Time-Course Responses of Apple Leaf Endophytes to the Infection of Gymnosporangium yamadae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Sampling
2.2. DNA Extraction and PCR Amplification
2.3. Library Preparation for Illumina MiSeq Sequencing
2.4. Bioinformatics Analysis
3. Results
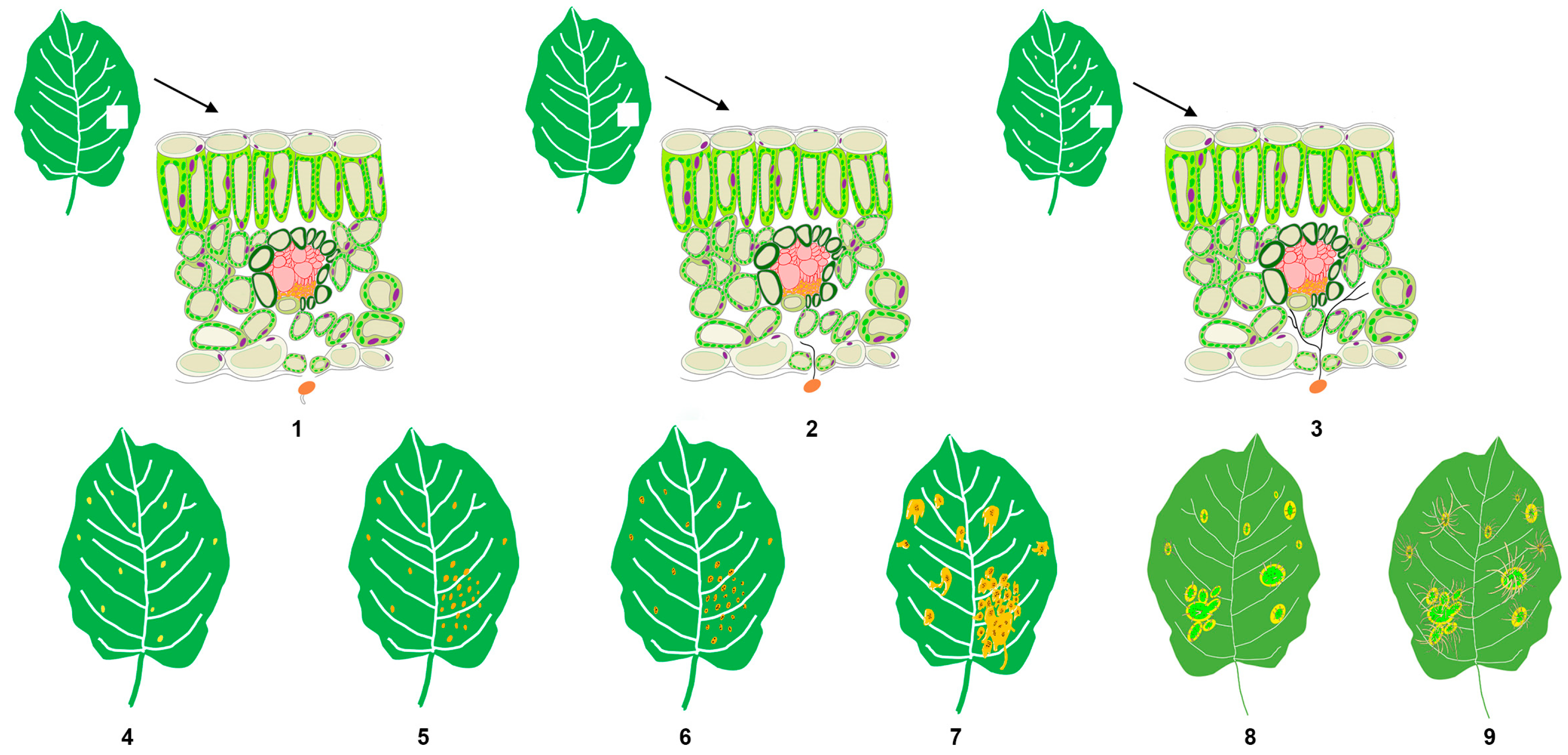
3.1. A Series of Symptoms Change during Apple Rust Infection
3.2. Composition of Bacterial and Fungal Communities in the Endophytes of Apple Leaves
3.3. Changes in Endophytic Bacterial and Fungal Communities during Disease Progression in Apple Leaves
3.4. Microbial Correlation of Diseased and Healthy Samples
3.5. Microbial Functional Allocation at Different Stages of the Disease Progression in Apple Leaves
4. Discussion
4.1. Diversity and Structure Dynamics during Disease Progression
4.2. Shift in Composition of Leaves Endophytic Microbial Communities with G. yamadae Infection
4.3. Small Changes in Host Microbiome Composition Predict Disease Outcomes Earlier Than External Symptom Variation
4.4. Contrary to the Characteristics of Healthy Leaves with More Beneficial Endophytes, the Harmful Flora Became the Characteristic Flora in the Experimental Group
4.5. Changes in the Co-Occurrence of the Endophytic Microbial Community with the Enlargement of the Course of Disease
4.6. Endophytes Contend with Gymnosporangium yamadae through a Series of Metabolic Processes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kern, F.D. A Revised Taxonomic Account of Gymnosporangium. Mycologia 1973, 66, 206. [Google Scholar]
- Lu, Y.F.; Chen, Q.; Bu, Y.F.; Luo, R.; Hao, S.X.; Zhang, J.; Tian, J.; Yao, Y.C. Flavonoid accumulation plays an important role in the rust resistance of Malus plant leaves. Front. Plant Sci. 2017, 8, 1286. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Liu, F.; Li, Y.M.; Cai, L. Inferring phylogeny and speciation of Gymnosporangium species and their coevolution with host plants. Sci Rep. 2016, 6, 29339. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.G.; Zhao, D.Y. Development orientation and trend of the apple industry under the new situation. Chin. Acad. Agric. Sci. 2019, 1, 1–7. (In Chinese) [Google Scholar]
- Liu, B.; Zhang, Y.; He, D.J.; Li, Y.X. Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 2018, 10, 11. [Google Scholar] [CrossRef]
- Yun, H.Y.; Andrew, M.; Amy, R. First report of Gymnosporangium yamadae, Japanese apple rust, on Malus from North America. Plant Dis. 2009, 93, 430. [Google Scholar] [CrossRef]
- Cao, B.; Tian, C.M.; Liang, Y.M. Gymnosporangium huanglongense sp. nov. from western China. Mycotaxon 2016, 131, 375–383. [Google Scholar] [CrossRef]
- Xu, H.W.; Zhu, Y.P. Disease epidemics and control techniques of pear rust in Hangzhou area. China Fruit 1997, 1, 10–12. (In Chinese) [Google Scholar]
- Shaw, M. The physiology and host-parasite relations of the rusts. Annu. Rev. Phytopathol. 1963, 1, 259–294. [Google Scholar] [CrossRef]
- Pereira, L.B.; Thomazella, D.P.; Teixeira, P.J. Plant-microbiome crosstalk and disease development. Curr. Opin. Plant Biol. 2023, 72, 102351. [Google Scholar] [CrossRef]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef]
- Matsumoto, H.; Fan, X.Y.; Wang, Y.; Kusstatscher, P.; Duan, J.; Wu, S.; Chen, S.L.; Qiao, K.; Wang, Y.L.; Ma, B.; et al. Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 2021, 7, 60–72. [Google Scholar] [CrossRef]
- Chang, Y.; Xia, X.; Sui, L.; Kang, Q.; Lu, Y.; Li, L. Endophytic colonization of entomopathogenic fungi increases plant disease resistance by changing the endophytic bacterial community. J. Basic Microbiol. 2021, 61, 1098–1112. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, J.; Zhang, H.; Ji, G.; Zeng, L.; Li, Y.; Yu, C.; Dilantha-Femando, W.G.; Chen, W. Bacterial blight induced shifts in endophytic microbiome of rice leaves and the enrichment of specific bacterial strains with pathogen antagonism. Front. Plant Sci. 2020, 11, 963. [Google Scholar] [CrossRef] [PubMed]
- Kõiv, V.; Roosaare, M.; Vedler, E.; Kivistik, P.A.; Toppi, K.; Schryer, D.W.; Remm, T.T.; Mäe, A. Microbial population dynamics in response to Pectobacterium atrosepticum infection in potato tubers. Sci. Rep. 2015, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Liu, X.; Chen, J.; Wang, B.; Li, Y.; Ye, Y.; Ma, W.; Ma, L. Effects on community composition and function Pinus massoniana Infected by Bursaphelenchus xylophilus. BMC Microbiol. 2022, 22, 157. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Xiong, C.; Gao, C.; Tsui, C.-K.M.; Wang, M.M.; Zhou, X.; Zhang, A.M.; Cai, L. Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome 2021, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; Hollander, M.D. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, L.; Guillerm-Erckelboudt, A.Y.; Gazengel, K.; Linglin, J.; Ourry, M.; Glory, P.; Sarnigue, A.; Daval, S.; Manzanares-Dauleux, M.J.; Mougel, C. Temporal dynamics of bacterial and fungal communities during the infection of Brassica rapa roots by the protist Plasmodiophora brassicae. PLoS ONE 2019, 14, e0204195. [Google Scholar] [CrossRef] [PubMed]
- Morrison, E.W.; Kasson, M.T.; Heath, J.J.; Garnas, J.R. Pathogen and endophyte assemblages co-vary with beech bark disease progression, tree decline, and regional climate. Front. For. Glob. Change 2021, 4, 673099. [Google Scholar] [CrossRef]
- Tao, S.Q.; Auer, L.; Morin, E.; Liang, Y.M.; Duplessis, S. Transcriptome analysis of apple leaves infected by the rust fungus Gymnosporangium yamadae at two sporulation stages. Mol. Plant-Microbe Interact. 2020, 33, 444–461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Cao, B.; Pan, Y.M.; Tao, S.Q.; Zhang, N.L. Metabolite-mediated responses of phyllosphere microbiota to rust infection in two Malus species. Microbiol. Spectr. 2023, 11, e03831-22. [Google Scholar] [CrossRef] [PubMed]
- Busby, P.E.; Peay, K.G.; Newcombe, G. Common foliar fungi of Populus trichocarpa modify Melampsora rust disease severity. New Phytol. 2016, 209, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Balvočiūtė, M.; Huson, D.H.; Silva, R.D.P. Greengenes, NCBI and OTT—How do these taxonomies compare? BMC Genom. 2017, 18, 114. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Knight, R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. GigaScience 2013, 2, 16. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Caspi, R.; Altman, T.; Billington, R.; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014, 42, D459–D471. [Google Scholar] [CrossRef]
- Lorrain, C.; Gonçalves dos Santos, K.C.; Germain, H.; Hecker, A.; Duplessis, S. Advances in understanding obligate biotrophy in rust fungi. New Phytol. 2019, 222, 1190–1206. [Google Scholar] [CrossRef]
- Dou, D.L.; Zhou, J.M. Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 2012, 12, 484–495. [Google Scholar] [CrossRef]
- Bakker, P.A.; Pieterse, C.M.; de Jonge, R.; Berendsen, R.L. The soil-borne legacy. Cell 2018, 172, 1178–1180. [Google Scholar] [CrossRef]
- Xiong, C.; Singh, B.K.; He, J.Z.; Han, Y.L.; Li, P.P.; Wan, L.H.; Meng, G.Z.; Liu, S.Y.; Wang, J.T.; Wu, C.F.; et al. Plant developmental stage drives the differentiation in ecological role of the maize microbiome. Microbiome 2021, 9, 171. [Google Scholar] [CrossRef]
- Carrell, A.A.; Frank, A.C. Bacterial endophyte communities in the foliage of coast redwood and giant sequoia. Front. Microbiol. 2015, 6, 1008. [Google Scholar] [CrossRef]
- Pérez-García, A.; Romero, D.; De Vicente, A. Plant protection and growth stimulation by microorganisms: Biotechnological applications of Bacilli in agriculture. Curr. Opin. Biotechnol. 2011, 22, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Palaniyandi, S.A.; Yang, S.H.; Zhang, L.X.; Suh, J.W. Effects of actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 2013, 97, 9621–9636. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.H.; Wei, Z.; Shao, Z.Y.; Friman, V.P.; Cao, K.H.; Yang, T.J.; Kramer, J.; Wang, X.F.; Li, M.; Mei, X.L.; et al. Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat. Microbiol. 2020, 5, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Seybold, H.; Demetrowitsch, T.J.; Hassani, M.A.; Szymczak, S.; Reim, E.; Haueisen, J.; Lübbers, L.; Rühlemann, M.; Franke, A.; Schwarz, K.; et al. A fungal pathogen induces systemic susceptibility and systemic shifts in wheat metabolome and microbiome composition. Nat. Commun. 2020, 11, 1910. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.S.; Hachani, A.; Lin, J.S.; Filloux, A.; Lai, E.M. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 2014, 16, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Souza, D.P.; Oka, G.U.; Alvarez-Martinez, C.E.; Bisson-Filho, A.W.; Dunger, G.; Hobeika, L.; Cavalcante, N.S.; Alegria, M.C.; Barbosa, L.R.S.; Salinas, R.K.; et al. Bacterial killing via a type IV secretion system. Nat. Commun. 2015, 6, 6453. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.J.; Spakowicz, D.J.; Dalal, R.S.; Davis, J.H.; Lehr, N.A.; Dunican, B.F.; Orellana, E.A.; Narváez-Trujillo, A.; Strobel, S.A. Biosynthesis and genomic analysis of medium-chain hydrocarbon production by the endophytic fungal isolate Nigrograna mackinnonii E5202H. Appl. Microbiol. Biotechnol. 2015, 99, 3715–3728. [Google Scholar] [CrossRef] [PubMed]
- Stodůlková, E.; Man, P.; Kuzma, M.; Černý, J.; Císařová, I.; Kubátová, A.; Chudíčková, M.; Kolařík, M.; Flieger, M. A highly diverse spectrum of naphthoquinone derivatives produced by the endophytic fungus Biatriospora sp. CCF 4378. Folia Microbiol. 2015, 60, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Bez, C.; Esposito, A.; Thuy, H.D.; Hong, M.N.; Valè, G.; Licastro, D.; Bertani, I.; Piazza, S.; Venturi, V. The rice foot rot pathogen Dickeya zeae alters the in-field plant microbiome. Environ. Microbiol. 2021, 23, 7671–7687. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Banerjee, S.; Dini-Andreote, F.; Xu, Y.; Shen, Q.; Jousset, A.; Wei, Z. Small changes in rhizosphere microbiome composition predict disease outcomes earlier than pathogen density variations. ISME J. 2020, 16, 2448–2456. [Google Scholar] [CrossRef] [PubMed]
- Dini-Andreote, F. Endophytes: The second layer of plant defense. Trends Plant Sci. 2020, 25, 319–322. [Google Scholar] [CrossRef]
- Schlaberg, R. Microbiome diagnostics. Clin. Chem. 2020, 66, 68–76. [Google Scholar] [CrossRef]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef]
- Granados, G.M.; McTaggart, A.R.; Rodas, C.A.; Roux, J.; Wingfield, M.J. Species of Cryphonectriaceae occupy an endophytic niche in the Melastomataceae and are putative latent pathogens of Eucalyptus. Eur. J. Plant Pathol. 2020, 156, 273–283. [Google Scholar] [CrossRef]
- Sakalidis, M.L.; Hardy, G.E.S.; Burgess, T.I. Endophytes as potential pathogens of the baobab species Adansonia gregorii: A focus on the Botryosphaeriaceae. Fungal Ecol. 2011, 4, 1–14. [Google Scholar] [CrossRef]
- Sharma, D.K.; Sharma, M. Enumerations on seed-borne and post-harvest diseases of bitter gourd (Momordica charantia L.) and their management. Asia J. Pharm. Pharmacol. 2018, 4, 744–751. [Google Scholar] [CrossRef]
- Kairova, G.; Daulet, N.; Solomadin, M.; Sandybayev, N.; Orkara, S.; Beloussov, V.; Kerimbek, N.; Gritsenko, D.; Sapakhova, Z. Identification of apple varieties resistant to Fire Blight (Erwinia amylovora) Using Molecular Markers. Horticulturae 2023, 9, 1000. [Google Scholar] [CrossRef]
- Chen, J.J.; Tomes, S.; Gleave, A.P.; Hall, W.; Luo, Z.W.; Xu, J.; Yao, J.L. Significant improvement of apple (Malus domestica Borkh.) transgenic plant production by pre-transformation with a Baby boom transcription factor. Hortic. Res.-Engl. 2022, 9, uhab14. [Google Scholar] [CrossRef] [PubMed]
- Osdaghi, E.; Taghavi, S.M.; Fazliarab, A.; Elahifard, E.; Larmichhane, J.R. Characterization, geographic distribution and host range of Curtobacterium flaccumfaciens: An emerging bacterial pathogen in Iran. Crop Prot. 2015, 78, 185–192. [Google Scholar] [CrossRef]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Murray, T.D.; Schroeder, B.K.; Schneider, W.L.; Luster, D.G.; Sechler, A.; Rogers, E.E.; Subbotin, S.A. Rathayibacter toxicus, other Rathayibacter species inducing bacterial head blight of grasses, and the potential for livestock poisonings. Phytopathology 2017, 107, 804–815. [Google Scholar] [CrossRef]
- Kaplan, M.M. Novosphingobium aromaticivorans: A potential initiator of primary biliary cirrhosis. Am. J. Gastroenterol. 2004, 99, 2147–2149. [Google Scholar] [CrossRef]
- Singh, N.; Marwa, N.; Mishra, J.; Verma, P.C.; Rathaur, S.; Singh, N. Brevundimonas diminuta mediated alleviation of arsenic toxicity and plant growth promotion in Oryza sativa L. Ecotox. Environ. Saf. 2016, 125, 25–34. [Google Scholar] [CrossRef]
- Naqqash, T.; Imran, A.; Hameed, S.; Shahid, M.; Majeed, A.; Iqbal, J.; Hanif, M.K.; Ejaz, S.; Malik, K.A. First report of diazotrophic Brevundimonas spp. as growth enhancer and root colonizer of potato. Sci. Rep. 2020, 10, 14. [Google Scholar] [CrossRef]
- Ramette, A.; Frapolli, M.; Fischer-Le Saux, M.; Gruffaz, C.; Meyer, J.M.; Défago, G.; Sutra, L.; Moënne-Loccoz, Y. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 2011, 34, 180–188. [Google Scholar] [CrossRef]
- Santoyo, G.; Orozco-Mosqueda, M.D.; Govindappa, M. Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: A review. Biocontrol. Sci. Technol. 2012, 22, 855–872. [Google Scholar] [CrossRef]
- Daranas, N.; Rosello, G.; Cabrefiga, J.; Donati, I.; Frances, J.; Badosa, E.; Spinelli, F.; Montesinos, E.; Bonaterra, A. Biological control of bacterial plant diseases with Lactobacillus plantarum strains selected for their broad-spectrum activity. Ann. Appl. Biol. 2019, 174, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Siddharthan, N.; Balagurunathan, R.; Venkatesan, S.; Hemalatha, N. Bio-efficacy of Geobacillus thermodenitrificans PS41 against larvicidal, fungicidal, and plant growth-promoting activities. Environ. Sci. Pollut. Res. 2023, 30, 42596–42607. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Egamberdieva, D.; Lugtenberg, B.; Hagemann, M. Symbiotic Plant–Microbe Interactions: Stress Protection, Plant Growth Promotion, and Biocontrol by Stenotrophomonas; Springer: Cham, The Netherlands, 2010; pp. 445–460. [Google Scholar]
- Gaya, P.; Peiroten, A.; Landete, J.M. Transformation of plant isoflavones into bioactiIve isoflavones by lactic acid bacteria and bifidobacteria. J. Funct. Food 2017, 39, 198–205. [Google Scholar] [CrossRef]
- Inbar, J.; Chet, I. Evidence that chitinase produced by Aeromonas caviae is involved in the biological control of soil-borne plant pathogens by this bacterium. Soil Biol. Biochem. 1991, 23, 973–978. [Google Scholar] [CrossRef]
- Shi, Z.J.; Qi, X.L.; Zeng, X.A.; Lu, Y.J.; Zhou, J.L.; Cui, K.H.; Zhang, L.M. A newly isolated bacterium Comamonas sp. XL8 alleviates the toxicity of cadmium exposure in rice seedlings by accumulating cadmium. J. Hazard. Mater. 2021, 403, 9. [Google Scholar] [CrossRef] [PubMed]
- De Aquino, G.S.; Ventura, M.U.; Alexandrino, R.P.; Michelon, T.A.; Pescador, P.G.D.; Nicio, T.T.; Watanabe, V.S.; Diniz, T.G.; De Oliveira, A.M.; Hata, F.T. Plant-promoting rhizobacteria Methylobacterium komagatae increases crambe yields, root system and plant height. Ind. Crop. Prod. 2018, 121, 277–281. [Google Scholar] [CrossRef]
- Liu, W.X.; Wang, Q.L.; Hou, J.Y.; Tu, C.; Luo, Y.M.; Christie, P. Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 2016, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Nabti, E.; Bensidhoum, L.; Tabli, N.; Dahel, D.; Weiss, A.; Rothballer, M.; Schmid, M.; Hartmann, A. Growth stimulation of barley and biocontrol effect on plant pathogenic fungi by a Cellulosimicrobium sp. strain isolated from salt-affected rhizosphere soil in northwestern Algeria. Eur. J. Soil Biol. 2014, 61, 20–26. [Google Scholar] [CrossRef]
- Gos, F.M.W.R.; Savi, D.C.; Shaaban, K.A.; Thorson, J.S.; Aluizio, R.; Possiede, Y.M.; Rohr, J.; Glienke, C. Antibacterial activity of endophytic actinomycetes isolated from the medicinal plant Vochysia divergens (Pantanal, Brazil). Front. Microbiol. 2017, 8, 17. [Google Scholar] [CrossRef]
- Dif, G.; Belaouni, H.A.; Goudjal, Y.; Yekkour, A.; Djemouai, N.; Zitouni, A. Potential for plant growth promotion of Kocuria arsenatis Strain ST19 on tomato under salt stress conditions. S. Afr. J. Bot. 2021, 138, 94–104. [Google Scholar] [CrossRef]
- Faylon, P.S.; Ilao, R.I.; Reddy, M.S.; Sayyed, R.; Armanda, A. Recent Advances in Biofertilizers and Biofungicides (PGPR) for Sustainable Agriculture; Cambridge Scholars Publishing: Newcastle upon Tyne, UK, 2014. [Google Scholar]
- Deb, D.; Khan, A.; Dey, N. Phoma diseases: Epidemiology and control. Plant Pathol. 2020, 69, 1203–1217. [Google Scholar] [CrossRef]
- Stouffer, D.B.; Bascompte, J. Compartmentalization increases food-web persistence. Proc. Natl. Acad. Sci. USA 2011, 108, 3648–3652. [Google Scholar] [CrossRef] [PubMed]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- De Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [PubMed]
- Blumenstein, K.; Albrectsen, B.R.; Martín, J.A.; Hultberg, M.; Sieber, T.N.; Helander, M.; Witzell, J. Nutritional niche overlap potentiates the use of endophytes in biocontrol of a tree disease. BioControl 2015, 60, 655–667. [Google Scholar] [CrossRef]
- Wallis, C.M. Nutritional niche overlap analysis as a method to identify potential biocontrol fungi against trunk pathogens. Biocontrol 2021, 66, 559–571. [Google Scholar] [CrossRef]
- Carroll, G. Fungal endophytes in stems and leaves: From latent pathogen to mutualistic symbiont. Ecology 1998, 69, 2–9. [Google Scholar] [CrossRef]
- Freeman, S.; Rodriguez, J.R. Genetic conversion of a fungal plant pathogen to a nonpathogenic, endophytic mutualist. Science 1993, 260, 75–78. [Google Scholar] [CrossRef]
- Mejía, L.C.; Rojas, E.I.; Maynard, Z.; Bael, S.V.; Arnold, A.E.; Hebbar, P.; Samuels, G.J.; Robbins, N.; Herre, E.A. Endophytic fungi as biocontrol agents of Theobroma cacao pathogens. Biol. Control 2008, 46, 4–14. [Google Scholar] [CrossRef]
- Saikkonen, K.; Faeth, S.H.; Helander, M.; Sullivan, T.J. Fungal endophytes: A continuum of interactions with host plants. Ann. Rev. Ecol. Syst. 1998, 29, 319–343. [Google Scholar] [CrossRef]
- Joo, H.S.; Deyrup, S.T.; Shim, S.H. Endophyte-produced antimicrobials: A review of potential lead compounds with a focus on quorum-sensing disruptors. Phytochem. Rev. 2021, 20, 543–568. [Google Scholar] [CrossRef]
- Subramani, R.; Jayaprakashvel, M. Bacterial quorum sensing: Biofilm formation, survival behaviour and antibiotic resistance. Implic. Quor. Sens. Biofilm Form. Med. Agric. Food Ind. 2019, 21–37. [Google Scholar]
- Liu, J.M.; Wang, S.S.; Zheng, X.; Jin, N.; Lu, J.; Huang, Y.T.; Fan, B.; Wang, F.Z. Antimicrobial activity against phytopathogens and inhibitory activity on solanine in potatoes of the endophytic bacteria isolated from potato tubers. Front. Microbiol. 2020, 11, 570926. [Google Scholar] [CrossRef] [PubMed]
- Dalebroux, Z.D.; Swanson, M.S. ppGpp: Magic beyond RNA polymerase. Nat. Rev. Microbiol. 2012, 10, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Lüttmann, D.; Göpel, Y.; Görke, B. The phosphotransferase protein EIIANtr modulates the phosphate starvation response through interaction with histidine kinase PhoR in Escherichia coli. Mol. Microbiol. 2012, 86, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.M.; Ferreira, R.B.; Buckner, M.M.; Finlay, B.B. Quorum sensing in bacterial virulence. Microbiology 2010, 156, 2271–2282. [Google Scholar] [CrossRef]
- Ali, A.; Li, M.; Su, J.F.; Li, Y.F.; Wang, Z.; Bai, Y.H.; Ali, E.F.; Shaheen, S.M. Brevundimonas diminuta isolated from mines polluted soil immobilized cadmium (Cd2+) and zinc (Zn2+) through calcium carbonate precipitation: Microscopic and spectroscopic investigations. Sci. Total Environ. 2022, 813, 152668. [Google Scholar] [CrossRef]
- Wen, T.; Xie, P.H.; Liu, H.W.; Liu, T.; Zhao, M.L.; Yang, S.D.; Niu, G.Q.; Hale, L.; Singh, B.K.; Kowalchuk, G.A.; et al. Tapping the rhizosphere metabolites for the prebiotic control of soil-borne bacterial wilt disease. Nat. Commun. 2023, 14, 4497. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.R.; Gong, L.Q.; Jin, M.Y.; Wang, R.; Liu, R.; Gao, J.; Liu, M.D.; Huang, L.; Wang, G.Z.; Wang, D.; et al. Research advances in the structures and biological activities of secondary metabolites from Talaromyces. Front. Microbiol. 2022, 13, 984801. [Google Scholar] [CrossRef] [PubMed]
- Naraghi, L.; Heydari, A.; Rezaee, S.; Razavi, M. Biocontrol agent Talaromyces flavus stimulates the growth of cotton and potato. J. Plant Growth Regul. 2012, 31, 471–477. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Tao, S.; Liang, Y. Time-Course Responses of Apple Leaf Endophytes to the Infection of Gymnosporangium yamadae. J. Fungi 2024, 10, 128. https://doi.org/10.3390/jof10020128
Li Y, Tao S, Liang Y. Time-Course Responses of Apple Leaf Endophytes to the Infection of Gymnosporangium yamadae. Journal of Fungi. 2024; 10(2):128. https://doi.org/10.3390/jof10020128
Chicago/Turabian StyleLi, Yunfan, Siqi Tao, and Yingmei Liang. 2024. "Time-Course Responses of Apple Leaf Endophytes to the Infection of Gymnosporangium yamadae" Journal of Fungi 10, no. 2: 128. https://doi.org/10.3390/jof10020128
APA StyleLi, Y., Tao, S., & Liang, Y. (2024). Time-Course Responses of Apple Leaf Endophytes to the Infection of Gymnosporangium yamadae. Journal of Fungi, 10(2), 128. https://doi.org/10.3390/jof10020128




