Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, General Culture Conditions, and DNA Isolation
2.2. Genome Sequencing and Identification of azp BGC
2.3. Construction of Plasmid pJLH-RNAi-azpA for RNA-Mediated Silencing of azpA Gene
2.4. Construction of Plasmid pFC332-azpA for azpA Disruption by CRISPR-Cas9
2.5. Transformation of P. verrucosus FAE27 and Transformants Selection
2.6. RNA Extraction and qRT-PCR Experiments
2.7. Extraction of Red-Pigmented Metabolites and HPLC Analysis
3. Results
3.1. Identification and Characterization of azp BGC
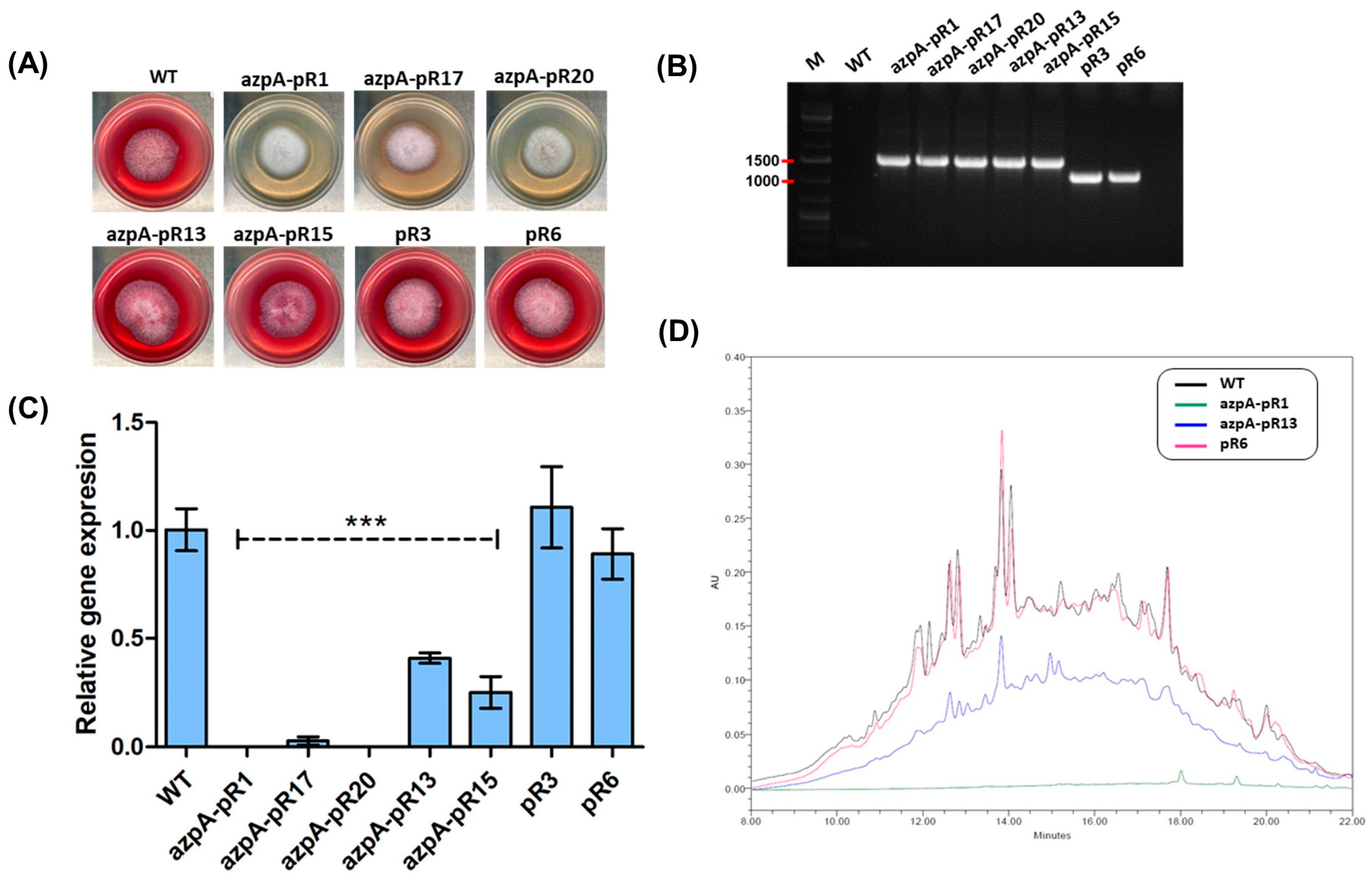
3.2. RNAi-Mediated Silencing of azpA in P. verrucosus FAE27
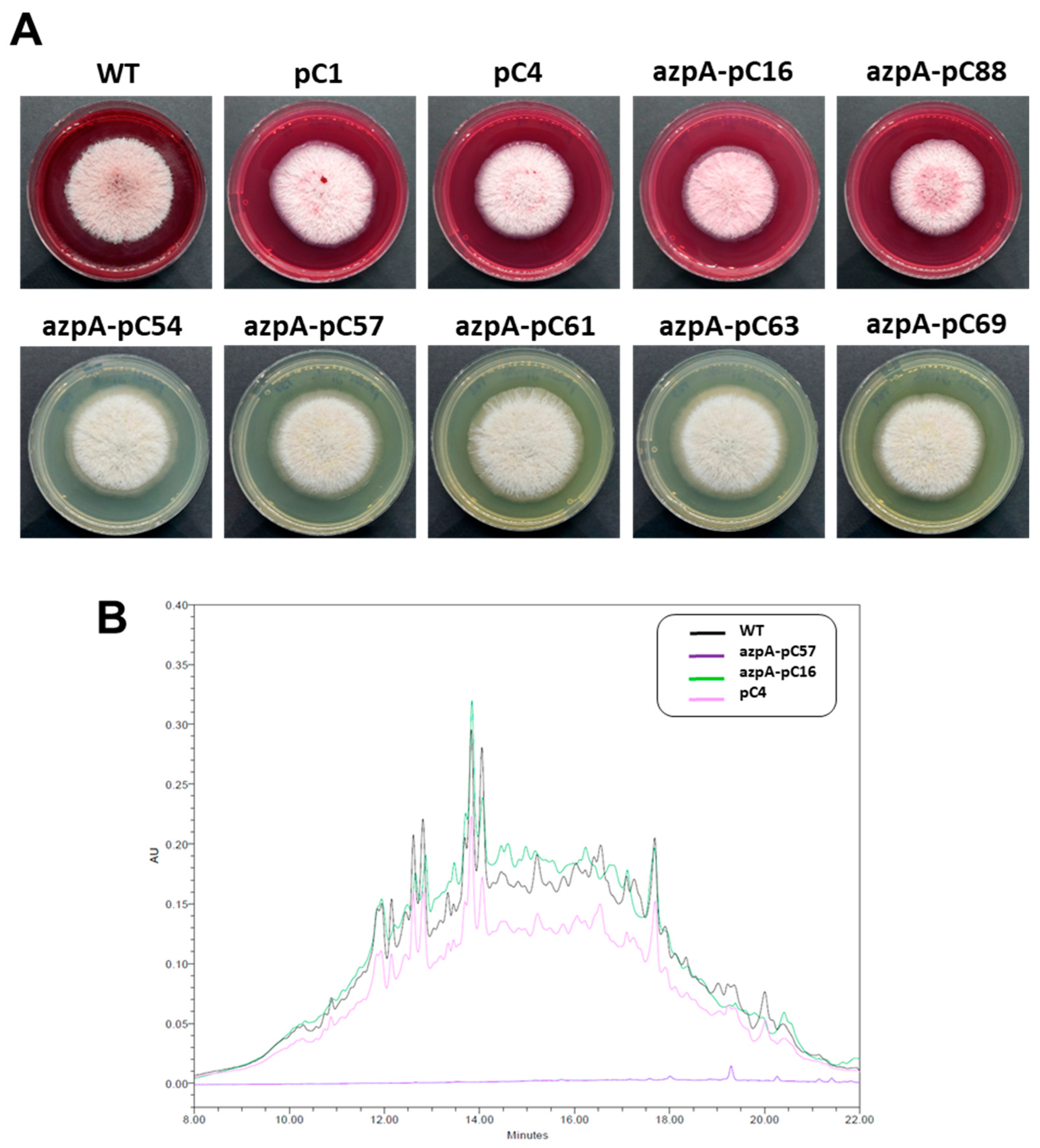
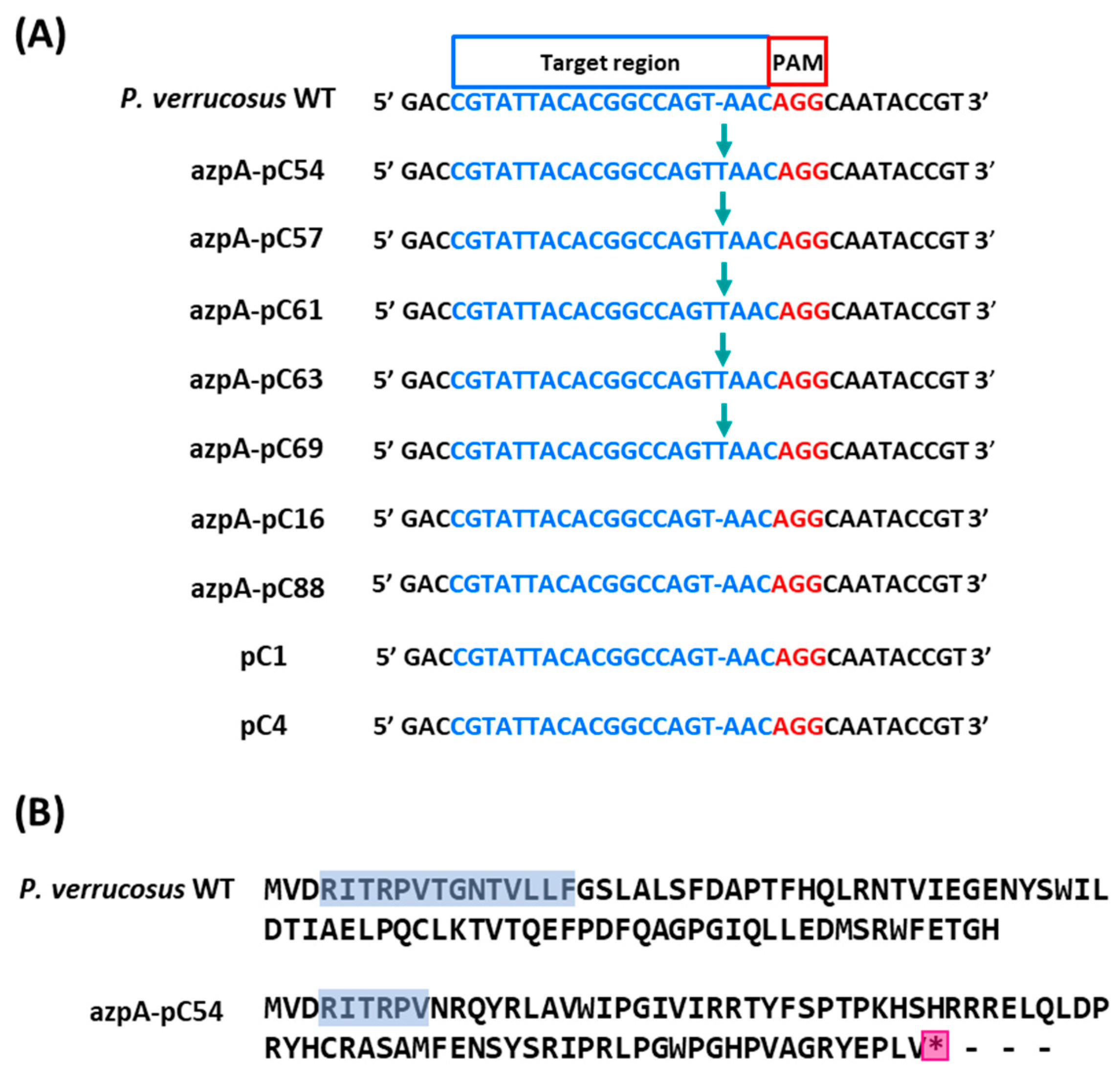
3.3. Disruption of azpA Gene in P. verrucosus FAE27 by CRISPR-Cas9
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, T.L.; Reichard, J.D.; Coleman, J.T.H.; Weller, T.J.; Thogmartin, W.E.; Reichert, B.E.; Bennett, A.B.; Broders, H.G.; Campbell, J.; Etchison, K.; et al. The scope and severity of white-nose syndrome on hibernating bats in North America. Conserv. Biol. 2021, 35, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Minnis, A.M.; Lindner, D.L. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol. 2013, 117, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, P.; Vásquez, G.; Gil-Durán, C.; Oliva, V.; Díaz, A.; Henríquez, M.; Álvarez, E.; Laich, F.; Chávez, R.; Vaca, I. Description of the first four species of the genus Pseudogymnoascus from Antarctica. Front. Microbiol. 2021, 12, 713189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Dong, C.; Chen, W.; Mou, Q.; Lu, X.; Han, Y.; Huang, J.; Liang, Z. The enigmatic Thelebolaceae (Thelebolales, Leotiomycetes): One new genus Solomyces and five new species. Front. Microbiol. 2020, 11, 572596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Shao, Q.Y.; Li, X.; Chen, W.H.; Liang, J.D.; Han, Y.F.; Huang, J.Z.; Liang, Z.Q. Culturable fungi from urban soils in China I: Description of 10 new taxa. Microbiol. Spectr. 2021, 9, e0086721. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.; van den Eynde, C.; Baert, F.; D’hooge, E.; De Pauw, R.; Normand, A.C.; Piarroux, R.; Stubbe, D. Remarkable fungal biodiversity on northern Belgium bats and hibernacula. Mycologia 2023, 115, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Han, Y.F.; Chen, W.H.; Tao, G. Additions to Thelebolales (Leotiomycetes, Ascomycota): Pseudogeomyces lindneri gen. et sp. nov. and Pseudogymnoascus campensis sp. nov. MycoKeys 2023, 95, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Li, X.; Chen, W.H.; Liang, J.D.; Han, Y.F. Culturable fungi from urban soils in China II, with the description of 18 novel species in Ascomycota (Dothideomycetes, Eurotiomycetes, Leotiomycetes and Sordariomycetes). MycoKeys 2023, 98, 167–220. [Google Scholar] [CrossRef]
- Furbino, L.E.; Godinho, V.M.; Santiago, I.F.; Pellizari, F.M.; Alves, T.M.; Zani, C.L.; Junior, P.A.; Romanha, A.J.; Carvalho, A.G.; Gil, L.H.; et al. Diversity patterns, ecology and biological activities of fungal communities associated with the endemic macroalgae across the Antarctic peninsula. Microb. Ecol. 2014, 67, 775–787. [Google Scholar] [CrossRef]
- Henríquez, M.; Vergara, K.; Norambuena, J.; Beiza, A.; Maza, F.; Ubilla, P.; Araya, I.; Chávez, R.; San-Martín, A.; Darias, J.; et al. Diversity of cultivable fungi associated with Antarctic marine sponges and screening for their antimicrobial, antitumoral and antioxidant potential. World J. Microbiol. Biotechnol. 2014, 30, 65–76. [Google Scholar] [CrossRef]
- Gonçalves, V.N.; Carvalho, C.R.; Johann, S.; Mendes, G.; Alves, T.M.; Zani, C.L.; Junior, P.A.S.; Murta, S.M.F.; Romanha, A.J.; Cantrell, C.L.; et al. Antibacterial, antifungal and antiprotozoal activities of fungal communities present in different substrates from Antarctica. Polar Biol. 2015, 38, 1143–1152. [Google Scholar] [CrossRef]
- Gomes, E.C.Q.; Godinho, V.M.; Silva, D.A.S.; de Paula, M.T.R.; Vitoreli, G.A.; Zani, C.L.; Alves, T.M.A.; Junior, P.A.S.; Murta, S.M.F.; Barbosa, E.C.; et al. Cultivable fungi present in Antarctic soils: Taxonomy, phylogeny, diversity, and bioprospecting of antiparasitic and herbicidal metabolites. Extremophiles 2018, 22, 381–393. [Google Scholar] [CrossRef]
- Purić, J.; Vieira, G.; Cavalca, L.B.; Sette, L.D.; Ferreira, H.; Vieira, M.L.C.; Sass, D.C. Activity of Antarctic fungi extracts against phytopathogenic bacteria. Lett. Appl. Microbiol. 2018, 66, 530–536. [Google Scholar] [CrossRef]
- Vieira, G.; Purić, J.; Morão, L.G.; Dos Santos, J.A.; Inforsato, F.J.; Sette, L.D.; Ferreira, H.; Sass, D.C. Terrestrial and marine Antarctic fungi extracts active against Xanthomonas citri subsp. citri. Lett. Appl. Microbiol. 2018, 67, 64–71. [Google Scholar] [CrossRef]
- Shi, T.; Li, X.Q.; Zheng, L.; Zhang, Y.H.; Dai, J.J.; Shang, E.L.; Yu, Y.Y.; Zhang, Y.T.; Hu, W.P.; Shi, D.Y. Sesquiterpenoids from the Antarctic fungus Pseudogymnoascus sp. HSX2#-11. Front. Microbiol. 2021, 12, 688202. [Google Scholar] [CrossRef]
- Antipova, T.V.; Zaitsev, K.V.; Zhelifonova, V.P.; Tarlachkov, S.V.; Grishin, Y.K.; Kochkina, G.A.; Vainshtein, M.B. The potential of arctic Pseudogymnoascus fungi in the biosynthesis of natural products. Fermentation 2023, 9, 702. [Google Scholar] [CrossRef]
- Shi, T.; Yu, Y.-Y.; Dai, J.-J.; Zhang, Y.-T.; Hu, W.-P.; Zheng, L.; Shi, D.-Y. New polyketides from the Antarctic fungus Pseudogymnoascus sp. HSX2#-11. Mar. Drugs 2021, 19, 168. [Google Scholar]
- Han, X.; Gao, H.; Lai, H.; Zhu, W.; Wang, Y. Anti-Aβ42 Aggregative polyketides from the Antarctic psychrophilic fungus Pseudogymnoascus sp. OUCMDZ-3578. J. Nat. Prod. 2023, 86, 882–890. [Google Scholar] [CrossRef]
- Hou, X.; Li, C.; Zhang, R.; Li, Y.; Li, H.; Zhang, Y.; Tae, H.S.; Yu, R.; Che, Q.; Zhu, T.; et al. Unusual tetrahydropyridoindole-containing tetrapeptides with human nicotinic acetylcholine receptors targeting activity discovered from Antarctica-derived psychrophilic Pseudogymnoascus sp. HDN17-933. Mar. Drugs 2022, 20, 593. [Google Scholar] [CrossRef]
- Shi, T.; Zheng, L.; Li, X.-Q.; Dai, J.-J.; Zhang, Y.-T.; Yu, Y.-Y.; Hu, W.-P.; Shi, D.-Y. Nitrogenous compounds from the Antarctic fungus Pseudogymnoascus sp. HSX2#-11. Molecules 2021, 26, 2636. [Google Scholar]
- Figueroa, L.; Jiménez, C.; Rodríguez, J.; Areche, C.; Chávez, R.; Henríquez, M.; de la Cruz, M.; Díaz, C.; Segade, Y.; Vaca, I. 3-Nitroasterric acid derivatives from an Antarctic sponge-derived Pseudogymnoascus sp. fungus. J. Nat. Prod. 2015, 78, 919–923. [Google Scholar] [CrossRef]
- Zhang, T.; Ren, P.; Chaturvedi, V.; Chaturvedi, S. Development of an agrobacterium-mediated transformation system for the cold-adapted fungi Pseudogymnoascus destructans and P. pannorum. Fungal Genet. Biol. 2015, 81, 73–81. [Google Scholar] [CrossRef]
- Díaz, A.; Villanueva, P.; Oliva, V.; Gil-Durán, C.; Fierro, F.; Chávez, R.; Vaca, I. Genetic transformation of the filamentous fungus Pseudogymnoascus verrucosus of Antarctic origin. Front. Microbiol. 2019, 10, 2675. [Google Scholar] [CrossRef]
- Lax, C.; Tahiri, G.; Patiño-Medina, J.A.; Cánovas-Márquez, J.T.; Pérez-Ruiz, J.A.; Osorio-Concepción, M.; Navarro, E.; Calo, S. The evolutionary significance of RNAi in the Fungal Kingdom. Int. J. Mol. Sci. 2020, 21, 9348. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.S.; Zhang, Z.; Liu, Y. RNA interference pathways in fungi: Mechanisms and functions. Annu. Rev. Microbiol. 2012, 66, 305–323. [Google Scholar] [CrossRef]
- Nakayashiki, H.; Nguyen, Q.B. RNA interference: Roles in fungal biology. Curr. Opin. Microbiol. 2008, 11, 494–502. [Google Scholar] [CrossRef]
- Van Leeuwe, T.M.; Arentshorst, M.; Ernst, T.; Alazi, E.; Punt, P.J.; Ram, A.F. Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi. Fungal Biol. Biotechnol. 2009, 6, 13. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N. Multiplex genome engineering using CRISPR-Cas systems. Science 2013, 339, 197–217. [Google Scholar] [CrossRef]
- O’Connell, M.R.; Oakes, B.L.; Sternberg, S.H.; Eastseletsky, A.; Kaplan, M.; Doudna, J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.V.; Currah, R.S. Two new species of Pseudogymnoascus with Geomyces anamorphs and their phylogenetic relationship with Gymnostellatospora. Mycologia 2006, 98, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.; Menezes, G.; Silva, T.; Bicas, J.; Oliveira, V.; Rosa, L. Antarctic fungi as producers of pigments. In Fungi of Antarctica; Rosa, L., Ed.; Springer: Cham, Switzerland, 2019; pp. 305–318. [Google Scholar]
- Mahuku, G. A simple extraction method suitable for PCR-based analysis of plant, fungal, and bacterial DNA. Plant Mol. Biol. Rep. 2004, 22, 71–81. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Ullán, R.V.; Godio, R.P.; Teijeira, F.; Vaca, I.; García-Estrada, C.; Feltrer, R.; Kosalková, K.; Martín, J.F. RNA-silencing in Penicillium chrysogenum and Acremonium chrysogenum: Validation studies using β-lactam genes expression. J. Microbiol. Methods 2008, 75, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Punt, P.J.; Oliver, R.P.; Dingemanse, M.A.; Pouwels, P.H.; van den Hondel, C.A. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 1987, 56, 117–124. [Google Scholar] [CrossRef]
- Gietz, R.D.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. In Methods in Enzymology; Guthrie, C., Fink, G.R., Eds.; Academic Press: Cambridge, MA, USA, 2002; Volume 350, pp. 87–96. [Google Scholar]
- Xie, S.; Shen, B.; Zhang, C.; Huang, X.; Zhang, Y. sgRNAcas9: A software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE 2014, 9, e100448. [Google Scholar] [CrossRef]
- Marcano, Y.; Montanares, M.; Gil-Durán, C.; González, K.; Levicán, G.; Vaca, I.; Chávez, R. PrlaeA affects the production of roquefortine C, mycophenolic acid, and andrastin A in Penicillium roqueforti, but it has little impact on asexual development. J. Fungi 2023, 9, 954. [Google Scholar] [CrossRef]
- Gil-Durán, C.; Palma, D.; Marcano, Y.; Palacios, J.-L.; Martínez, C.; Rojas-Aedo, J.F.; Levicán, G.; Vaca, I.; Chávez, R. CRISPR/Cas9-mediated disruption of the pcz1 gene and its impact on growth, development, and penicillin production in Penicillium rubens. J. Fungi 2023, 9, 1010. [Google Scholar] [CrossRef]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Schumann, U.; Smith, N.A.; Wang, M.B. A fast and efficient method for preparation of high-quality RNA from fungal mycelia. BMC Res. Notes 2013, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, B.; Karki, S.; Chiu, S.H.; Kim, H.-J.; Suh, J.-W.; Nam, B.; Yoon, Y.-M.; Chen, C.-C.; Kwon, H.-J. Genetic localization and in vivo characterization of a Monascus azaphilone pigment biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 2013, 97, 6337–6345. [Google Scholar] [CrossRef] [PubMed]
- Zabala, A.O.; Xu, W.; Chooi, Y.H.; Tang, Y. Characterization of a silent azaphilone gene cluster from Aspergillus niger ATCC 1015 reveals a hydroxylation-mediated pyran-ring formation. Chem. Biol. 2012, 19, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, C.; Flon, V.; Mann, S.; Leleu, S.; Prado, S.; Franck, X. Biosynthesis of azaphilones: A review. Nat. Prod. Rep. 2021, 38, 1058–1071. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Shao, Y.; Chen, F. Monascus pigments. Appl. Microbiol. Biotechnol. 2012, 96, 1421–1440. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, R.; Liu, Q.; He, Y.; He, K.; Ding, X.; Kang, L.; Guo, X.; Xie, N.; Zhou, Y.; et al. Orange, red, yellow: Biosynthesis of azaphilone pigments in Monascus fungi. Chem. Sci. 2017, 8, 4917–4925. [Google Scholar] [CrossRef]
- Chen, C.; Tao, H.; Chen, W.; Yang, B.; Zhou, X.; Luo, X.; Liu, Y. Recent advances in the chemistry and biology of azaphilones. RSC Adv. 2020, 10, 10197–10220. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, B.; Du, X.; Li, P.; Liang, B.; Cheng, X.; Du, L.; Huang, D.; Wang, L.; Wang, S. Complete genome sequence and transcriptomics analyses reveal pigment biosynthesis and regulatory mechanisms in an industrial strain, Monascus purpureus YY-1. Sci. Rep. 2015, 5, 8331. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Feng, Y.; Molnár, I.; Chen, F. Nature and nurture: Confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments. Nat. Prod. Rep. 2019, 36, 561–572. [Google Scholar] [CrossRef]
- Stadler, M.; Fournier, J. Pigment chemistry, taxonomy and phylogeny of the Hypoxyloideae (Xylariaceae). Rev. Iberoam. Micol. 2006, 23, 160–170. [Google Scholar] [CrossRef]
- Kuhnert, E.; Surup, F.; Sir, E.B.; Lambert, C.; Hyde, K.D.; Hladki, A.I.; Romero, A.I.; Stadler, M. Lenormandins A—G, new azaphilones from Hypoxylon lenormandii and Hypoxylon jaklitschii sp. nov., recognised by chemotaxonomic data. Fungal Div. 2015, 71, 165–184. [Google Scholar] [CrossRef]
- Yang, M.Y.; Wang, Y.X.; Chang, Q.H.; Li, L.F.; Liu, Y.F.; Cao, F. Cytochalasans and azaphilones: Suitable chemotaxonomic markers for the Chaetomium species. Appl. Microbiol. Biotechnol. 2021, 105, 8139–8155. [Google Scholar] [CrossRef]
- Osmanova, N.; Schultze, W.; Ayoub, N. Azaphilones: A class of fungal metabolites with diverse biological activities. Phytochem. Rev. 2010, 9, 315–342. [Google Scholar] [CrossRef]
- Sousa, T.F.; de Araújo Júnior, M.B.; Peres, E.G.; Souza, M.P.; da Silva, F.M.A.; de Medeiros, L.S.; de Souza, A.D.L.; de Souza, A.Q.L.; Yamagishi, M.E.B.; da Silva, G.F.; et al. Discovery of dual PKS involved in sclerotiorin biosynthesis in Penicillium meliponae using genome mining and gene knockout. Arch. Microbiol. 2023, 205, 75. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.R.; Han, S.; Shin, S.C.; Yeom, S.C.; Kim, H.J. Improved natural food colorant production in the filamentous fungus Monascus ruber using CRISPR-based engineering. Food Res. Int. 2023, 167, 112651. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Yang, Q.; Xue, Z.; Liu, Y. RNA interference in fungi: Pathways, functions, and applications. Eukaryot. Cell 2011, 10, 1148–1155. [Google Scholar] [CrossRef]
- Moriwaki, A.; Ueno, M.; Arase, S.; Kihara, J. RNA-mediated gene silencing in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 2007, 269, 85–89. [Google Scholar] [CrossRef]
- Hu, Y.; Hao, X.; Lou, J.; Zhang, P.; Pan, J.; Zhu, X. A PKS gene, pks-1, is involved in chaetoglobosin biosynthesis, pigmentation and sporulation in Chaetomium globosum. Sci. China Life Sci. 2012, 55, 1100–1108. [Google Scholar] [CrossRef]
- Hu, Y.; Hao, X.; Chen, L.; Akhberdi, O.; Yu, X.; Liu, Y.; Zhu, X. Gα-cAMP/PKA pathway positively regulates pigmentation, chaetoglobosin A biosynthesis and sexual development in Chaetomium globosum. PLoS ONE 2018, 13, e0195553. [Google Scholar] [CrossRef]
- Alhawatema, M.S.; Gebril, S.; Cook, D.; Creamer, R. RNAi-mediated down-regulation of a melanin polyketide synthase (pks1) gene in the fungus Slafractonia leguminicola. World J. Microbiol. Biotechnol. 2017, 33, 179. [Google Scholar] [CrossRef]
- Voigt, O.; Knabe, N.; Nitsche, S.; Erdmann, E.A.; Schumacher, J.; Gorbushina, A.A. An advanced genetic toolkit for exploring the biology of the rock-inhabiting black fungus Knufia petricola. Sci Rep. 2020, 10, 12021. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Yilmaz, N.; Thrane, U.; Rasmussen, K.B.; Houbraken, J.; Samson, R.A. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE 2013, 8, e84102. [Google Scholar] [CrossRef]
- Fernández, P.; Fañanás, F.J.; Rodríguez, F. Nitrogenated azaphilone derivatives through a silver-catalysed reaction of imines from ortho-alkynylbenzaldehydes. Chemistry 2017, 23, 3002–3006. [Google Scholar] [CrossRef] [PubMed]
- Sledzinski, P.; Nowaczyk, M.; Olejniczak, M. Computational tools and supporting CRISPR-Cas experiments. Cells 2020, 9, 1288. [Google Scholar] [CrossRef] [PubMed]
- Chibucos, M.C.; Crabtree, J.; Nagaraj, S.; Chaturvedi, S.; Chaturvedi, V. Draft genome sequences of human pathogenic fungus Geomyces pannorum sensu lato and bat white nose syndrome pathogen Geomyces (Pseudogymnoascus) destructans. Genome Announc. 2013, 1, e01045-13. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Drees, K.P.; Foster, J.T.; Lindner, D.L. Extreme sensitivity to ultraviolet light in the fungal pathogen causing white-nose syndrome of bats. Nat. Commun. 2018, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Davy, C.M.; Donaldson, M.E.; Bandouchova, H.; Breit, A.M.; Dorville, N.A.S.; Dzal, Y.A.; Kovacova, V.; Kunkel, E.L.; Martínková, N.; Norquay, K.J.O.; et al. Transcriptional host-pathogen responses of Pseudogymnoascus destructans and three species of bats with white-nose syndrome. Virulence 2020, 11, 781–794. [Google Scholar] [CrossRef]
- Kim, S.; Lee, R.; Jeon, H.; Lee, N.; Park, J.; Moon, H.; Shin, J.; Min, K.; Kim, J.E.; Yang, J.W.; et al. Identification of essential genes for the establishment of spray-induced gene silencing-based disease control in Fusarium graminearum. J. Agric. Food Chem. 2023, 71, 19302–19311. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, K.; Li, M.; Hu, H.; Zhang, X.; Liu, J.; Pan, H.; Zhang, Y. SsAGM1-mediated uridine diphosphate-N-acetylglucosamine synthesis is essential for development, stress response, and pathogenicity of Sclerotinia sclerotiorum. Front. Microbiol. 2022, 13, 938784. [Google Scholar] [CrossRef]
- Del-Cid, A.; Gil-Durán, C.; Vaca, I.; Rojas-Aedo, J.F.; García-Rico, R.O.; Levicán, G.; Chávez, R. Identification and functional analysis of the mycophenolic acid gene cluster of Penicillium roqueforti. PLoS ONE 2016, 11, e0147047. [Google Scholar] [CrossRef]
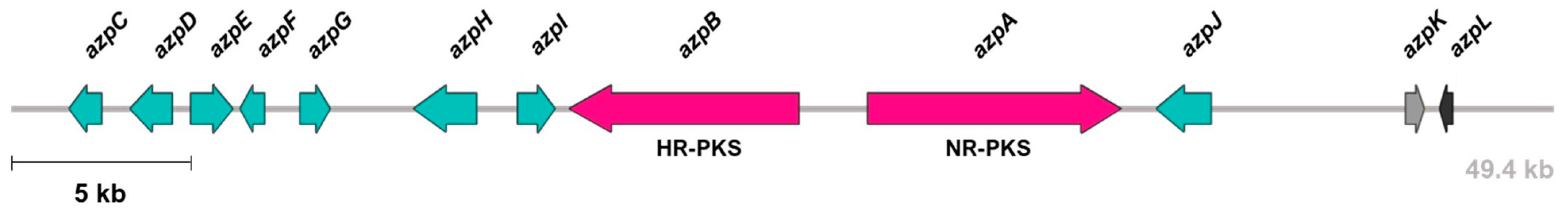
| Closest Characterized Homologues | |||||
|---|---|---|---|---|---|
| Protein Name | Size (Aminoacids) | Putative Function | Protein Name (Organism) | GenBank Accession Number | Identity (%) |
| AzpA | 2698 | Non-reducing polyketide synthase | Conidial yellow pigment biosynthesis polyketide synthase (Monascus pilosus) | AGN71604 | 61 |
| AzpB | 2315 | Highly reducing polyketide synthase | Polyketide synthase (Aspergillus niger) | EHA28244 | 45 |
| AzpC | 374 | Ketoreductase | Aldehyde reductase (Monascus pilosus) | AGN71608 | 50 |
| AzpD | 455 | O-acetyltransferase | Acetyltransferase (Monascus pilosus) | AGN71607 | 43 |
| AzpE | 445 | FAD monooxygenase | Monooxygenase (Phoma sp.) | QCO93109 | 53 |
| AzpF | 268 | Serine hydrolase | Amino oxidase/esterase (Monascus pilosus) | AGN71609 | 58 |
| AzpG | 364 | Enoyl reductase | Putative quinone-oxidoreductase-like protein (Monascus pilosus) | AGN71610 | 50 |
| AzpH | 644 | FAD oxidase | Isoamyl alcohol oxidase (Penicillium expansum) | AIG62142 | 36 |
| AzpI | 368 | Cytochrome P450 | BuaG cytochrome P450 (Aspergillus burnettii) | QBE85647 | 49 |
| AzpJ | 482 | FAD oxidase | FAD oxidase (Aspergillus niger) | EHA28243 | 47 |
| AzpK | 216 | Transporter | AflT transporter (Aspergillus flavus) | AAS90069 | 45 |
| AzpL | 119 | Transcription factor | Putative citrinin biosynthesis transcriptional activator CtnR (Monascus pilosus) | AGN71605 | 51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma, D.; Oliva, V.; Montanares, M.; Gil-Durán, C.; Travisany, D.; Chávez, R.; Vaca, I. Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus. J. Fungi 2024, 10, 157. https://doi.org/10.3390/jof10020157
Palma D, Oliva V, Montanares M, Gil-Durán C, Travisany D, Chávez R, Vaca I. Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus. Journal of Fungi. 2024; 10(2):157. https://doi.org/10.3390/jof10020157
Chicago/Turabian StylePalma, Diego, Vicente Oliva, Mariana Montanares, Carlos Gil-Durán, Dante Travisany, Renato Chávez, and Inmaculada Vaca. 2024. "Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus" Journal of Fungi 10, no. 2: 157. https://doi.org/10.3390/jof10020157
APA StylePalma, D., Oliva, V., Montanares, M., Gil-Durán, C., Travisany, D., Chávez, R., & Vaca, I. (2024). Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus. Journal of Fungi, 10(2), 157. https://doi.org/10.3390/jof10020157







