Peroxin MoPex22 Regulates the Import of Peroxisomal Matrix Proteins and Appressorium-Mediated Plant Infection in Magnaporthe oryzae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Gene Deletion and Complementation
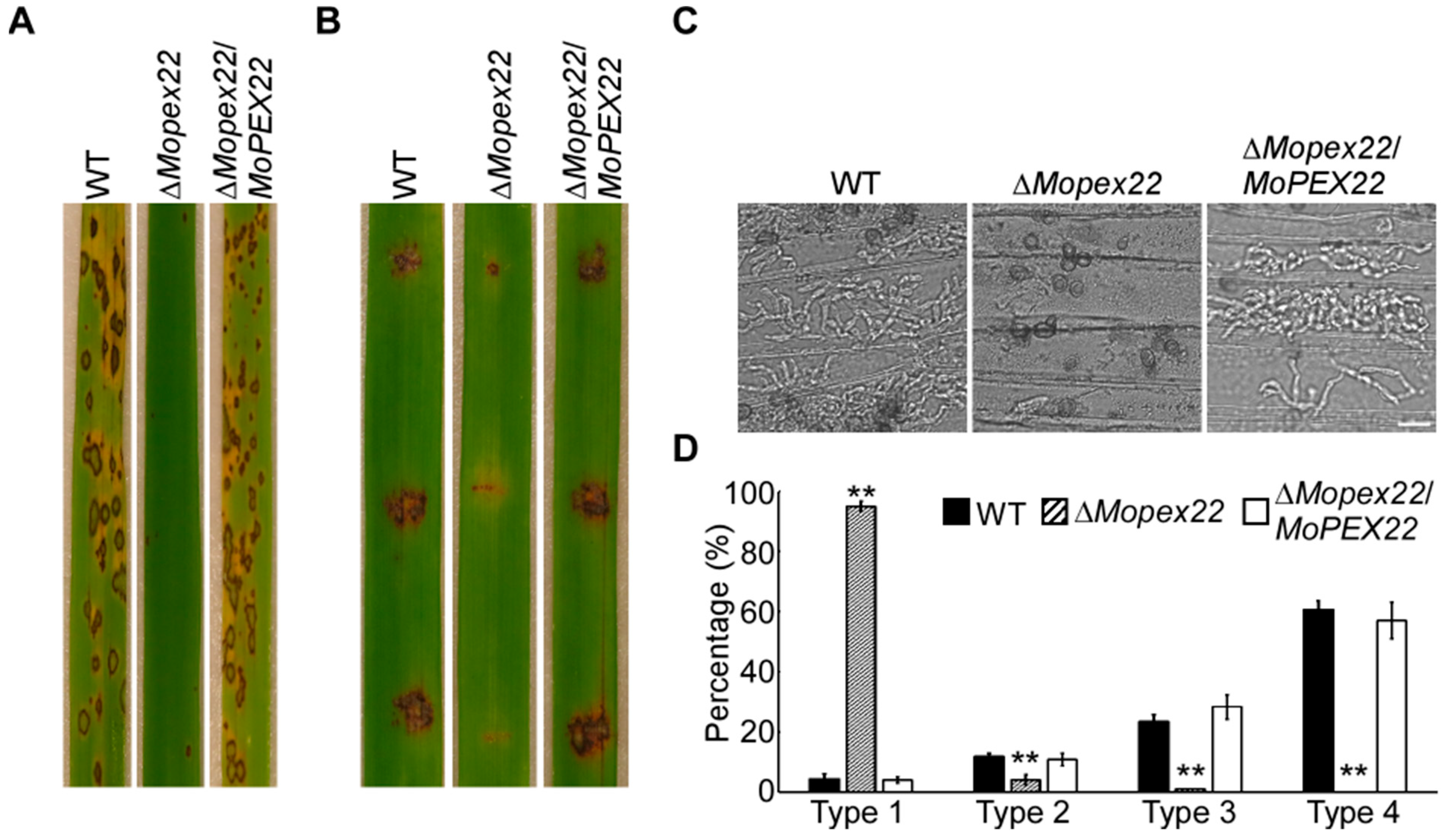
2.3. Virulence Assays and Infection Process Observation
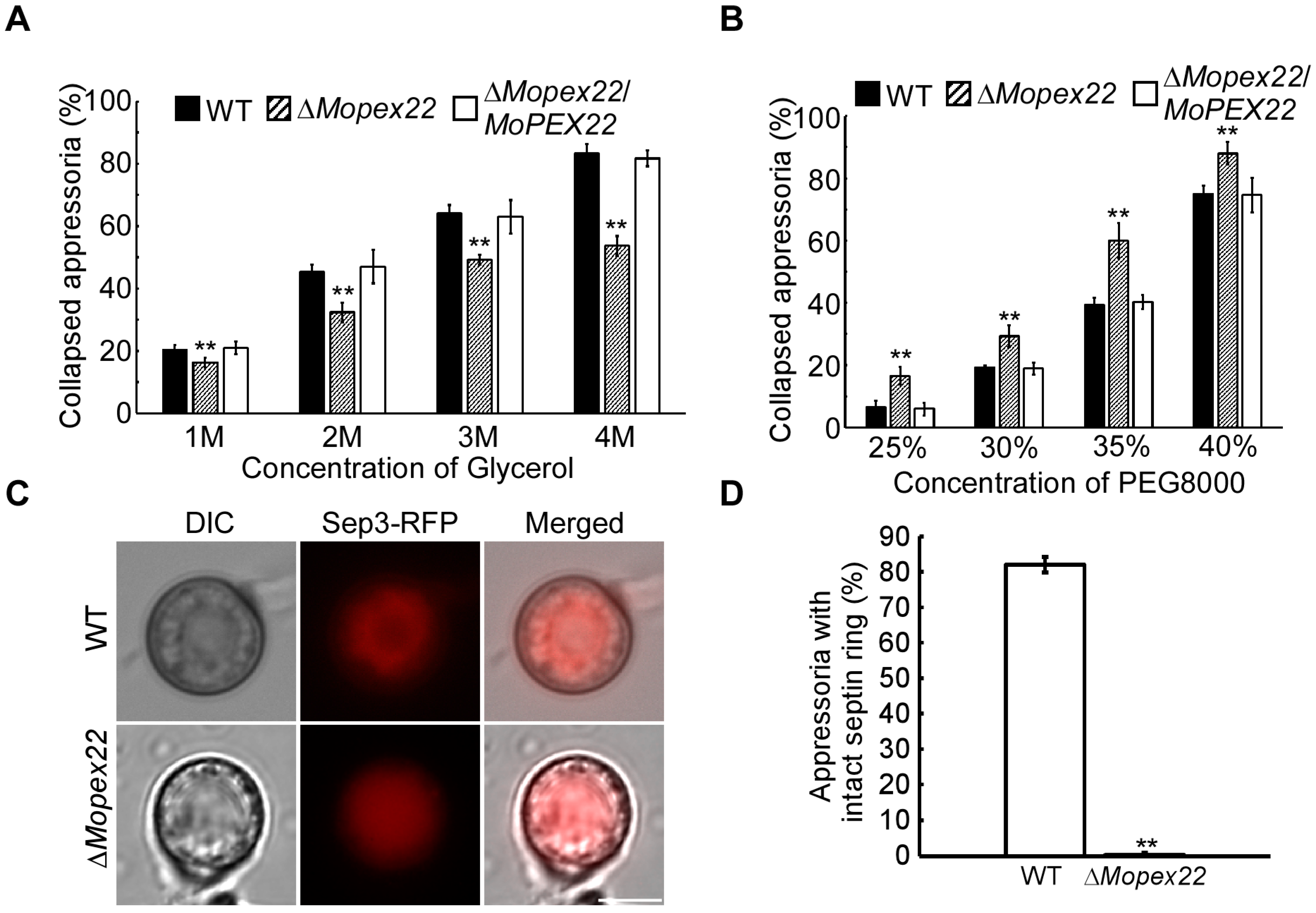
2.4. Appressorium Turgor Assay
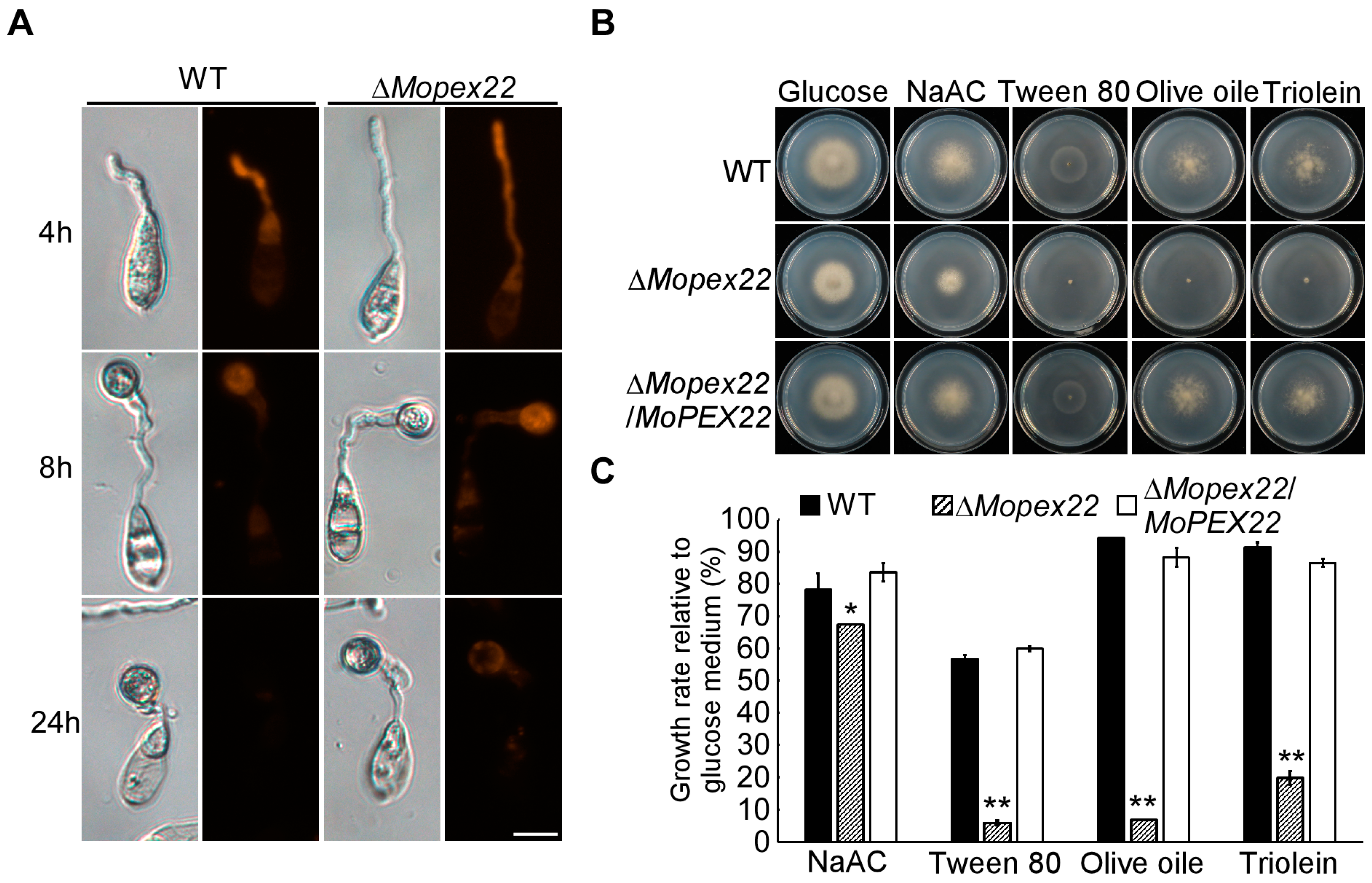
2.5. Measurement of Lipid Mobilization
2.6. Assay for Fatty Acid Utilization
2.7. Statistical Analysis
3. Results
3.1. MoPex22 Is Important for the Growth of M. oryzae
3.2. MoPex22 Is Important for Appressorium-Mediated Penetration
3.3. MoPex22 Regulates Appressorium Development
3.4. MoPex22 Is Involved in Lipid Metabolism in M. oryzae
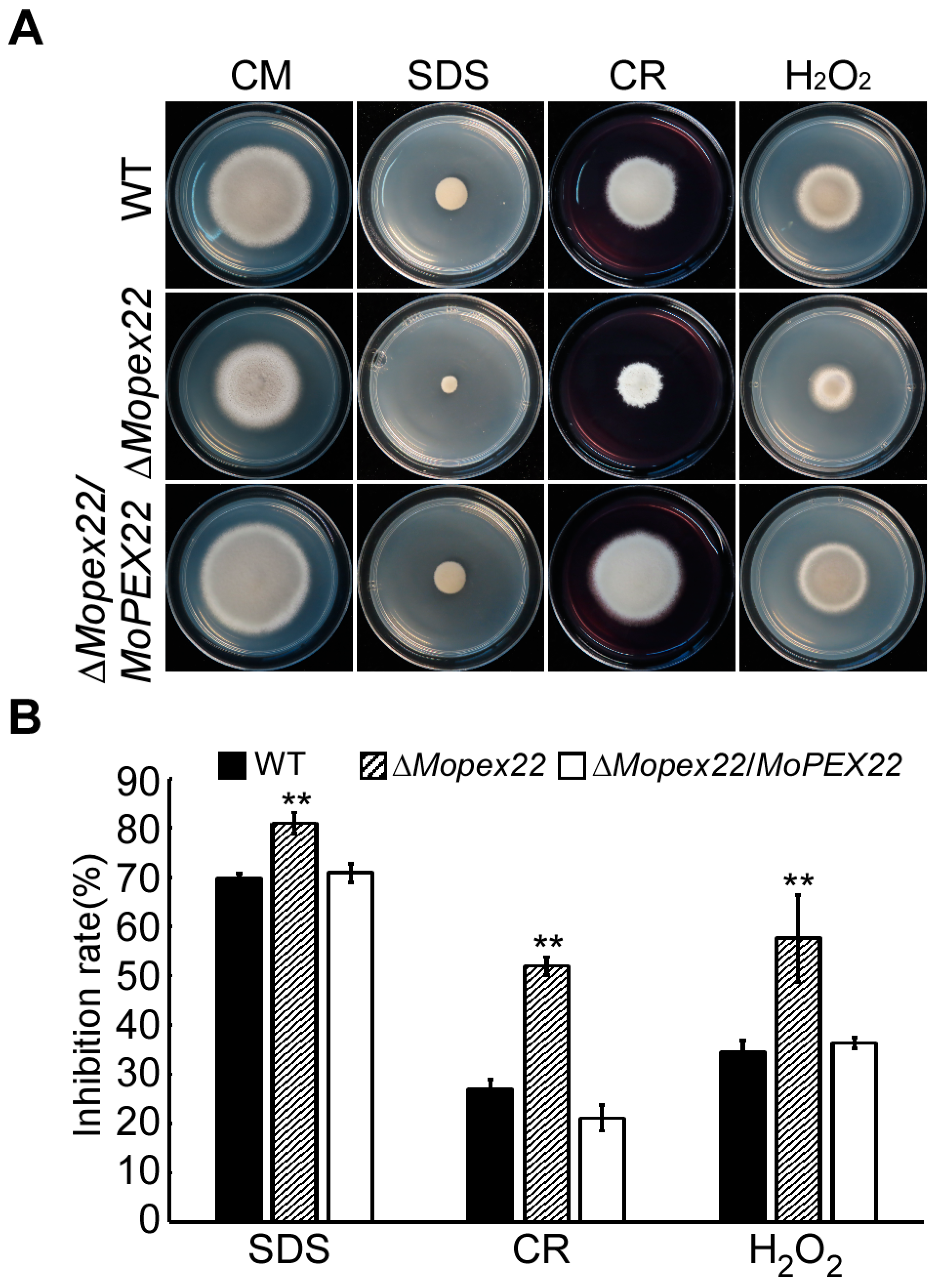
3.5. Deletion of MoPEX22 Reduced the Tolerance of the Mutant to SDS, CR, and H2O2
3.6. MoPex22 Localizes to Peroxisomes and Regulates the Import of Matrix Proteins into Peroxisomes
3.7. MoPex22 Anchors MoPex4 to Peroxisomes
3.8. MoPex22 Regulates the Localization of MoPex5
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghatak, A.; Willocquet, L.; Savary, S.; Kumar, J. Variability in aggressiveness of rice blast (Magnaporthe oryzae) isolates originating from rice leaves and necks: A case of pathogen specialization? PLoS ONE 2013, 8, e66180. [Google Scholar] [CrossRef]
- Ryder, L.S.; Talbot, N.J. Regulation of appressorium development in pathogenic fungi. Curr. Opin. Plant Biol. 2015, 26, 8–13. [Google Scholar] [CrossRef]
- Veneault-Fourrey, C.; Barooah, M.; Egan, M.; Wakley, G.; Talbot, N.J. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006, 312, 580–583. [Google Scholar] [CrossRef]
- Kershaw, M.J.; Talbot, N.J. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. USA 2009, 106, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- de Jong, J.C.; MCCormack, B.J.; Smirnoff, N.; Talbot, N.J. Glycerol generates turgor in rice blast. Nature 1997, 389, 244–245. [Google Scholar] [CrossRef]
- Kawamura, C.; Moriwaki, J.; Kimura, N.; Fujita, Y.; Fuji, S.; Hirano, T.; Koizumi, S.; Tsuge, T. The melanin biosynthesis genes of Alternaria alternata can restore pathogenicity of the melanin-deficient mutants of Magnaporthe grisea. Mol. Plant Microbe Interact. 1997, 10, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Ryder, L.S.; Dagdas, Y.F.; Kershaw, M.J.; Venkataraman, C.; Madzvamuse, A.; Yan, X.; Cruz-Mireles, N.; Soanes, D.M.; Oses-Ruiz, M.; Styles, V.; et al. A sensor kinase controls turgor-driven plant infection by the rice blast fungus. Nature 2019, 574, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Dagdas, Y.F.; Yoshino, K.; Dagdas, G.; Ryder, L.S.; Bielska, E.; Steinberg, G.; Talbot, N.J. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 2012, 336, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Sakulkoo, W.; Oses-Ruiz, M.; Oliveira Garcia, E.; Soanes, D.M.; Littlejohn, G.R.; Hacker, C.; Correia, A.; Valent, B.; Talbot, N.J. A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 2018, 359, 1399–1403. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Dagdas, Y.F.; Gupta, Y.K.; Mentlak, T.A.; Yi, M.; Martinez-Rocha, A.L.; Saitoh, H.; Terauchi, R.; Talbot, N.J.; Valent, B. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 2013, 4, 1996. [Google Scholar] [CrossRef]
- Wanders, R.J. Peroxisomes, lipid metabolism, and peroxisomal disorders. Mol. Genet. Metab. 2004, 83, 16–27. [Google Scholar] [CrossRef]
- Hynes, M.J.; Murray, S.L.; Khew, G.S.; Davis, M.A. Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics 2008, 178, 1355–1369. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, C.X.; Na, Y.T.; Ren, D.D.; Zhang, C.H.; He, Y.F.; Wang, Y.W.; Xiang, S.; Ren, W.H.; Jiang, Y.N.; et al. Compartmentalization of melanin biosynthetic enzymes contributes to self-defense against intermediate compound scytalone in Botrytis cinerea. Mbio 2021, 12, e00007-21. [Google Scholar] [CrossRef]
- Fransen, M.; Lismont, C. Redox signaling from and to peroxisomes: Progress, challenges, and prospects. Antioxid. Redox Sign 2019, 30, 95–112. [Google Scholar] [CrossRef]
- Titorenko, V.I.; Rachubinski, R.A. The life cycle of the peroxisome. Nat. Rev. Mol. Cell Bio 2001, 2, 357–368. [Google Scholar] [CrossRef]
- Lametschwandtner, G.; Brocard, C.; Fransen, M.; Van Veldhoven, P.; Berger, J.; Hartig, A. The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J. Biol. Chem. 1998, 273, 33635–33643. [Google Scholar] [CrossRef]
- Stanley, W.A.; Filipp, F.V.; Kursula, P.; Schuller, N.; Erdmann, R.; Schliebs, W.; Sattler, M.; Wilmanns, M. Recognition of a functional peroxisome type 1 target by the dynamic import receptor Pex5p. Mol. Cell 2006, 24, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Stein, K.; Schell-Steven, A.; Erdmann, R.; Rottensteiner, H. Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: Implications for the first steps in PTS2 protein import. Mol. Cell Biol. 2002, 22, 6056–6069. [Google Scholar] [CrossRef]
- Nair, D.M.; Purdue, P.E.; Lazarow, P.B. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J. Cell Biol. 2004, 167, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Lazarow, P.B. The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 2006, 1763, 1599–1604. [Google Scholar] [CrossRef] [PubMed]
- Gatto, G.J.; Geisbrecht, B.V.; Gould, S.J.; Berg, J.M. Peroxisomal targeting signal-1 recognition by the TPR domains of human Pex5. Nat. Struct. Biol. 2000, 7, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- Dammai, V.; Subramani, S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 2001, 105, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Zolman, B.K.; Monroe-Augustus, M.; Silva, I.D.; Bartel, B. Identification and functional characterization of peroxin4 and the interacting protein peroxin22. Plant Cell 2005, 17, 3422–3435. [Google Scholar] [CrossRef]
- Williams, C.; van den Berg, M.; Panjikar, S.; Stanley, W.A.; Distel, B.; Wilmanns, M. Insights into ubiquitin-conjugating enzyme/co-activator interactions from the structure of the Pex4p:Pex22p complex. Embo J. 2012, 31, 391–402. [Google Scholar] [CrossRef]
- Platta, H.W.; El Magraoui, F.; Schlee, D.; Grunau, S.; Girzalsky, W.; Erdmann, R. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 2007, 177, 197–204. [Google Scholar] [CrossRef]
- Williams, C.; van den Berg, M.; Sprenger, R.R.; Distel, B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 2007, 282, 22534–22543. [Google Scholar] [CrossRef]
- El Magraoui, F.; Schrötter, A.; Brinkmeier, R.; Kunst, L.; Mastalski, T.; Müller, T.; Marcus, K.; Meyer, H.E.; Girzalsky, W.; Erdmann, R.; et al. The cytosolic domain of Pex22p stimulates the Pex4p-dependent ubiquitination of the PTS1-receptor. PLoS ONE 2014, 9, e105894. [Google Scholar] [CrossRef]
- Min, K.; Son, H.; Lee, J.; Choi, G.J.; Kim, J.C.; Lee, Y.W. Peroxisome function is required for virulence and survival of Fusarium graminearum. Mol. Plant Microbe Interact. 2012, 25, 1617–1627. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, S.Y.; Ju, Z.Z.; Zhang, C.Q.; Tang, G.F.; Wang, J.; Wen, Z.Y.; Chen, W.; Ma, Z.H. Contribution of peroxisomal docking machinery to mycotoxin biosynthesis, pathogenicity and pexophagy in the plant pathogenic fungus. Environ. Microbiol. 2018, 20, 3224–3245. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.J.; Wang, L.N.; Sun, S.H.; Liu, A.X.; Liang, Y.C.; Yu, J.F.; Dong, H.S. FgPEX1 and FgPEX10 are required for the maintenance of woronin bodies and full virulence of Fusarium graminearum. Curr. Genet. 2019, 65, 1383–1396. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, L.N.; Liang, Y.C.; Yu, J.F. FgPEX4 is involved in development, pathogenicity, and cell wall integrity in Fusarium graminearum. Curr. Genet. 2019, 65, 747–758. [Google Scholar] [CrossRef]
- Wang, L.N.; Zhang, L.; Liu, C.J.; Sun, S.H.; Liu, A.X.; Liang, Y.C.; Yu, J.F. The roles of FgPEX2 and FgPEX12 in virulence and lipid metabolism in Fusarium graminearum. Fungal Genet. Biol. 2020, 135, 103288. [Google Scholar] [CrossRef]
- Li, L.; Wang, J.; Zhang, Z.; Wang, Y.; Liu, M.; Jiang, H.; Chai, R.; Mao, X.; Qiu, H.; Liu, F.; et al. MoPex19, which is essential for maintenance of peroxisomal structure and woronin bodies, is required for metabolism and development in the rice blast fungus. PLoS ONE 2014, 9, e85252. [Google Scholar] [CrossRef]
- Goh, J.; Jeon, J.; Kim, K.S.; Park, J.; Park, S.Y.; Lee, Y.H. The Pex7-mediated peroxisomal import system is required for fungal development and pathogenicity in Magnaporthe oryzae. PLoS ONE 2011, 6, e28220. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, Z.; Wang, Y.L.; Li, L.; Chai, R.Y.; Mao, X.Q.; Jiang, H.; Qiu, H.P.; Du, X.F.; Lin, F.C.; et al. PTS1 peroxisomal import pathway plays shared and distinct roles to PTS2 pathway in development and pathogenicity of Magnaporthe oryzae. PLoS ONE 2013, 8, e55554. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Li, L.; Zhang, Z.; Qiu, H.P.; Li, D.M.; Fang, Y.; Jiang, H.; Chai, R.Y.; Mao, X.Q.; Wang, Y.L.; et al. One of three Pex11 family members is required for peroxisomal proliferation and full virulence of the rice blast fungus. PLoS ONE 2015, 10, e0134249. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Wang, Z.; Liu, C.Y. Roles of peroxisomes in the rice blast fungus. Biomed. Res. Int. 2016, 2016, 9343417. [Google Scholar] [CrossRef] [PubMed]
- Kubo, Y.; Fujihara, N.; Harata, K.; Neumann, U.; Robin, G.P.; O’Connell, R. Colletotrichum orbiculare FAM1 encodes a novel woronin body-associated Pex22 peroxin required for appressorium-mediated plant infection. mBio 2015, 6, e01305–e01315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, C.; Wang, M.; Tao, Y.; Liang, Y.; Yu, J. Peroxin FgPEX22-like is involved in FgPEX4 tethering and Fusarium graminearum pathogenicity. Front. Microbiol. 2021, 12, 756292. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Pan, H.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Leung, H.; Borromeo, E.S.; Bernardo, M.A.; Notteghem, J.L. Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology 1988, 78, 1227–1233. [Google Scholar] [CrossRef]
- Chao, C.C.T.; ELLINGBOE, A.H. Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can. J. Bot. 1991, 69, 2130–2134. [Google Scholar] [CrossRef]
- Bolton, M.D.; Thomma, B.P.H.J. Plant Fungal Pathogens: Methods and Protocols; Humana Press: New York, NY, USA, 2012; p. xvi. 648p. [Google Scholar] [CrossRef]
- Bruno, K.S.; Tenjo, F.; Li, L.; Hamer, J.E.; Xu, J.R. Cellular localization and role of kinase activity of Pmk1 in Magnaporthe grisea. Eukaryot. Cell 2004, 3, 1525–1532. [Google Scholar] [CrossRef]
- Thines, E.; Weber, R.W.S.; Talbot, N.J. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 2000, 12, 1703–1718. [Google Scholar] [CrossRef] [PubMed]
- Ryder, L.S.; Dagdas, Y.F.; Mentlak, T.A.; Kershaw, M.J.; Thornton, C.R.; Schuster, M.; Chen, J.; Wang, Z.; Talbot, N.J. NADPH oxidases regulate septin-mediated cytoskeletal remodeling during plant infection by the rice blast fungus. Proc. Natl. Acad. Sci. USA 2013, 110, 3179–3184. [Google Scholar] [CrossRef]
- Foster, A.J.; Ryder, L.S.; Kershaw, M.J.; Talbot, N.J. The role of glycerol in the pathogenic lifestyle of the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2017, 19, 1008–1016. [Google Scholar] [CrossRef]
- Chumley, F.G.; Valent, B. Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Mol. Plant Microbe Interact. 1990, 3, 135–143. [Google Scholar] [CrossRef]
- Falter, C.; Reumann, S. The essential role of fungal peroxisomes in plant infection. Mol. Plant Pathol. 2022, 23, 781–794. [Google Scholar] [CrossRef]
- Hettema, E.H.; Erdmann, R.; van der Klei, I.; Veenhuis, M. Evolving models for peroxisome biogenesis. Curr. Opin. Cell Biol. 2014, 29, 25–30. [Google Scholar] [CrossRef]
- Jansen, R.L.M.; Santana-Molina, C.; van den Noort, M.; Devos, D.P.; van der Klei, I.J. Comparative genomics of peroxisome biogenesis proteins: Making sense of the Pex proteins. Front. Cell Dev. Biol. 2021, 9, 654163. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Soanes, D.M.; Kershaw, M.J.; Talbot, N.J. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta-oxidation during appressorium-mediated plant infection. Mol. Plant Microbe Interact. 2007, 20, 475–491. [Google Scholar] [CrossRef]
- Fujihara, N.; Sakaguchi, A.; Tanaka, S.; Fujii, S.; Tsuji, G.; Shiraishi, T.; O’Connell, R.; Kubo, Y. Peroxisome biogenesis factor Pex13 is required for appressorium-mediated plant infection by the anthracnose fungus. Mol. Plant Microbe Interact. 2010, 23, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Bhambra, G.K.; Wang, Z.Y.; Soanes, D.M.; Wakley, G.E.; Talbot, N.J. Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol. Microbiol. 2006, 61, 46–60. [Google Scholar] [CrossRef]
- Geoghegan, I.; Steinberg, G.; Gurr, S. The role of the fungal cell wall in the infection of plants. Trends Microbiol. 2017, 25, 957–967. [Google Scholar] [CrossRef]
- Zhu, S.; Yan, Y.; Qu, Y.; Wang, J.; Feng, X.; Liu, X.; Lin, F.; Lu, J. Role refinement of melanin synthesis genes by gene knockout reveals their functional diversity in Pyricularia oryzae strains. Microbiol. Res. 2021, 242, 126620. [Google Scholar] [CrossRef]
- Fujii, I.; Mori, Y.; Watanabe, A.; Kubo, Y.; Tsuji, G.; Ebizuka, Y. Enzymatic synthesis of 1,3,6,8-tetrahydroxynaphthalene solely from malonyl coenzyme A by a fungal iterative type I polyketide synthase Pks1. Biochemistry 2000, 39, 8853–8858. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Takano, Y.; Furusawa, I.; Okuno, T. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 2001, 13, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- van den Bosch, H.; Schutgens, R.B.; Wanders, R.J.; Tager, J.M. Biochemistry of peroxisomes. Annu. Rev. Biochem. 1992, 61, 157–197. [Google Scholar] [CrossRef]
- Kershaw, M.J.; Basiewicz, M.; Soanes, D.M.; Yan, X.; Ryder, L.S.; Csukai, M.; Oses-Ruiz, M.; Valent, B.; Talbot, N.J. Conidial morphogenesis and septin-mediated plant infection require Smo1, a Ras GTPase-activating protein in Magnaporthe oryzae. Genetics 2019, 211, 151–167. [Google Scholar] [CrossRef]
- Li, L.; Chen, X.; Zhang, S.; Yang, J.; Chen, D.; Liu, M.; Zhang, H.; Zheng, X.; Wang, P.; Peng, Y.; et al. MoCAP proteins regulated by MoArk1-mediated phosphorylation coordinate endocytosis and actin dynamics to govern development and virulence of Magnaporthe oryzae. PLoS Genet. 2017, 13, e1006814. [Google Scholar] [CrossRef]
- Schrader, M.; Fahimi, H.D. Peroxisomes and oxidative stress. Biochim. Biophys. Acta 2006, 1763, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, S.; Liu, Y.; Ma, C. Redox regulated peroxisome homeostasis. Redox Biol. 2015, 4, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Li, L.; Chai, R.Y.; Qiu, H.P.; Zhang, Z.; Wang, Y.L.; Liu, X.H.; Lin, F.C.; Sun, G.C. Pex13 and Pex14, the key components of the peroxisomal docking complex, are required for peroxisome formation, host infection and pathogenicity-related morphogenesis in Magnaporthe oryzae. Virulence 2019, 10, 292–314. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, R.; Lu, K.; Yang, L.; Jiang, J.; Li, L. Peroxin MoPex22 Regulates the Import of Peroxisomal Matrix Proteins and Appressorium-Mediated Plant Infection in Magnaporthe oryzae. J. Fungi 2024, 10, 143. https://doi.org/10.3390/jof10020143
Chen R, Lu K, Yang L, Jiang J, Li L. Peroxin MoPex22 Regulates the Import of Peroxisomal Matrix Proteins and Appressorium-Mediated Plant Infection in Magnaporthe oryzae. Journal of Fungi. 2024; 10(2):143. https://doi.org/10.3390/jof10020143
Chicago/Turabian StyleChen, Rangrang, Kailun Lu, Lina Yang, Jihong Jiang, and Lianwei Li. 2024. "Peroxin MoPex22 Regulates the Import of Peroxisomal Matrix Proteins and Appressorium-Mediated Plant Infection in Magnaporthe oryzae" Journal of Fungi 10, no. 2: 143. https://doi.org/10.3390/jof10020143
APA StyleChen, R., Lu, K., Yang, L., Jiang, J., & Li, L. (2024). Peroxin MoPex22 Regulates the Import of Peroxisomal Matrix Proteins and Appressorium-Mediated Plant Infection in Magnaporthe oryzae. Journal of Fungi, 10(2), 143. https://doi.org/10.3390/jof10020143



