The Dynamic Interaction between Oil Palm and Phytophthora palmivora in Bud Rot Disease: Insights from Transcriptomic Analysis and Network Modelling
Abstract
1. Introduction
2. Materials and Methods
2.1. RNAseq Data
2.2. Analysis of Differentially Expressed Genes of P. palmivora and Oil Palm
2.3. Annotation of Differentially Expressed Genes (DEGs) of P. palmivora and Oil Palm and Bioinformatic Identification of Effectors
2.4. Construction of Co-Expression Networks of P. palmivora and Oil Palm
2.5. Gene Validation by qRT-PCR
3. Results
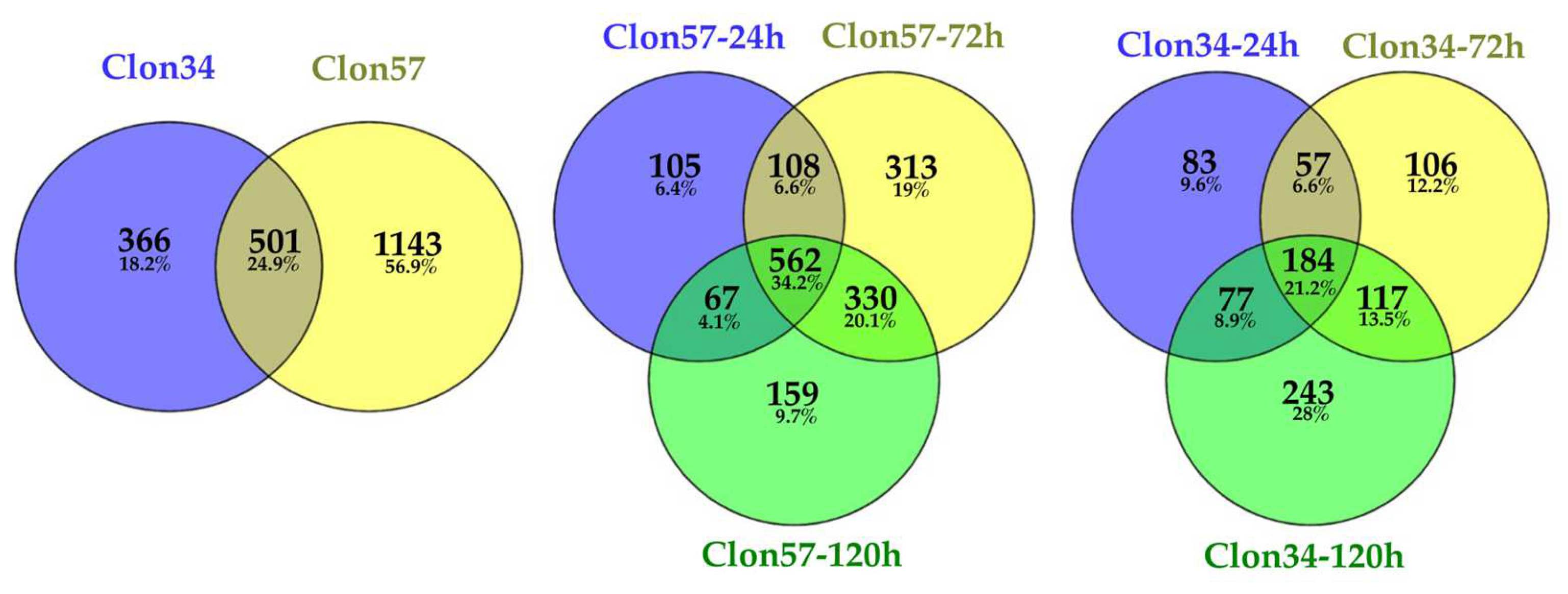
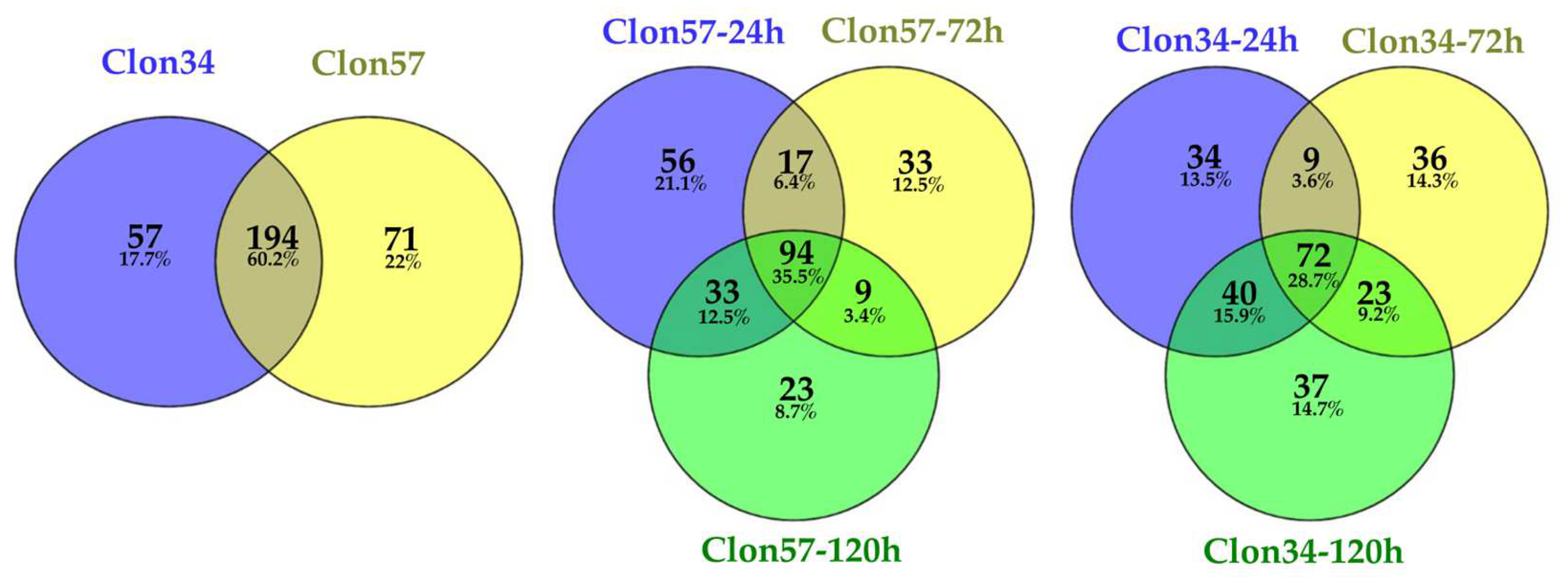
3.1. Oil Palm and Phytophthora Palmivora Transcriptome Analysis
3.2. Bioinformatic Identification of P. palmivora Effectors
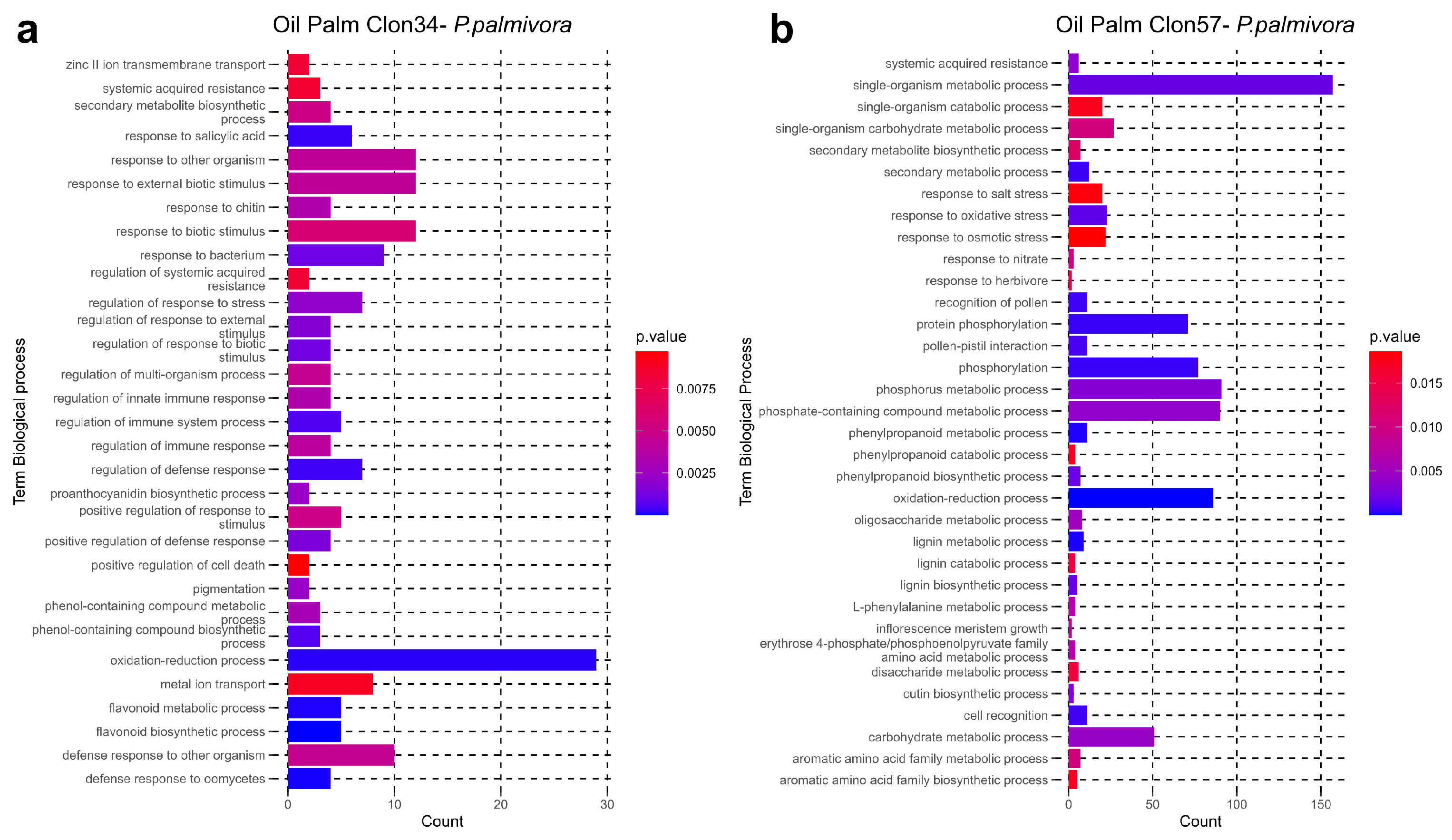
3.3. GO Enrichment of P. palmivora Genes in the Interaction with Clon34 and Clon57
3.4. Gene Co-Expression Network Analysis of P. palmivora
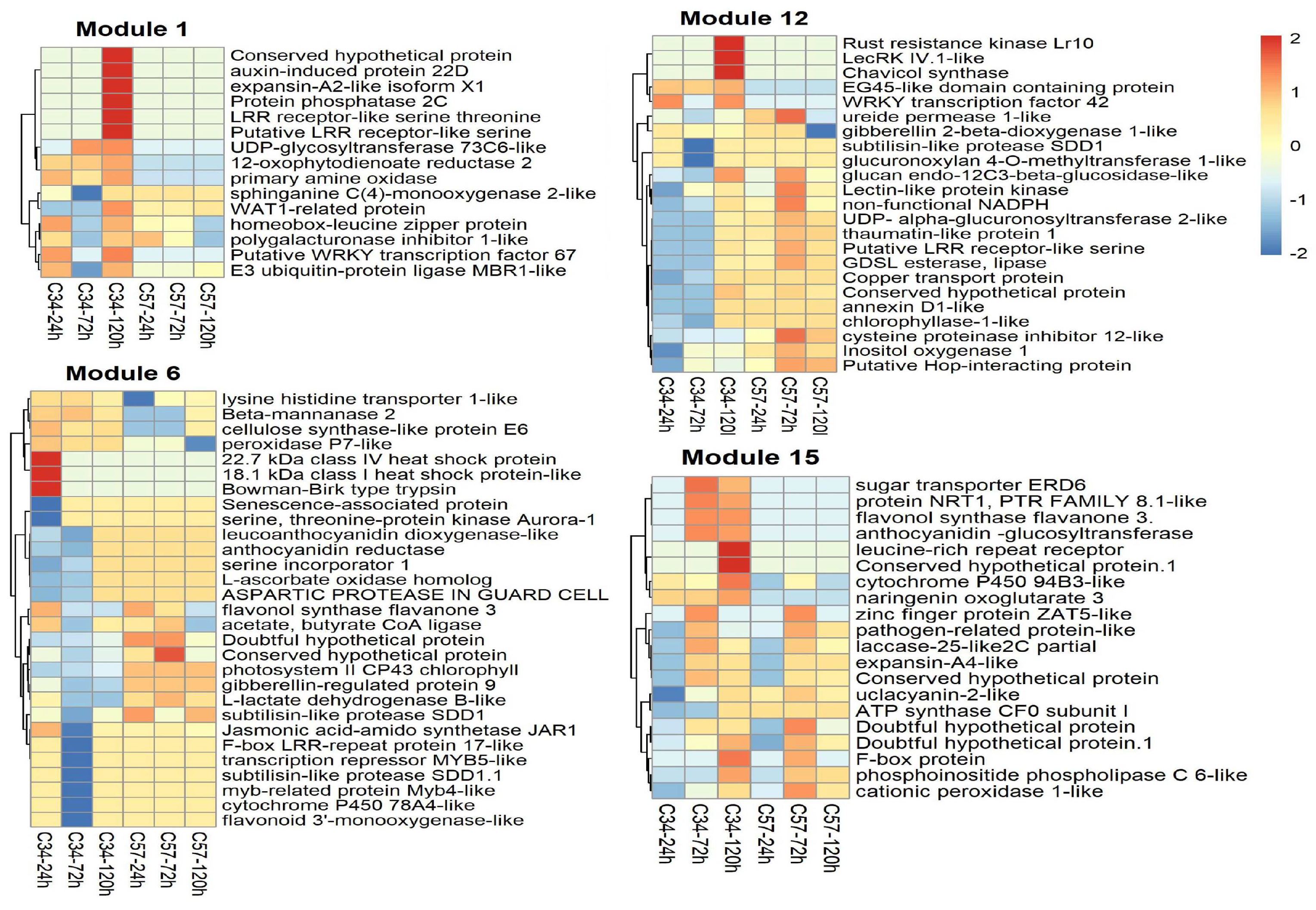
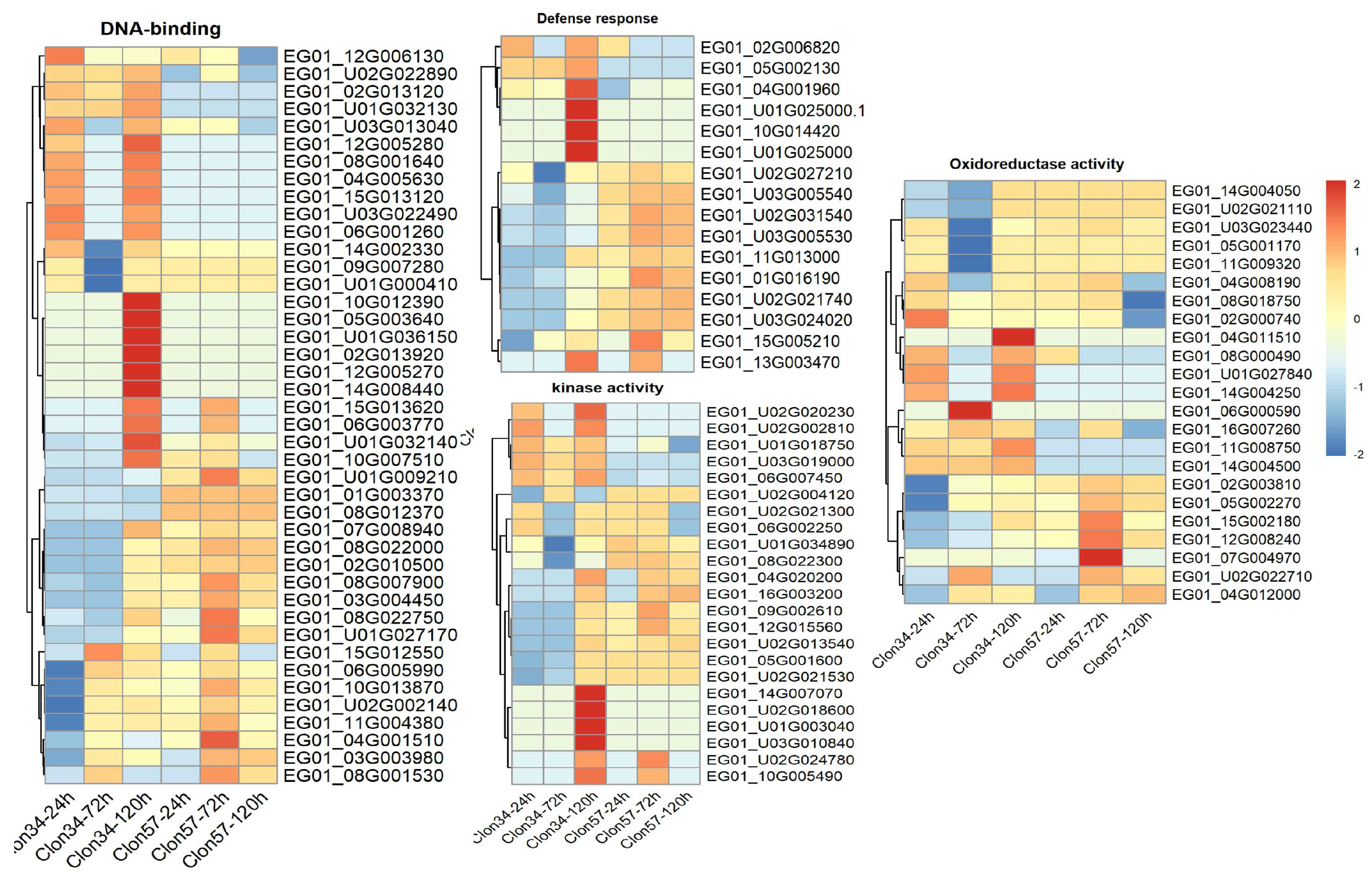
3.5. Gene Co-Expression Networks and Gene Ontology Analysis of Oil Palm Genotypes
3.6. Central Genes and Modularity of Co-Expression Networks in Oil Palm
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarria, G.A.; Martinez, G.; Varon, F.; Drenth, A.; Guest, D.I. Histopathological studies of the process of Phytophthora palmivora infection in oil palm. Eur. J. Plant Pathol. 2016, 145, 39–51. [Google Scholar] [CrossRef]
- Navia, E.A.; Ávila, R.A.; Daza, E.E.; Restrepo, E.F.; Romero, H.M. Assessment of tolerance to bud rot in oil palm under field conditions. Eur. J. Plant Pathol. 2014, 140, 711–720. [Google Scholar] [CrossRef]
- Afandi, D.; Basyuni, M.; Putri, L.; Chalil, D.; Wati, R.; Siregar, E.; Syahputra, I. Molecular performances of oil palm Elaeis guineensis tolerance to Ganoderma sp. J. Phys. Conf. Ser. 2018, 1116, 052001. [Google Scholar] [CrossRef]
- Avila-Mendez, K.; Rodrigo, Á.; Araque, L.; Romero, H.M. Simultaneous transcriptome analysis of oil palm clones and Phytophthora palmivora reveals oil palm defense strategies. PLoS ONE 2019, 14, e0222774. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Pathogen perception and responses in plant immunity. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Khavkin, E. Plant—Pathogen molecular dialogue: Evolution, mechanisms and agricultural implementation. Russ. J. Plant Physiol. 2021, 68, 197–211. [Google Scholar] [CrossRef]
- Rao, X.; Dixon, R.A. Co-expression networks for plant biology: Why and how. Acta Biochim. Biophys. Sin. 2019, 51, 981–988. [Google Scholar] [CrossRef]
- Ávila-Méndez, K.; Avila-Diazgranados, R.; Pardo, A.; Herrera, M.; Sarria, G.; Romero, H.M. Response of in vitro obtained oil palm and interspecific OxG hybrids to inoculation with Phytophthora palmivora. For. Pathol. 2019, 49, e12486. [Google Scholar] [CrossRef]
- Singh, R.; Ong-Abdullah, M.; Low, E.-T.L.; Manaf, M.A.A.; Rosli, R.; Nookiah, R.; Ooi, L.C.-L.; Ooi, S.-E.; Chan, K.-L.; Halim, M.A.; et al. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 2013, 500, 335–339. [Google Scholar] [CrossRef]
- Gil, J.; Herrera, M.; Duitama, J.; Sarria, G.; Restrepo, S.; Romero, H.M. Genomic variability of Phytophthora palmivora isolates from different oil palm cultivation regions in Colombia. Phytopathology 2020, 110, 1553–1564. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P. EffectorP 3.0: Prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. MPMI 2022, 35, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Dodds, P.N.; Singh, K.B.; Taylor, J.M. ApoplastP: Prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 2018, 217, 1764–1778. [Google Scholar] [CrossRef] [PubMed]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Kleinberg, J.M. Hubs, authorities, and communities. ACM Comput. Surv. 1999, 31, 5-es. [Google Scholar] [CrossRef]
- Wasserman, S.; Faust, K. Social Network Analysis: Methods and Applications; Cambridge University Press: Cambridge, UK, 1994. [Google Scholar]
- Brandes, U. A faster algorithm for betweenness centrality. J. Math. Sociol. 2001, 25, 163–177. [Google Scholar] [CrossRef]
- West, D.B. Introduction to Graph Theory; Prentice Hall: Upper Saddle River, NJ, USA, 2001; Volume 2. [Google Scholar]
- Carella, P.; Gogleva, A.; Tomaselli, M.; Alfs, C.; Schornack, S. Phytophthora palmivora establishes tissue-specific intracellular infection structures in the earliest divergent land plant lineage. Proc. Natl. Acad. Sci. USA 2018, 115, E3846–E3855. [Google Scholar] [CrossRef] [PubMed]
- Evangelisti, E.; Gogleva, A.; Hainaux, T.; Doumane, M.; Tulin, F.; Quan, C.; Yunusov, T.; Floch, K.; Schornack, S. Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors. BMC Biol. 2017, 15, 39. [Google Scholar] [CrossRef]
- Gavrin, A.; Rey, T.; Torode, T.A.; Toulotte, J.; Chatterjee, A.; Kaplan, J.L.; Evangelisti, E.; Takagi, H.; Charoensawan, V.; Rengel, D. Developmental modulation of root cell wall architecture confers resistance to an oomycete pathogen. Curr. Biol. 2020, 30, 4165–4176.e4165. [Google Scholar] [CrossRef] [PubMed]
- Zuñiga, C.; Peacock, B.; Liang, B.; McCollum, G.; Irigoyen, S.C.; Tec-Campos, D.; Marotz, C.; Weng, N.-C.; Zepeda, A.; Vidalakis, G.; et al. Linking metabolic phenotypes to pathogenic traits among “Candidatus Liberibacter asiaticus” and its hosts. NPJ Syst. Biol. Appl. 2020, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gogleva, A.; Drost, H.-G.; Schornack, S. SecretSanta: Flexible pipelines for functional secretome prediction. Bioinformatics 2018, 34, 2295–2296. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef]
- Stuer, N.; Van Damme, P.; Goormachtig, S.; Van Dingenen, J. Seeking the interspecies crosswalk for filamentous microbe effectors. Trends Plant Sci. 2023, 28, 1045–1059. [Google Scholar] [CrossRef]
- Al-Shahrour, F.; Díaz-Uriarte, R.; Dopazo, J. FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 2004, 20, 578–580. [Google Scholar] [CrossRef]
- Pham, J.; Stam, R.; Heredia, V.M.; Csukai, M.; Huitema, E. An NMRA-like protein regulates gene expression in Phytophthora capsici to drive the infection cycle on tomato. Mol. Plant-Microbe Interact. 2018, 31, 665–677. [Google Scholar] [CrossRef]
- Jupe, J.; Stam, R.; Howden, A.J.; Morris, J.A.; Zhang, R.; Hedley, P.E.; Huitema, E. Phytophthora capsici-tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 2013, 14, R63. [Google Scholar] [CrossRef]
- Avrova, A.O.; Boevink, P.C.; Young, V.; Grenville-Briggs, L.J.; van West, P.; Birch, P.R.J.; Whisson, S.C. A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell Microbiol. 2008, 10, 2271–2284. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Sharif, Y.; Zafar, M.H.; Ali, H.; Khan, K.A. Role of primary metabolites in plant defense against pathogens. Microb. Pathog. 2019, 137, 103728. [Google Scholar] [CrossRef]
- Von Groll, U.; Berger, D.; Altmann, T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 2002, 14, 1527–1539. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Survila, M.; Davidsson, P.R.; Pennanen, V.; Kariola, T.; Broberg, M.; Sipari, N.; Heino, P.; Palva, E.T. Peroxidase-generated apoplastic ROS impair cuticle integrity and contribute to DAMP-elicited defenses. Front. Plant Sci. 2016, 7, 1945. [Google Scholar] [CrossRef] [PubMed]
- Ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Gong, Z.-H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; An, Y.; Xu, P.; Xiao, J. Functioning of PPR proteins in organelle RNA metabolism and chloroplast biogenesis. Front. Plant Sci. 2021, 12, 627501. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bouwmeester, K.; Beseh, P.; Shan, W.; Govers, F. Phenotypic analyses of Arabidopsis T-DNA insertion lines and expression profiling reveal that multiple L-type lectin receptor kinases are involved in plant immunity. Mol. Plant-Microbe Interact. 2014, 27, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Hunter, K.; Kimura, S.; Rokka, A.; Tran, H.C.; Toyota, M.; Kukkonen, J.P.; Wrzaczek, M. CRK2 enhances salt tolerance by regulating callose deposition in connection with PLD α 1. Plant Physiol. 2019, 180, 2004–2021. [Google Scholar] [CrossRef]
- Abuqamar, S.; Ajeb, S.; Sham, A.; Enan, M.R.; Iratni, R. A mutation in the expansin-like A 2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana. Mol. Plant Pathol. 2013, 14, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Mosher, S.; Seybold, H.; Rodriguez, P.; Stahl, M.; Davies, K.A.; Dayaratne, S.; Morillo, S.A.; Wierzba, M.; Favery, B.; Keller, H.; et al. The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 2013, 73, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; Van Damme, E.J. Lectin domains at the frontiers of plant defense. Front. Plant Sci. 2014, 5, 397. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149. [Google Scholar] [CrossRef]
- Su, T.; Xu, Q.; Zhang, F.-C.; Chen, Y.; Li, L.-Q.; Wu, W.-H.; Chen, Y.-F. WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 2015, 167, 1579–1591. [Google Scholar] [CrossRef]
- Wu, B.; Gao, L.; Gao, J.; Xu, Y.; Liu, H.; Cao, X.; Zhang, B.; Chen, K. Genome-wide identification, expression patterns, and functional analysis of UDP glycosyltransferase family in peach (Prunus persica L. Batsch). Front. Plant Sci. 2017, 8, 389. [Google Scholar] [CrossRef]
- Cai, J.; Jiang, Y.; Ritchie, E.S.; Macho, A.P.; Yu, F.; Wu, D. Manipulation of plant metabolism by pathogen effectors: More than just food. FEMS Microbiol. Rev. 2023, 47, fuad007. [Google Scholar] [CrossRef]
- Wang, W.; Jiao, F. Effectors of Phytophthora pathogens are powerful weapons for manipulating host immunity. Planta 2019, 250, 413–425. [Google Scholar] [CrossRef]
- Morkunas, I.; Ratajczak, L. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plant. 2014, 36, 1607–1619. [Google Scholar] [CrossRef]
- Wang, W.; Liu, J.; Mishra, B.; Mukhtar, M.S.; McDowell, J.M. Sparking a sulfur war between plants and pathogens. Trends Plant Sci. 2022, 27, 1253–1265. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Shin, N.-H.; Trang, D.T.; Hong, W.-J.; Kang, K.; Chuluuntsetseg, J.; Moon, J.-K.; Yoo, Y.-H.; Jung, K.-H.; Yoo, S.-C. Rice senescence-induced receptor-like kinase (OsSRLK) is involved in phytohormone-mediated chlorophyll degradation. Int. J. Mol. Sci. 2019, 21, 260. [Google Scholar] [CrossRef]
- Peng, J.; Aluthmuhandiram, J.V.; Chethana, K.T.; Zhang, Q.; Xing, Q.; Wang, H.; Liu, M.; Zhang, W.; Li, X.; Yan, J. An NmrA-Like Protein, Lws1, Is Important for Pathogenesis in the Woody Plant Pathogen Lasiodiplodia theobromae. Plants 2022, 11, 2197. [Google Scholar] [CrossRef] [PubMed]
- Bradley, E.L.; Ökmen, B.; Doehlemann, G.; Henrissat, B.; Bradshaw, R.E.; Mesarich, C.H. Secreted glycoside hydrolase proteins as effectors and invasion patterns of plant-associated fungi and oomycetes. Front. Plant Sci. 2022, 13, 853106. [Google Scholar] [CrossRef] [PubMed]
- Ökmen, B.; Bachmann, D.; De Wit, P.J. A conserved GH17 glycosyl hydrolase from plant pathogenic Dothideomycetes releases a DAMP causing cell death in tomato. Mol. Plant Pathol. 2019, 20, 1710–1721. [Google Scholar] [CrossRef]
- Seidl, M.F.; Van den Ackerveken, G.; Govers, F.; Snel, B. A domain-centric analysis of oomycete plant pathogen genomes reveals unique protein organization. Plant Physiol. 2011, 155, 628–644. [Google Scholar] [CrossRef] [PubMed]
- Abrahamian, M.; Ah-Fong, A.M.; Davis, C.; Andreeva, K.; Judelson, H.S. Gene expression and silencing studies in Phytophthora infestans reveal infection-specific nutrient transporters and a role for the nitrate reductase pathway in plant pathogenesis. PLoS Path. 2016, 12, e1006097. [Google Scholar] [CrossRef]
- Ah-Fong, A.M.V.; Kagda, M.S.; Abrahamian, M.; Judelson, H.S. Niche-specific metabolic adaptation in biotrophic and necrotrophic oomycetes is manifested in differential use of nutrients, variation in gene content, and enzyme evolution. PLoS Path. 2019, 15, e1007729. [Google Scholar] [CrossRef]
- Fagard, M.; Launay, A.; Clément, G.; Courtial, J.; Dellagi, A.; Farjad, M.; Krapp, A.; Soulié, M.-C.; Masclaux-Daubresse, C. Nitrogen metabolism meets phytopathology. J. Exp. Bot. 2014, 65, 5643–5656. [Google Scholar] [CrossRef]
- Menino, J.F.; Saraiva, M.; Gomes-Rezende, J.; Sturme, M.; Pedrosa, J.; Castro, A.G.; Ludovico, P.; Goldman, G.H.; Rodrigues, F. P. brasiliensis virulence is affected by SconC, the negative regulator of inorganic sulfur assimilation. PLoS ONE 2013, 8, e74725. [Google Scholar] [CrossRef]
- Franza, T.; Expert, D. Role of iron homeostasis in the virulence of phytopathogenic bacteria: An ‘a la carte’menu. Mol. Plant Pathol. 2013, 14, 429–438. [Google Scholar] [CrossRef]
- Dellagi, A.; Segond, D.; Rigault, M.; Fagard, M.; Simon, C.; Saindrenan, P.; Expert, D. Microbial siderophores exert a subtle role in Arabidopsis during infection by manipulating the immune response and the iron status. Plant Physiol. 2009, 150, 1687–1696. [Google Scholar] [CrossRef]
- Olea, F.; Pérez-García, A.; Cantón, F.R.; Rivera, M.E.; Cañas, R.; Avila, C.; Cazorla, F.M.; Cánovas, F.M.; de Vicente, A. Up-regulation and localization of asparagine synthetase in tomato leaves infected by the bacterial pathogen Pseudomonas syringae. Plant Cell Physiol. 2004, 45, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Seifi, H.S.; Van Bockhaven, J.; Angenon, G.; Höfte, M. Glutamate Metabolism in Plant Disease and Defense: Friend or Foe? MPMI 2013, 26, 475–485. [Google Scholar] [CrossRef]
- Chojak-Koźniewska, J.; Kuźniak, E.; Zimny, J. The effects of combined abiotic and pathogen stress in plants: Insights from salinity and Pseudomonas syringae pv lachrymans interaction in cucumber. Front. Plant Sci. 2018, 9, 1691. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef]
- Gorshkov, V.; Tsers, I. Plant susceptible responses: The underestimated side of plant–pathogen interactions. Biol. Rev. 2022, 97, 45–66. [Google Scholar] [CrossRef]
- Chowdhury, J.; Henderson, M.; Schweizer, P.; Burton, R.A.; Fincher, G.B.; Little, A. Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp. hordei. New Phytol. 2014, 204, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Berka, M.; Kopecká, R.; Berková, V.; Brzobohatý, B.; Černý, M. Regulation of heat shock proteins 70 and their role in plant immunity. J. Exp. Bot. 2022, 73, 1894–1909. [Google Scholar] [CrossRef]
- Fernie, A.R. Evolution: An early role for flavonoids in defense against oomycete infection. Curr. Biol. 2019, 29, R688–R690. [Google Scholar] [CrossRef]
- Hu, Z.; Zhong, X.; Zhang, H.; Luo, X.; Wang, Y.; Wang, Y.; Liu, T.; Zhang, Y.; Wang, X.; An, H.; et al. GhMYB18 confers Aphis gossypii Glover resistance through regulating the synthesis of salicylic acid and flavonoids in cotton plants. Plant Cell Rep. 2023, 42, 355–369. [Google Scholar] [CrossRef] [PubMed]






| Effector Category | Number of Proteins |
|---|---|
| Signal peptide genes (Secret Santa) | 80 |
| Apoplastic effectors (ApoplastP) | 30 |
| With signal peptide | 23 |
| Without signal peptide | 7 |
| Citoplasmatic effectors (EffectorP 3.0) | 29 |
| With signal peptide | 7 |
| Without signal peptide | 22 |
| RxLR effectors | 13 |
| Possible Elicitors | |
| Elicitines | 16 |
| Cellulose-binding domain, fungal | 2 |
| Carbohydrate binding proteins | 8 |
| NPP1 (necrosis and ethylene-inducing protein) | 4 |
| Proteases | |
| Serine protease | 5 |
| Cysteine protease | 2 |
| Cell wall degrading enzymes (CWDE) | |
| Glycosyl hydrolase | 8 |
| Endoglucanase | 6 |
| Pectinesterase | 3 |
| Others | |
| Cysteine-rich proteins | 2 |
| ABC transporters | 12 |
| Kinase | 5 |
| Oxidase | 4 |
| Hypothetical proteins | 28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Gaona, M.; Botero-Rozo, D.; Araque, L.; Romero, H.M. The Dynamic Interaction between Oil Palm and Phytophthora palmivora in Bud Rot Disease: Insights from Transcriptomic Analysis and Network Modelling. J. Fungi 2024, 10, 164. https://doi.org/10.3390/jof10030164
García-Gaona M, Botero-Rozo D, Araque L, Romero HM. The Dynamic Interaction between Oil Palm and Phytophthora palmivora in Bud Rot Disease: Insights from Transcriptomic Analysis and Network Modelling. Journal of Fungi. 2024; 10(3):164. https://doi.org/10.3390/jof10030164
Chicago/Turabian StyleGarcía-Gaona, Mariandrea, David Botero-Rozo, Leonardo Araque, and Hernán Mauricio Romero. 2024. "The Dynamic Interaction between Oil Palm and Phytophthora palmivora in Bud Rot Disease: Insights from Transcriptomic Analysis and Network Modelling" Journal of Fungi 10, no. 3: 164. https://doi.org/10.3390/jof10030164
APA StyleGarcía-Gaona, M., Botero-Rozo, D., Araque, L., & Romero, H. M. (2024). The Dynamic Interaction between Oil Palm and Phytophthora palmivora in Bud Rot Disease: Insights from Transcriptomic Analysis and Network Modelling. Journal of Fungi, 10(3), 164. https://doi.org/10.3390/jof10030164








