, B.A. Double lives: Transfer of fungal endophytes from leaves to woody substrates. PeerJ 2020, 8, e9341. [Google Scholar] [CrossRef]
- McGrann, G.R.; Andongabo, A.; Sjökvist, E.; Trivedi, U.; Dussart, F.; Kaczmarek, M.; Mackenzie, A.; Fountaine, J.M.; Taylor, J.M.; Paterson, L.J. The genome of the emerging barley pathogen Ramularia collo-cygni. BMC Genom. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Batzer, J.C.; Mueller, D.S. Soybean fungal endophytes Alternaria and Diaporthe spp. are differentially impacted by fungicide application. Plant Dis. 2020, 104, 52–59. [Google Scholar] [CrossRef]
- Arnold, A.E.; Mejía, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar] [CrossRef]
- Nelson, J.M.; Hauser, D.A.; Hinson, R.; Shaw, A.J. A novel experimental system using the liverwort Marchantia polymorpha and its fungal endophytes reveals diverse and context-dependent effects. New Phytol. 2018, 218, 1217–1232. [Google Scholar] [CrossRef]
- Sadoral, J.P.; Cumagun, C.J.R. Observations on the Potential of an Endophytic Fungus Associated with Cacao Leaves against Phytophthora palmivora. Microbiol. Res. 2021, 12, 528–538. [Google Scholar] [CrossRef]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [PubMed]
- Janzen, D.H. Why fruits rot, seeds mold, and meat spoils. Am. Nat. 1977, 111, 691–713. [Google Scholar] [CrossRef]
- Peris, J.E.; Rodríguez, A.; Peña, L.; Fedriani, J.M. Fungal infestation boosts fruit aroma and fruit removal by mammals and birds. Sci. Rep. 2017, 7, 5646. [Google Scholar] [CrossRef]






| Species | Isolate | Host | Location | GenBank Accession No. | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TUB2 | ITS | TEF1-α | RPB2 | ACT | CHS-1 | LSU | |||||
| Cercospora chrysanthemoides | CPC 20529 | Chrysanthemoides monilifera | South Africa | — | — | KC005813 | — | — | — | — | [30] |
| Colletotrichum brisbanense | CBS 292.67 | Capsicum annuum | Australia | — | — | — | — | — | JQ948952 | — | [31] |
| Colletotrichum citri | CBS 134233 | Citrus aurantifolia | China | — | — | — | — | — | KY856138 | — | [32] |
| Colletotrichum cosmi | CBS 853.73 | Cosmos sp. | Netherlands | — | — | — | — | — | JQ948935 | — | [31] |
| Colletotrichum eriobotryae | BCRC FU31138 | Eriobotrya japonica | Taiwan | — | — | — | — | — | MN191653 | — | [33] |
| Colletotrichum fioriniae | En25-3 | Vitis sp. ‘Clinton’ | Canada | — | MZ127182 | — | OK431476 | — | OK380951 | This study. | |
| Colletotrichum fioriniae | CBS 128517 | Fiorinia externa | USA | — | — | — | — | — | JQ948953 | — | [31] |
| Colletotrichum laticiphilum | CBS 112989 | Hevea brasiliensis | India | — | — | — | — | — | JQ948950 | — | [31] |
| Colletotrichum nymphaeae | CBS 515.78 | Nymphaea alba | Netherlands | — | — | — | — | — | JQ948858 | — | [31] |
| Colletotrichum simmondsii | CBS 122122 | Carica papaya | Australia | — | — | — | — | — | JQ948937 | — | [31] |
| Colletotrichum spaethianum | CBS 167.49 | Hosta sieboldiana | Germany | — | — | — | — | — | GU228297 | — | [34] |
| Colletotrichum walleri | CBS 125472 | Coffea sp. | Vietnam | — | — | — | — | — | JQ948936 | — | [31] |
| Colletotrichum wanningense | CGMCC 3.18936 | Hevea brasiliensis | China | — | — | — | — | — | MZ352012 | — | Liu et al. (Direct Submission) |
| Diaporthe aff. gulyae | En20-4 | Vitis sp. unknown | Canada | OK383388 | MZ127185 | — | — | — | OK380954 | This study | |
| Diaporthe alleghaniensis | CBS 495.72 | Betula alleghaniensis | Canada | KC843228 | NR_103696 | KC343733 | — | — | — | — | [35] |
| Diaporthe alnea | CBS 146.46 | Alnus sp. | Unknown | KC343976 | NR_147525 | KC343734 | — | — | — | — | [35] |
| Diaporthe angelicae | CBS 111592 | Heracleum sphondylium | Austria | KC343995 | KC343027 | KC343753 | — | — | — | — | [35] |
| Diaporthe arctii | DP0482 | Arctium lappa | Austria | KJ610891 | KJ590736 | KJ590776 | — | — | — | — | [36] |
| Diaporthe betulae | CFCC 50470 | Betula platyphylla | China | KT733021 | KT732951 | KT733017 | — | — | — | — | [37]) |
| Diaporthe betulae | CFCC 50469 | Betula platyphylla | China | KT733020 | NR_147578 | KT733016 | — | — | — | — | [37] |
| Diaporthe bicincta | CBS 121004 | Juglans sp. | USA | KC344102 | NR_147526 | KC343860 | — | — | — | — | [35] |
| Diaporthe biguttusis | CGMCC 3.17081 | Lithocarpus glaber | China | KF576306 | NR_147533 | KF576257 | — | — | — | — | [38] |
| Diaporthe celastrina | CBS 139.27 | Celastrus scandens | Unknown | KC344015 | NR_152457 | KC343773 | — | — | — | — | [35] |
| Diaporthe celeris | CBS 143349 | Vitis vinifera | UK | MG281190 | MG281017 | MG281538 | — | — | — | — | [39] |
| Diaporthe charlesworthii | BRIP 54884m | Rapistrum rugosum | Australia | KJ197268 | NR_147538 | KJ197250 | — | — | — | — | [40] |
| Diaporthe chensiensis | CFCC 52567 | Abies chensiensis | China | MH121584 | NR_165876 | MH121544 | — | — | — | — | [41] |
| Diaporthe cotoneastri | CBS 439.82 | Cotoneaster sp. | UK | JX275437 | MH861511 | GQ250341 | — | — | — | — | [42,43,44] |
| Diaporthe cucurbitae | DAOM 42078 | Cucumis sp. | Canada | KP118848 | KM453210 | KM453211 | — | — | — | — | [36] |
| Diaporthe cuppatea | CBS 117499 | Aspalathus linearis | South Africa | KC344025 | MH863021 | KC343783 | — | — | — | — | [35,42] |
| Diaporthe ellipicola | CGMCC 3.17084 | Lithocarpus glaber | China | KF576294 | NR_147531 | KF576245 | — | — | — | — | [38] |
| Diaporthe eres | En61-3 | Vitis sp. ‘Isabella’ | Canada | OK383392 | MZ127189 | — | — | — | OK380958 | This study | |
| Diaporthe eres | En26-4 | Vitis sp. ‘Clinton’ | Canada | OK383390 | MZ127187 | — | — | — | OK380956 | This study | |
| Diaporthe eres | En26-5 | Vitis sp. ‘Clinton’ | Canada | OK383391 | MZ127188 | — | — | — | OK380957 | This study | |
| Diaporthe eres | En25-6 | Vitis sp. ‘Clinton’ | Canada | OK383389 | MZ127186 | — | — | — | MZ127186 | This study | |
| Diaporthe eres | AR5193 | Ulmus sp. | Germany | KJ420799 | KJ210529 | KJ210550 | — | — | — | — | [45] |
| Diaporthe guangxiensis | JZB320094 | Vitis vinifera | China | MK500168 | MK335772 | MK523566 | — | — | — | — | [46] |
| Diaporthe gulyae | BRIP 54025 | Helianthus annuus | Australia | KJ197271 | NR_111615 | JN645803 | — | — | — | — | [40,47,48] |
| Diaporthe hispaniae | CBS 143351 | Vitis vinifera | Spain | MG281296 | MG281123 | MG281644 | — | — | — | — | [39] |
| Diaporthe hungariae | CBS 143353 | Vitis vinifera | Hungary | MG281299 | MG281126 | MG281647 | — | — | — | — | [39] |
| Diaporthe lusitanicae | CBS 123212 | Foeniculum vulgare | Portugal | KC344104 | MH863279 | KC343862 | — | — | — | — | [35,42], |
| Diaporthe mahothocarpus | CGMCC 3.15181 | Lithocarpus glabra | China | KF576312 | KC153096 | KC153087 | — | — | — | — | |
| Diaporthe maritima | DAOMC 25056 | Picea rubens | Canada | KU574615 | NR_152463 | KU552023 | — | — | — | — | [49] |
| Diaporthe momicola | MFLUCC 16-0113 | Prunus persica | China | KU557587 | NR_172386 | KU557631 | — | — | — | — | [50] |
| Diaporthe myracrodruonis | URM 7972 | Astronium urundeuva | Brazil | MK205291 | NR_163320 | MK213408 | — | — | — | — | [15] |
| Diaporthe neoarctii | CBS 109490 | Ambrosia trifida | USA | KC344113 | NR_111854 | KC343871 | — | — | — | — | [35,47] |
| Diaporthe novem | CBS 127270 | Glycine max | Croatia | KC344124 | MH864503 | KC343882 | — | — | — | — | [35,42] |
| Diaporthe novem | CBS 127271 | Glycine max | Croatia | KC344125 | MH864504 | KC343883 | — | — | — | — | [35,42] |
| Diaporthe padina | CFCC 52590 | Prunus padus | China | MH121604 | NR_165879 | MH121567 | — | — | — | — | [41] |
| Diaporthe phaseolorum | AR4203 | Phaseolus vulgaris | USA | KJ610893 | KJ590738 | KJ590739 | — | — | — | — | [36] |
| Diaporthe rosicola | MFLU 17-0646 | Rosa sp. | UK | MG843877 | NR_157515 | MG829270 | — | — | — | — | [51] |
| Diaporthe shennongjiaensis | CNUCC 201905 | Juglans regia | China | MN227012 | MN216229 | MN224672 | — | — | — | — | [52] |
| Diaporthe sinensis | ZJUP0033-4 | Amaranthus sp. | China | MK660447 | MK637451 | MK660449 | — | — | — | — | [53] |
| Diaporthe sp. | En01-1 | Vitis riparia | Canada | OK383387 | MZ127184 | — | — | — | OK380953 | This study | |
| Diaporthe vaccinii | CBS 160.32 | Oxycoccus macrocarpos | USA | KC344196 | NR_103701 | KC343954 | — | — | — | — | [35] |
| Diaporthe vacuae | CAA829 | Vaccinium corymbosum | Portugal | MK837928 | MK792306 | MK828077 | — | — | — | — | [54] |
| Diaporthe vacuae | CAA830 | Vaccinium corymbosum | Portugal | MK837931 | MK792309 | MK828080 | — | — | — | — | [54]) |
| Diaporthe vacuae | CAA1001 | Quercus suber | Portugal | MT309458 | MT237172 | MT309432 | — | — | — | — | [54] |
| Diaporthella corylina | CBS 121124 | Corylus sp. | China | KC343972 | KC343004 | KC343730 | — | — | — | — | [35] |
| Gnomoniopsis angolensis | CBS 145057 | Unknown plant sp. | Angola | — | MK047428 | — | MK047539 | — | — | — | [55] |
| Gnomoniopsis paraclavulata | BPI 877448 | Carpinus caroliniana | USA | — | EU254839 | — | EU219248 | — | — | [56] | |
| Gnomoniopsis paraclavulata | En61-1 | Vitis sp. ‘Isabella’ | Canada | OK383385 | MZ127179 | — | OK431474 | — | — | OK380949 | This study. |
| Gnomoniopsis racemula | BPI 871003 | Epilobium angustifolium | USA | — | EU254841 | — | EU219241 | — | — | — | [56] |
| Gnomoniopsis rosae | CBS 145085 | Rosa sp. | New Zealand | — | NR_161142 | — | MK047547 | — | — | — | [55] |
| Gnomoniopsis smithogilvyi | CBS 130190 | Castanea sp. | Australia | — | MH865607 | — | JQ910648 | — | — | — | [30,42] |
| Melanconis alni | AR 3748 | Alnus viridis | Austria | — | EU199195 | — | EU199153 | — | — | — | [57] |
| Nemania aenea | N110C | Unknown | Unknown | — | AJ390427 | — | — | — | — | — | [58] |
| Nemania aenea | nem046 | Centaurea stoebe | USA | — | EF589887 | — | — | — | — | — | [59] |
| Nemania aenea var. aureolatum | ATCC 60819 | Quercus sp. | Switzerland | — | AF201704 | — | — | — | — | — | [60] |
| Nemania aureolutea | En25-1 | Vitis sp. ‘Clinton’ | Canada | OK383386 | MZ127183 | — | OK431477 | OK431472 | — | OK380952 | This study |
| Nemania aureolutea | MAR101219 | Quercus canariensis | Spain | — | MW136058 | — | — | — | — | — | [61] |
| Nemania chestersii | N23A | Unknown | Unknown | — | AJ390430 | — | — | — | — | — | [58] |
| Nemania serpens | NC0348 | Lecanora oreinoides | USA | — | JQ761380 | — | — | — | — | — | [62] |
| Nemania serpens | NC0276 | Diploschistes rampoddensis | USA | — | JQ761314 | — | — | — | — | — | [62] |
| Nemania serpens | N112C | Unknown | Unknown | — | AJ390436 | — | — | — | — | — | [58] |
| Nemania serpens | CBS 533.72 | Corylus avellana | Netherlands | — | FN428829 | — | — | — | — | — | [63] |
| Nemania serpens | CBS 679.86 | Betula sp. | Switzerland | — | KU683765 | — | — | — | — | — | [62] |
| Nemania serpens var. macrospora | N21A | Unknown | Unknown | — | AJ390433 | — | — | — | — | — | [58] |
| Nemania serpens var. macrospora | ATCC 60823 | Unknown | Unknown | — | AF201707 | — | — | — | — | — | [60] |
| Nemania serpens var. serpens | CBS 659.70 | Soil | Canada | — | MH859890 | — | — | — | — | — | [42] |
| Nemania sp. | Cor 16 | Nephroma laevigatum | France | — | MG916993 | — | — | — | — | — | [64] |
| Nemania sp. | Cor 15 | Nephroma laevigatum | France | — | MG916992 | — | — | — | — | — | [64] |
| Nemania sp. | Gir 10 | Nephroma laevigatum | France | — | MG917014 | — | — | — | — | — | [64] |
| Nemania sp. | Gir 9 | Nephroma laevigatum | France | — | MG917013 | — | — | — | — | — | [64] |
| Neophaeomoniella eucalyptigena | CBS 145093 | Eucalyptus pilularis | Australia | — | NR_161148 | MK047569 | — | — | — | — | [55] |
| Neophaeomoniella niveniae | CBS 131316 | Nivenia stokoei | South Africa | — | JQ044435 | MN861682 | — | — | — | — | [65,66] |
| Neophaeomoniella niveniae | STE-U 7959 | Olea europaea subsp. cuspidata | South Africa | — | MT791053 | MT787396 | — | — | — | — | [67] |
| Neophaeomoniella niveniae | En61-2 | Vitis sp. ‘Isabella’ | Canada | OK383384 | MZ127178 | — | — | — | OK380948 | This study | |
| Neophaeomoniella zymoides | CBS 121168 | Prunus salicina | South Africa | — | GQ154600 | MN861679 | — | — | — | — | [65,68] |
| Neophaeomoniella zymoides | STE-U 7960 | Olea europaea subsp. cuspidata | South Africa | — | MT791054 | MT787397 | — | — | — | — | [67] |
| Phaeomoniella chlamydospora | IBVD01 | Vitis vinifera | Brazil | — | KP213118 | KP213113 | — | — | — | — | [69] |
| Ramularia collo-cygni | CBS 101180 | Hordeum vulgare | Austria | — | NR_154944 | — | KX288543 | KX287666 | — | — | [70] |
| Ramularia eucalypti | CBS 120726 | Corymbia grandifolia | Italy | — | KJ504792 | — | KJ504663 | KF253635 | — | — | [71,72] |
| Ramularia glennii | CBS 129441 | Homo sapiens | Netherlands | — | MH865235 | — | KJ504433 | KJ504640 | — | — | [42,71] |
| Ramularia haroldporteri | CPC 16296 | Unidentified plant | South Africa | — | NR_154911 | — | KJ504637 | KJ504430 | — | — | [71] |
| Ramularia heraclei | CBS 108969 | Heracleum sphondylium | Netherlands | — | NR_154948 | — | KX288578 | KX287702 | — | — | [70] |
| Ramularia hydrangeae-macrophyllae | CBS 122273 | Hydrangea macrophylla | New Zealand | — | NR_145125 | — | KX288592 | KX287716 | — | — | [70] |
| Ramularia lamii var. lamii | CBS 108970 | Lamium album | Netherlands | — | NR_154949 | — | KX288620 | KX287744 | — | — | [70] |
| Ramularia mali | CBS 129581 | Malus sp. | Italy | — | NR_156582 | — | KJ504649 | MH876894 | — | — | [42,71] |
| Ramularia osterici | CPC 10750 | Ostericum koreanum | South Korea | — | NR_154950 | — | KX288642 | KX287765 | — | — | [70] |
| Ramularia pratensis var. pratensis | CPC 11294 | Rumex crispus | South Korea | — | EU019284.2 | — | KT216537 | KF903599 | — | — | [73,74,75] |
| Ramularia sp. | En60-1 | Vitis sp. ‘Marechal foch’ | Canada | OK383393 | MZ127180 | — | OK431478 | OK431473 | — | OK380959 | This study |
| Ramularia stellenboschensis | CBS 130600 | Protea sp. | South Africa | — | NR_145101 | — | KX288676 | KX287798 | — | — | [42,70] |
| Ramularia vallisumbrosae | CBS 272.38 | Narcissus ‘Golden Spur’ | UK | — | NR_154953 | — | KX288698 | KX288699 | — | — | [70] |
| Sphaerulina amelanchier | En26-1 | Vitis sp. ‘Clinton’ | Canada | — | MZ127181 | OK431475 | — | — | OK380950 | This study. | |
| Sphaerulina amelanchier | CBS 135110 | Amelanchier sp. | Netherlands | — | — | KF253543 | — | — | — | — | [72] |
| Sphaerulina gei | CBS 102318 | Geum urbanum | Netherlands | — | — | KF253560 | — | — | — | — | [72] |
| Sphaerulina hyperici | CBS 102313 | Hypericum sp. | Netherlands | — | — | KF253563 | — | — | — | — | [72] |
| Sphaerulina pelargonii | CBS 138857 | Pelargonium sp. | South Africa | — | — | KP004506 | — | — | — | — | [72] |
| Sphaerulina rhabdoclinis | CBS 102195 | Pseudotsuga menziesii | Germany | — | — | KF253578 | — | — | — | — | [72] |
| Sphaerulina tirolensis | CBS 109018 | Rubus idaeus | Austria | — | — | KF253585 | — | — | — | — | [72] |
| Sphaerulina westendorpii | CBS 109002 | Rubus sp. | Netherlands | — | — | KF253588 | — | — | — | — | [72] |
| Xylaria longipes | CBS 347.37 | Unknown | Unknown | — | MH855925 | — | — | — | — | — | [42] |
| Zymoseptoria halophila | CBS 128854 | Hordeum vulgare | Iran | — | MH865126 | — | KX348110 | KF253946 | — | — | [30,42,72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, S.; Wright, A.H.; Tanney, J.B.; Renaud, J.B.; Sumarah, M.W. Fungal Endophytes: Discovering What Lies within Some of Canada’s Oldest and Most Resilient Grapevines. J. Fungi 2024, 10, 105. https://doi.org/10.3390/jof10020105
Ali S, Wright AH, Tanney JB, Renaud JB, Sumarah MW. Fungal Endophytes: Discovering What Lies within Some of Canada’s Oldest and Most Resilient Grapevines. Journal of Fungi. 2024; 10(2):105. https://doi.org/10.3390/jof10020105
Chicago/Turabian StyleAli, Shawkat, A. Harrison Wright, Joey B. Tanney, Justin B. Renaud, and Mark W. Sumarah. 2024. "Fungal Endophytes: Discovering What Lies within Some of Canada’s Oldest and Most Resilient Grapevines" Journal of Fungi 10, no. 2: 105. https://doi.org/10.3390/jof10020105
APA StyleAli, S., Wright, A. H., Tanney, J. B., Renaud, J. B., & Sumarah, M. W. (2024). Fungal Endophytes: Discovering What Lies within Some of Canada’s Oldest and Most Resilient Grapevines. Journal of Fungi, 10(2), 105. https://doi.org/10.3390/jof10020105






