Diversity of Rock-Inhabiting Fungi in Tarragona Province, Spain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites
2.2. Sampling Design
2.3. Fungal Isolation
2.4. Phenotypic Characterization
2.5. DNA Extraction, Amplification, and Sequencing
2.6. Fungal Identification and Phylogenetic Analyses
2.7. Physiological Characterization of the Strains of Interest
3. Results
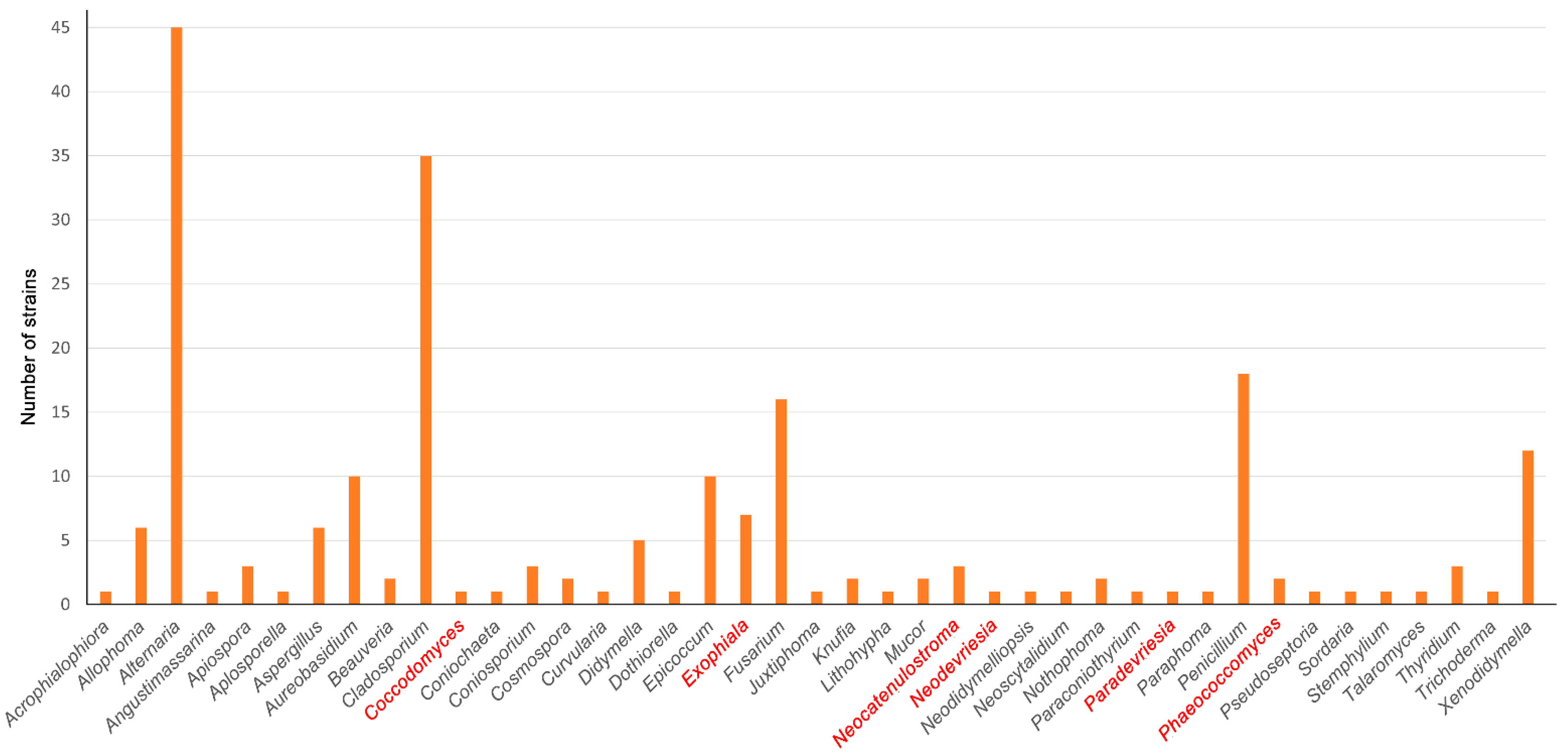
3.1. Phenotypic and Molecular Identification of the Fungal Strains
3.2. Phylogeny
3.2.1. Order Dothideales
3.2.2. Genus Exophiala
3.2.3. Order Capnodiales
3.3. Taxonomy
3.4. Physiology
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scheerer, S.; Ortega-Morales, O.; Gaylarde, C. Microbial deterioration of stone monuments-an updated overview. In Advances in Applied Microbiology, 1st ed.; Elesvier Inc.: Amsterdam, The Netherlands, 2009; pp. 97–139. [Google Scholar] [CrossRef]
- Sterflinger, K.; Krumbein, W.E. Multiple stress factors affecting growth of rock-inhabiting black fungi. Bot. Acta 1995, 108, 490–496. [Google Scholar] [CrossRef]
- Zak, J.C.; Wildman, H.G. Fungi in stressful environments. In Biodiversity of Fungi Inventory and Monitoring Methods; Mueller, G.M., Bills, G.F., Foster, M.S., Eds.; Elsevier Academic Press: Boston, MA, USA, 2004; pp. 513–531. [Google Scholar] [CrossRef]
- Liu, B.; Fu, R.; Wu, B.; Liu, X.; Xiang, M. Rock-inhabiting fungi: Terminology, diversity, evolution and adaptation mechanisms. Mycology 2021, 13, 1–31. [Google Scholar] [CrossRef]
- Berti, L.; Marvasi, M.; Perito, B. Characterization of the community of black meristematic fungi inhabiting the external white marble of the Florence Cathedral. J. Fungi 2023, 9, 665. [Google Scholar] [CrossRef]
- Choe, Y.-H.; Kim, M.; Woo, J.; Lee, M.J.; Lee, J.I.; Lee, E.J.; Lee, Y.K. Comparing rock-inhabiting microbial communities in different rock types from a high arctic polar desert. FEMS Microbiol. Ecol. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Isola, D.; Zucconi, L.; Onofri, S.; Caneva, G.; De Hoog, G.S.; Selbmann, L. Extremotolerant rock inhabiting black fungi from Italian monumental sites. Fungal Divers. 2016, 76, 75–96. [Google Scholar] [CrossRef]
- Ruibal, C.; Platas, G.; Bills, G. High diversity and morphological convergence among melanised fungi from rock formations in the Central Mountain System of Spain. Persoonia 2008, 21, 93–110. [Google Scholar] [CrossRef]
- Ruibal, C.; Selbmann, L.; Avci, S.; Martin-Sanchez, P.M.; Gorbushina, A.A. Roof-inhabiting cousins of rock-inhabiting fungi: Novel melanized microcolonial fungal species from photocatalytically reactive subaerial surfaces. Life 2018, 8, 30. [Google Scholar] [CrossRef]
- Santo, A.P.; Cuzman, O.A.; Petrocchi, D.; Pinna, D.; Salvatici, T.; Perito, B. Black on white: Microbial growth darkens the external marble of florence cathedral. Appl. Sci. 2021, 11, 6163. [Google Scholar] [CrossRef]
- Tiquia-Arashiro, S.; Grube, M. Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S.M., Grube, M., Eds.; Springer Nature Switzerland AG: Gewerbestrasse, Switzerland, 2019; pp. 39–57. [Google Scholar] [CrossRef]
- Wollenzien, U.; de Hoog, G.; Krumbein, W.; Urzí, C. On the isolation of microcolonial fungi occurring on and in marble and other calcareous rocks. Sci. Total. Environ. 1995, 167, 287–294. [Google Scholar] [CrossRef]
- Sterflinger, K.; De Baere, R.; de Hoog, G.; De Wachter, R.; Krumbein, W.E.; Haase, G. Coniosporium perforans and C. apollinis, two new rock-inhabiting fungi isolated from marble in the Sanctuary of Delos (Cyclades, Greece). Antonie van Leeuwenhoek 1997, 72, 349–363. [Google Scholar] [CrossRef]
- Cappitelli, F.; Nosanchuk, J.D.; Casadevall, A.; Toniolo, L.; Brusetti, L.; Florio, S.; Principi, P.; Borin, S.; Sorlini, C. Synthetic consolidants attacked by melanin-producing fungi: Case study of the biodeterioration of Milan (Italy) cathedral marble treated with acrylics. Appl. Environ. Microbiol. 2007, 73, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Gorbushina, A.A.; Lyalikova, N.N.; Vlasov, D.Y.; Khizhnyak, T.V. Microbial communities on the monuments of Moscow and St. Petersburg: Biodiversity and trophic relations. Microbiology 2002, 71, 350–356. [Google Scholar] [CrossRef]
- Marfenina, O.E.; Makarova, N.V.; Ivanova, A.E. Opportunistic fungi in soils and surface air of a megalopolis (for the Tushino Region, Moscow). Microbiology 2011, 80, 870–876. [Google Scholar] [CrossRef]
- Viles, H.A.; Gorbushina, A.A. Soiling and microbial colonisation on urban roadside limestone: A three year study in Oxford, England. Build. Environ. 2003, 38, 1217–1224. [Google Scholar] [CrossRef]
- May, E. Microbes on building stone—For good or ill? Culture 2003, 24, 5–8. [Google Scholar]
- Warscheid, T.; Braams, J. Biodeterioration of stone: A review. Int. Biodeterior. Biodegrad. 2000, 46, 343–368. [Google Scholar] [CrossRef]
- Kurup, V.P.; Shen, H.-D.; Banerjee, B. Respiratory fungal allergy. Microbes Infect. 2000, 2, 1101–1110. [Google Scholar] [CrossRef]
- Marvasi, M.; Donnarumma, F.; Frandi, A.; Mastromei, G.; Sterflinger, K.; Tiano, P.; Perito, B. Black microcolonial fungi as deteriogens of two famous marble statues in Florence, Italy. Int. Biodeterior. Biodegrad. 2012, 68, 36–44. [Google Scholar] [CrossRef]
- Páramo-Aguilera, L.; Ortega-Morales, B.O.; Narváez-Zapata, J.A. Culturable fungi associated with urban stone surfaces in Mexico City. Electron. J. Biotechnol. 2012, 15, 1–17. [Google Scholar] [CrossRef]
- Suihko, M.-L.; Alakomi, H.-L.; Gorbushina, A.; Fortune, I.; Marquardt, J.; Saarela, M. Characterization of aerobic bacterial and fungal microbiota on surfaces of historic Scottish monuments. Syst. Appl. Microbiol. 2007, 30, 494–508. [Google Scholar] [CrossRef]
- Ametrano, C.G.; Muggia, L.; Grube, M. Extremotolerant black fungi from rocks and lichens. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S.M., Grube, M., Eds.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; pp. 39–57. [Google Scholar] [CrossRef]
- de Hoog, G.; Beguin, H.; Batenburg-van de Vegte, W.H. Phaeotheca triangularis, a new meristematic black yeast from a humidifier. Antonie van Leeuwenhoek 1997, 71, 289–295. [Google Scholar] [CrossRef]
- Sterflinger, K. Black yeasts and meristematic fungi: Ecology, diversity and identification. In Biodiversity and Ecophysiology of Yeasts the Yeast Handbook; Péter, G., Rosa, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar] [CrossRef]
- Vicente, V.; Attili-Angelis, D.; Pie, M.; Queiroz-Telles, F.; Cruz, L.; Najafzadeh, M.; de Hoog, G.; Zhao, J.; Pizzirani-Kleiner, A. Environmental isolation of black yeast-like fungi involved in human infection. Stud. Mycol. 2008, 61, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Prenafeta-Boldú, F.X.; Summerbell, R.; De Hoog, G.S. Fungi growing on aromatic hydrocarbons: Biotechnology’s unexpected encounter with biohazard? FEMS Microbiol. Rev. 2006, 30, 109–130. [Google Scholar] [CrossRef] [PubMed]
- de Hoog, G.S.; Ende AHGG van den Uijthof, J.M.J.; Untereiner, W.A. Nutritional physiology of type isolates of currently accepted species of Exophiala and Phaeococcomyces. Antonie van Leeuwenhoek 1995, 68, 43–49. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, G.S.; Zeng, J.S.; Harrak, M.J.; Sutton, D.A. Exophiala xenobiotica sp. nov., an opportunistic black yeast inhabiting environments rich in hydrocarbons. Antonie van Leeuwenhoek 2006, 90, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Prenafeta-Boldú, F.X.; Kuhn, A.; Luykx, D.M.; Anke, H.; van Groenestijn, J.W.; de Bont, J.A. Isolation and characterization of fungi growing on volatile aromatic hydrocarbons as their sole carbon and energy source. Mycol. Res. 2001, 105, 477–484. [Google Scholar] [CrossRef]
- Isola, D.; Scano, A.; Orrù, G.; Prenafeta-Boldú, F.X.; Zucconi, L. Hydrocarbon-contaminated sites: Is there something more than Exophiala xenobiotica? New insights into black fungal diversity using the long cold incubation method. J. Fungi 2021, 7, 817. [Google Scholar] [CrossRef] [PubMed]
- de Hoog, G.S.; Yurlova, N.A. Conidiogenesis, nutritional physiology and taxonomy of Aureobasidium and Hormonema. Antonie van Leeuwenhoek 1994, 65, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Cordero, R.J.; Casadevall, A. Functions of fungal melanin beyond virulence. Fungal Biol. Rev. 2017, 31, 99–112. [Google Scholar] [CrossRef]
- Sterflinger, K.; de Hoog, G.S.; Haase, G. Phylogeny and ecology of meristematic ascomycetes. Stud. Mycol. 1999, 43, 5–22. [Google Scholar]
- Wheeler, M.H.; Bell, A.A. Melanins and their importance in pathogenic fungi. Curr. Top Med. Mycol. 1988, 2, 338–387. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Hermanides-Nijhof, E.J. The black yeasts and allied hyphomycetes. Stud. Mycol. 1977, 15, 151–222. [Google Scholar]
- Staley, J.T.; Palmer, F.; Adams, J.B. Microcolonial fungi: Common inhabitants on desert rocks? Science 1982, 215, 1093–1095. [Google Scholar] [CrossRef] [PubMed]
- Gorbushina, A.A.; Krumbein, W.E.; Hamman, C.H.; Panina, L.; Soukharjevski, S.; Wollenzien, U. Role of black fungi in color change and biodeterioration of antique marbles. Geomicrobiol. J. 1993, 11, 205–221. [Google Scholar] [CrossRef]
- Slepecky, R.A.; Starmer, W.T. Phenotypic plasticity in fungi: A review with observations on Aureobasidium pullulans. Mycologia 2009, 101, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Gorbushina, A.A. Life on the rocks. Environ. Microbiol. 2007, 9, 1613–1631. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Perfect, J.; de Hoog, G.S. Black Molds and Melanized Yeasts Pathogenic to Humans. Cold Spring Harb. Perspect. Med. 2015, 5, a019570. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.M.; Moreno, L.F.; Stielow, B.; Muszewska, A.; Hainaut, M.; Gonzaga, L.; Abouelleil, A.; Patané, J.S.L.; Priest, M.; Souza, R.; et al. Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota). Stud. Mycol. 2017, 86, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Abdollahzadeh, J.; Groenewald, J.; Coetzee, M.; Wingfield, M.; Crous, P. Evolution of lifestyles in Capnodiales. Stud. Mycol. 2020, 95, 381–414. [Google Scholar] [CrossRef]
- Crous, P.; Schoch, C.; Hyde, K.; Wood, A.; Gueidan, C.; de Hoog, G.; Groenewald, J. Phylogenetic lineages in the Capnodiales. Stud. Mycol. 2009, 64, 17–47. [Google Scholar] [CrossRef]
- Schoch, C.; Crous, P.; Groenewald, J.; Boehm, E.; Burgess, T.; de Gruyter, J.; de Hoog, G.; Dixon, L.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef]
- Sun, W.; Su, L.; Yang, S.; Sun, J.; Liu, B.; Fu, R.; Wu, B.; Liu, X.; Cai, L.; Guo, L.; et al. Unveiling the hidden diversity of rock-inhabiting fungi: Chaetothyriales from China. J. Fungi 2020, 6, 187. [Google Scholar] [CrossRef]
- Egidi, E.; de Hoog, G.S.; Isola, D.; Onofri, S.; Quaedvlieg, W.; de Vries, M.; Verkley, G.J.M.; Stielow, J.B.; Zucconi, L.; Selbmann, L. Phylogeny and taxonomy of meristematic rock-inhabiting black fungi in the Dothideomycetes based on multi-locus phylogenies. Fungal Divers. 2014, 65, 127–165. [Google Scholar] [CrossRef]
- Cuscó, A.; Catozzi, C.; Viñes, J.; Sanchez, A.; Francino, O. Microbiota profiling with long amplicons using Nanopore sequencing: Full-length 16S rRNA gene and the 16S-ITS-23S of the rrn operon. F1000Research 2019, 7, 1755. [Google Scholar] [CrossRef]
- Nagano, Y.; Miura, T.; Tsubouchi, T.; Lima, A.O.; Kawato, M.; Fujiwara, Y.; Fujikura, K. Cryptic fungal diversity revealed in deep-sea sediments associated with whale-fall chemosynthetic ecosystems. Mycology 2020, 11, 263–278. [Google Scholar] [CrossRef]
- Luo, Y.; Wei, X.; Yang, S.; Gao, Y.-H.; Luo, Z.-H. Fungal diversity in deep-sea sediments from the Magellan seamounts as revealed by a metabarcoding approach targeting the ITS2 regions. Mycology 2020, 11, 214–229. [Google Scholar] [CrossRef]
- Zhang, Z.-F.; Liu, Y.; Li, M. Pacific Biosciences Single-Molecule Real-Time (SMRT) Sequencing Reveals High Diversity of Basal Fungal Lineages and Stochastic Processes Controlled Fungal Community Assembly in Mangrove Sediments. 2020. Available online: https://www.researchsquare.com/article/rs-97364/v1 (accessed on 4 January 2024). [CrossRef]
- Wu, B.; Hussain, M.; Zhang, W.; Stadler, M.; Liu, X.; Xiang, M. Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 2019, 10, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Beck, H.E.; Zimmermann, N.E.; McVicar, T.R.; Vergopolan, N.; Berg, A.; Wood, E.F. Present and future köppen-geiger climate classification maps at 1-km resolution. Sci. Data 2018, 5, 180214. [Google Scholar] [CrossRef]
- Ringer, S. Regarding the action of hydrate of soda, hydrate of ammonia, and hydrate of potash on the ventricle of the frog’s heart. J. Physiol. 1882, 3, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Hawksworth, D.; Kirk, P.; Sutton, B.; Pegler, D. Ainsworth & Bisby’s Dictionary of the Fungi, 8th ed.; Prensa de la Universidad de Oxford, Ed.; CAB International: Wallingford, UK, 1996. [Google Scholar]
- Onions, A.; Pitt, J. Appendix: Media. In Filamentous Fungi; Hawksworth, D., Kirsop, B., Eds.; Cambridge University Press: Cambridge, UK, 1988; Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/jobm.3620300217 (accessed on 15 May 2023).
- Atlas, R.M. Handbook of Microbiological Media, 4th ed.; CRC Press Taylor & Francis Group: Washington, DC, USA, 2010; 2036p. [Google Scholar] [CrossRef]
- Jarvis, B. Comparison of an improved rose bengal-chlortetracycline agar with other media for the selective isolation and enumeration of moulds and yeasts in foods. J. Appl. Bacteriol. 1973, 36, 723–727. [Google Scholar] [CrossRef]
- King, A.D.; Hocking, A.D.; Pitt, J.I. Dichloran-rose bengal medium for enumeration and isolation of molds from foods. Appl. Environ. Microbiol. 1979, 37, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi, 2nd ed.; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2010. [Google Scholar]
- Reiss, J. Ein selektives kulturmedium für der Nachweiss von Aspergillus flavus. Zbl. Bokt. Hyg. I. Abt. Orig. 1972, 220, 564–566. [Google Scholar]
- Crous, P.W.; Verkley, G.J.M.; Groenewald, J.Z.; Samson, R.A. Fungal Biodiversity; CBS Laboratory Manual Series; The Westerdijk Fungal Biodiversity Institute: Utrecht, The Netherlands, 2009. [Google Scholar]
- Kornerup, A.; Wanscher, J. Methuen Handbook of Colour, 3rd ed.; Eyre Methuen: London, UK, 1978; 252p. [Google Scholar]
- Smith, H.; Wingfield, M.J.; Crous, P.W.; Coutinho, I.A. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. S. Afr. J. Bot. 1996, 62, 86–88. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications, 2nd ed.; Innis, M., Gelfand, D., Sninsky, J., White, T., Eds.; Academic Press, Inc.: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 1997, 35, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 9, 2044–2049. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Cohen, S.D. A protocol for direct sequencing of multiple gene specific PCR products from Discula umbrinella, a fungal endophyte, utilizing bufferless precast electrophoresis. J. Microbiol. Methods 2005, 61, 131–135. [Google Scholar] [CrossRef]
- Takara Bio Inc. EmeraldAmp® GT PCR Master Mix; Takara Bio Europe SAS: Saint-Germain-en-Laye, France, 2012; p. 1. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: Enabling high-impact science for phylogenetics researchers with limited resources. In Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the Campus and Beyond, Chicago, IL, USA, 16–20 July 2012; Available online: https://dl.acm.org/doi/abs/10.1145/2335755.2335836 (accessed on 14 May 2023).
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.A. MrModeltest Version 2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Metropolis, N.; Ulam, S. The monte carlo method. J. Am. Stat. Assoc. 1949, 44, 335–341. Available online: https://www.jstor.org/stable/2280232 (accessed on 15 May 2023). [CrossRef]
- van der Walt, J.P.; Yarrow, D. Methods for the isolation, maintenance, classification and identification of yeasts. In The Yeasts; Elsevier Science Publishers B.V.: Amsterdam, The Netherlands, 1984; pp. 45–104. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Guého, E.; Masclaux, F.; Gerrits van den Ende, A.H.G.; Kwon-Chung, K.J.; Mcginnis, M.R. Nutritional physiology and taxonomy of human-pathogenic Cladosporium-Xylohypha species. Med. Mycol. 1995, 33, 339–347. [Google Scholar] [CrossRef]
- Wollenzien, U.; de Hoog, G.S.; Krumbein, W.; Uijthof, J.M.J. Sarcinomyces petricola, a new microcolonial fungus from marble in the Mediterranean basin. Antonie van Leeuwenhoek 1997, 71, 281–288. [Google Scholar] [CrossRef]
- Uribe Gutiérrez, L.A. Caracterización Fisiológica de Levaduras Aisladas de la Filósfera de Mora; Pontificia Universidad Javeriana: Bogotá, Columbia, 2007; Available online: https://repository.javeriana.edu.co/bitstream/handle/10554/8298/tesis276.pdf;jsessionid=C4CC63BB7E5E435DF74B4DCD756156A5?sequence=1 (accessed on 11 January 2023).
- Schwarz, P.; Lortholary, O.; Dromer, F.; Dannaoui, E. Carbon assimilation profiles as a tool for identification of Zygomycetes. J. Clin. Microbiol. 2007, 45, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, E.; Stchigel, A.M.; Cano, J.; Sutton, D.A.; Fothergill, A.W.; Chander, J.; Salas, V.; Rinaldi, M.G.; Guarro, J. Diversidad filogenética del hongo mucoral emergente Apophysomyces: Propuesta de tres nuevas especies. Rev. Iberoam. Micol. 2010, 27, 80–89. [Google Scholar] [CrossRef]
- Crous, P.; Osieck, E.; Jurjevi, Ž.; Boers, J.; Van Iperen, A.; Starink-Willemse, M.; Dima, B.; Balashov, S.; Bulgakov, T.; Johnston, P.; et al. Fungal Planet description sheets: 1284–1382. Persoonia 2021, 47, 178–374. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; Nilsson, R.H.; Bhunjun, C.S.; de Farias, A.R.G.; Sun, Y.-R.; Wijesinghe, S.N.; Raza, M.; Bao, D.-F.; Lu, L.; Tibpromma, S.; et al. The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Divers. 2022, 114, 327–386. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, H.F.; Song, Z.X.; Du, X.Y.; Deng, J.X. Identification and characterization of Gonatobotryum apiculatum causing leaf spot and blight on Sinowilsonia henryi. Mycobiology 2020, 48, 70–74. [Google Scholar] [CrossRef]
- Bates, S.T.; Reddy, G.S.N.; Garcia-Pichel, F. Exophiala crusticola anam. nov. (affinity Herpotrichiellaceae), a novel black yeast from biological soil crust in the Western United States. Int. J. Syst. Evol. Microbiol. 2006, 56, 2697–2702. [Google Scholar] [CrossRef]
- Li, D.M.; Li, R.Y.; De Hoog, G.; Wang, Y.X.; Wang, D.L. Exophiala asiatica, a new species from a fatal case in China. Med. Mycol. 2009, 47, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.; Wingfield, M.; Cheewangkoon, R.; Carnegie, A.; Burgess, T.; Summerell, B.; Edwards, J.; Taylor, P.; Groenewald, J. Foliar pathogens of eucalypts. Stud. Mycol. 2019, 94, 125–298. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.; Braun, U.; Groenewald, J. Mycosphaerella is polyphyletic. Stud. Mycol. 2007, 58, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Shivas, R.G.; Wingfield, M.J.; Summerell, B.A.; Rossman, A.Y.; Alves, J.L.; Adams, G.C.; Barreto, R.W.; Bell, A.; Coutinho, M.L.; et al. Fungal Planet description sheets: 128–153. Persoonia 2012, 29, 146–201. [Google Scholar] [CrossRef] [PubMed]
- Kasprzyk, I.; Grinn-Gofroń, A.; Ćwik, A.; Kluska, K.; Cariñanos, P.; Wójcik, T. Allergenic fungal spores in the air of urban parks. Aerobiologia 2021, 37, 39–51. [Google Scholar] [CrossRef]
- Egbuta, M.A.; Mwanza, M.M.; Babalola, O.O. Health risks associated with exposure to filamentous fungi. Int. J. Environ. Res. Public Health 2017, 14, 719. [Google Scholar] [CrossRef]
- Dionne, B.; Neff, L.; Lee, S.A.; Sutton, D.A.; Wiederhold, N.P.; Lindner, J.; Fan, H.; Jakeman, B. Pulmonary fungal infection caused by Neoscytalidium dimidiatum. J. Clin. Microbiol. 2015, 53, 2381–2384. [Google Scholar] [CrossRef]
- Salehi, M.; Zibafar, E.; Mahmoudi, S.; Hashemi, S.; Gatmiri, S.; Shoar, M.G.; Manshadi, S.D.; Jahanbin, B.; Alizadeh, R.; Hosseinpour, L.; et al. First report of invasive pulmonary infection by Didymella microchlamydospora and successful treatment with voriconazole. Clin. Microbiol. Infect. 2019, 25, 392–393. [Google Scholar] [CrossRef] [PubMed]
- Stricker, S.M.; Gossen, B.D.; McDonald, M.R. Risk assessment of secondary metabolites produced by fungi in the genus Stemphylium. Can. J. Microbiol. 2021, 67, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Gaona-Álvarez, C.; González-Velasco, C.; Morais-Foruria, F.; Alastruey-Izquierdo, A. Unusual aetiology of keratitis in a patient with bullous keratopathy. Enfermedades Infecc. Y Microbiol. Clin. 2020, 38, 84–85. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Hermoso, D.; Valenzuela-Lopez, N.; Rivero-Menendez, O.; Alastruey-Izquierdo, A.; Guarro, J.; Cano-Lira, J.F.; Stchigel, A.M. Diversity of coelomycetous fungi in human infections: A 10-y experience of two European reference centres. Fungal Biol. 2019, 123, 341–349. [Google Scholar] [CrossRef]
- Ruibal, C.; Gueidan, C.; Selbmann, L.; Gorbushina, A.; Crous, P.; Groenewald, J.; Muggia, L.; Grube, M.; Isola, D.; Schoch, C.; et al. Phylogeny of rock-inhabiting fungi related to Dothideomycetes. Stud. Mycol. 2009, 64, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Coleine, C.; Stajich, J.E.; de Los Ríos, A.; Selbmann, L. Beyond the extremes: Rocks as ultimate refuge for fungi in drylands. Mycologia 2021, 113, 108–133. [Google Scholar] [CrossRef]
- Ramanauskas, R.; Juzeliûnas, E.; Buèinskienë, D.; Lugauskas, A.; Peèiulytë, D. Investigation of microbiologically influenced corrosion 1. Characterization of natural outdoor conditions in Lithuania. Chemija 2005, 16, 25–34. [Google Scholar]
- De Leo, F.; Urzì, C.; de Hoog, G.S. Two Coniosporium species from rock surfaces. Stud. Mycol. 1999, 1999, 70–79. [Google Scholar]
- Crous, P.; Luangsa-Ard, J.; Wingfield, M.; Carnegie, A.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.; Baseia, I.; Cano-Lira, J.; et al. Fungal Planet description sheets: 785–867. Persoonia 2018, 41, 238–417. [Google Scholar] [CrossRef]
- Hughes, S.J. Conidiophores, conidia, and classification. Can. J. Bot. 1953, 31, 577–659. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Binder, M.; Groenewald, J.Z.; Summerell, B.A.; Carnegie, A.J.; Burgess, T.I.; Crous, P.W. Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 2014, 33, 1–40. [Google Scholar] [CrossRef] [PubMed]
- von Arx, J.A.; Müller, E. Über die neue Ascomycetengattung Aulographina. Sydowia 1960, 14, 330–333. [Google Scholar]
- Wang, M.-M.; Shenoy, B.D.; Li, W.; Cai, L. Molecular phylogeny of Neodevriesia, with two new species and several new combinations. Mycologia 2017, 109, 965–974. [Google Scholar] [CrossRef]
- Crous, P.W.; Schumacher, R.K.; Wingfield, M.J.; Lombard, L.; Giraldo, A.; Christensen, M.; Gardiennet, A.; Nakashima, C.; Pereira, O.L.; Smith, A.; et al. Fungal systematics and evolution: FUSE 1. Sydowia 2015, 67, 118. [Google Scholar] [CrossRef]














| Parameters | Calafell | Els Pallaresos | Montbrió del Camp | Reus |
|---|---|---|---|---|
| Lineal distance to the shore (km) | 0 | 5.7 | 7.4 | 9.0 |
| Average annual temperature (°C) | 15.9 | 16 | 15.6 | 15.0 |
| Minimum average annual temperature (°C) | 8.4 | 8 | 7.7 | 6.3 |
| Maximum average annual temperature (°C) | 24.2 | 24 | 24.3 | 24 |
| Average annual rainfall (mm) | 602 | 513 | 550 | 525 |
| Relative humidity: lower value (%) | 64.87 | 67.8 | 61.57 | 57.27 |
| Relative humidity: highest value (%) | 74.60 | 72.6 | 75.41 | 75.69 |
| Prevailing wind direction | S/W | S/W | S/W | S/W |
| Town | Sample Name | Coordinates (UTM) * | Source |
|---|---|---|---|
| Calafell | C1 | 31T 79,177.10 61,802.90 | Darkened concrete fence of a garden house |
| C2 | 31T 79,134.60 61,766.60 | Darkened concrete wall | |
| C3 | 31T 79,796.60 61,222.00 | Darkened concrete wall | |
| C4 | 31T 78,841.90 61,117.50 | Blackened block wall | |
| C5 | 31T 78,584.00 61,023.10 | Darkened concrete wall | |
| Montbrió del Camp | M1 | 31T 32,720.20 54,291.00 | Darkened concrete wall |
| M2 | 31T 32,658.70 54,252.30 | Darkened brick wall | |
| M3 | 31T 32,414.20 54,091.30 | Darkened concrete wall | |
| M4 | 31T 32,413.80 54,090.90 | Darkened concrete wall | |
| M5 | 31T 32,378.60 54,109.80 | Darkened clay wall | |
| Els Pallaresos (Urban area) | P1 | 31T 54,937.40 59,758.10 | Darkened concrete wall |
| P2 | 31T 54,954.60 59,694.90 | Darkened brick wall | |
| P3 | 31T 54,952.50 59,638.90 | Darkened concrete wall | |
| P4 | 31T 55,523.50 59,652.30 | Darkened concrete fence of a home garden | |
| P5 | 31T 55,545.50 59,571.40 | Darkened metal railing of a small natural park | |
| Els Pallaresos (Industrial area) | S1 | 31T 54,978.50 59,782.70 | Darkened concrete wall |
| S2 | 31T 54,945.50 59,754.60 | Darkened metal fence near a tree | |
| S3 | 31T 54,916.20 59,764.70 | Blackened cement blocks wall | |
| S4 | 31T 54,899.30 59,817.50 | Darkened PVC pipe for pluvial drain | |
| S5 | 31T 54,946.80 59,861.60 | Darkened concrete wall | |
| Reus | R1 | 31T 49,684.80 57,714.70 | Darkened concrete wall |
| R2 | 31T 49,916.90 57,650.50 | Darkened concrete wall | |
| R3 | 31T 42,102.90 57,713.20 | Darkened concrete wall | |
| R4 | 31T 46,377.40 57,475.40 | Darkened brick wall | |
| R5 | 31T 41,270.60 58,092.00 | Blackened block wall |
| Locus | Primer | Sequence (5′→3′) * | Orientation | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|---|
| ITS/LSU | ITS5 | GGAAGTAAAAGTCGTAACAAGG | Forward | 53–55 | [66] |
| LR5 | ATCCTGAGGGAAACTTC | Reverse | 53–55 | [67] | |
| LSU | NL1 | GCATATCAATAAGCGGAGGAAAAG | Forward | 53–55 | [68] |
| NL4b | GGTCCGTGTTTCAAGACGG | Reverse | 53–55 | [68] | |
| rpb2 | fRpb2-5F | GGGGWGGAYCAGAAGAAG | Forward | 55–60 | [69] |
| fRpb2-7R | CCCATRGCTTGYTTRCCCAT | Reverse | 55–60 | [69] | |
| tub2 | T10 | ACGATAGGTTCACCTCCAGAC | Forward | 55–57 | [70] |
| Bt2a | GGTAACCAAATCGGTGCTGCTTTC | Forward | 55–57 | [71] | |
| Bt2b | ACCCTCAGTGTAGTGACCCTTGGC | Reverse | 55–57 | [71] | |
| tef1 | EF-1H | ATGGGTAAGGARGACAAGAC | Forward | 57 | [72] |
| EF-2T | GGAAGTACCAGTGATCATGTT | Reverse | 57 | [72] | |
| EF1-983F | GCYCCYGGHCAYCGTGAYTTYAT | Forward | 57 | [73] | |
| EF1-2218R | ATGACACCRACRGCRACRGTYTG | Reverse | 57 | [73] | |
| EF1-728F | CATCGAGAAGTTCGAGAAGG | Forward | 57 | [74] | |
| EF1-986R | TACTTGAAGGAACCCTTACC | Reverse | 57 | [74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sastoque, A.P.; Cano-Lira, J.F.; Stchigel, A.M. Diversity of Rock-Inhabiting Fungi in Tarragona Province, Spain. J. Fungi 2024, 10, 170. https://doi.org/10.3390/jof10030170
Sastoque AP, Cano-Lira JF, Stchigel AM. Diversity of Rock-Inhabiting Fungi in Tarragona Province, Spain. Journal of Fungi. 2024; 10(3):170. https://doi.org/10.3390/jof10030170
Chicago/Turabian StyleSastoque, Angie Paola, José Francisco Cano-Lira, and Alberto Miguel Stchigel. 2024. "Diversity of Rock-Inhabiting Fungi in Tarragona Province, Spain" Journal of Fungi 10, no. 3: 170. https://doi.org/10.3390/jof10030170
APA StyleSastoque, A. P., Cano-Lira, J. F., & Stchigel, A. M. (2024). Diversity of Rock-Inhabiting Fungi in Tarragona Province, Spain. Journal of Fungi, 10(3), 170. https://doi.org/10.3390/jof10030170








