Characterization of Microbotryum lychnidis-dioicae Secreted Effector Proteins, Their Potential Host Targets, and Localization in a Heterologous Host Plant
Abstract
1. Introduction
2. Materials and Methods
2.1. cDNA Library Construction
2.2. Cloning the Genes MVLG_02245, MVLG_06175, and MVLG_05122
2.3. Yeast Transformation
2.4. Yeast Secretion Trap Assay
2.5. Yeast Two-Hybrid Screening
2.6. Gibson Assembly to Construct MVLG_05122 Tagged with a Cyan Fluorescent Protein Gene and MVLG_06175 Tagged with a mCherry Protein Gene
2.7. Agrobacterium Transformation through Electroporation
2.8. Arabidopsis thaliana and Growth Conditions
2.9. Floral Dipping Transformation of Arabidopsis thaliana Mediated by Agrobacterium
2.10. Determination of Copy Number of Trans Genes in A. thaliana
2.11. Plant mRNA Extraction
2.12. qRT-PCR
2.13. Fluorescence Confocal Microscopy
2.14. Fusarium Infection Assay on Arabidopsis thaliana
3. Results
3.1. Bioinformatic Characterization of Putative Effectors
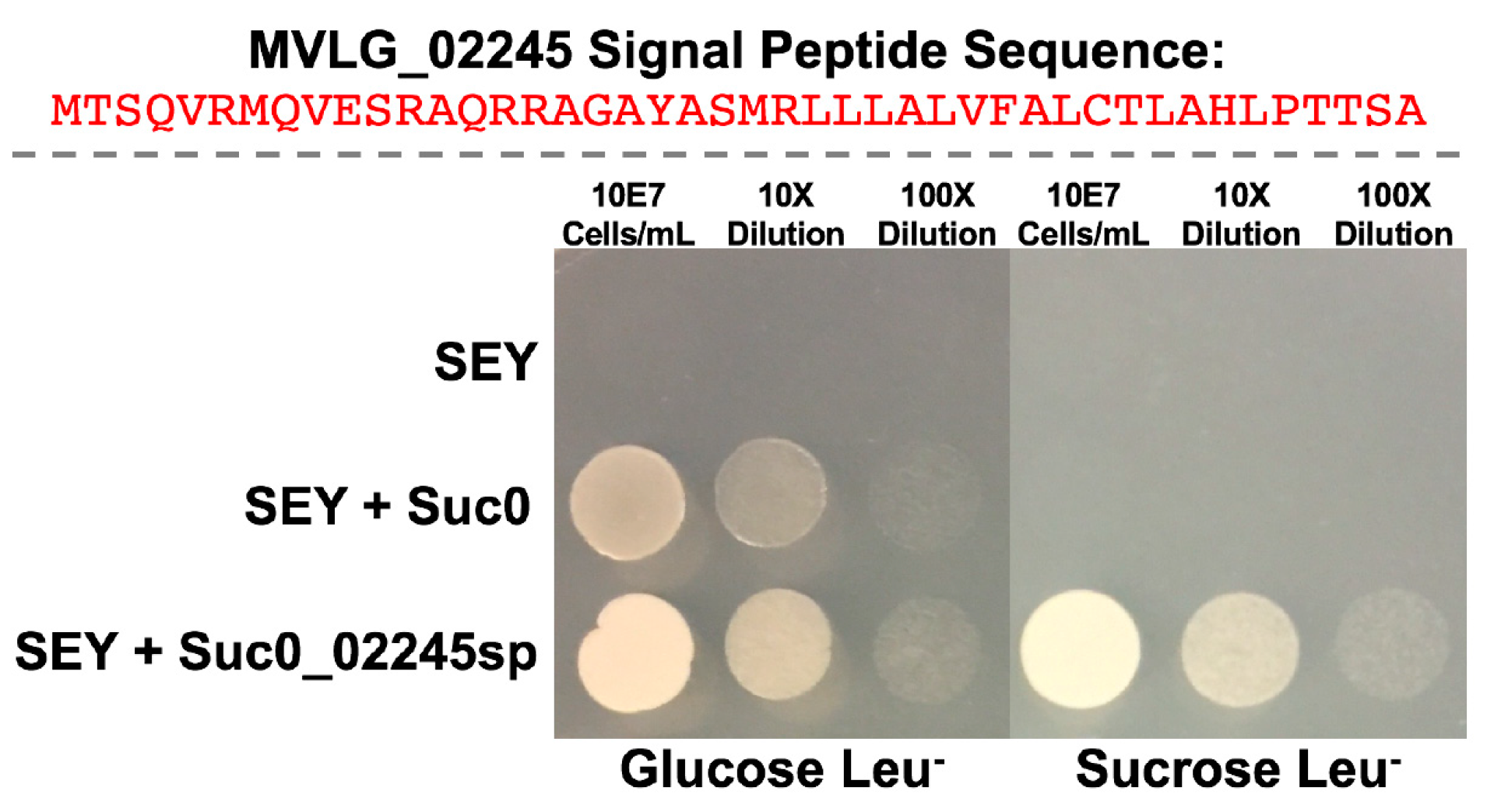
3.2. Yeast Secretion Trap Assay to Verify the Secretion of Effector Proteins
3.3. Yeast Two-Hybrid Screening to Reveal the Identities of Potential Plant Target Proteins
3.3.1. MVLG_06175 Interacts with Two Fungal Proteins and One Plant Host Protein
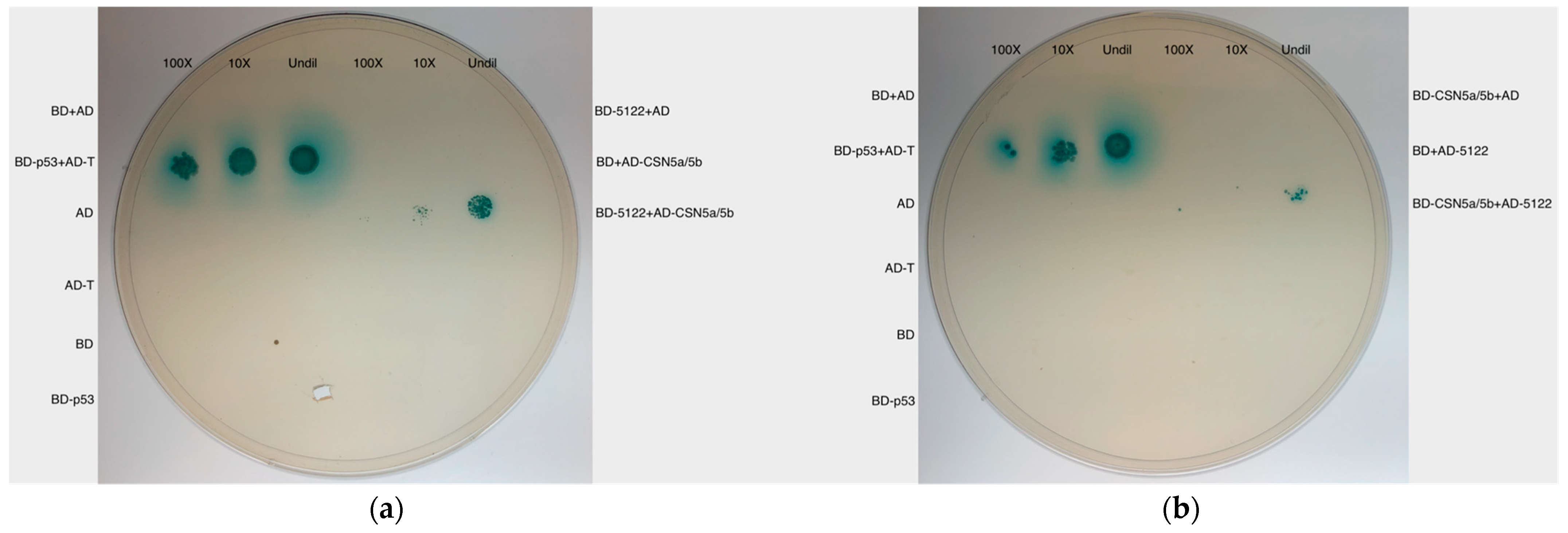
3.3.2. MVLG_05122 Interacts with Two Plant Host Proteins
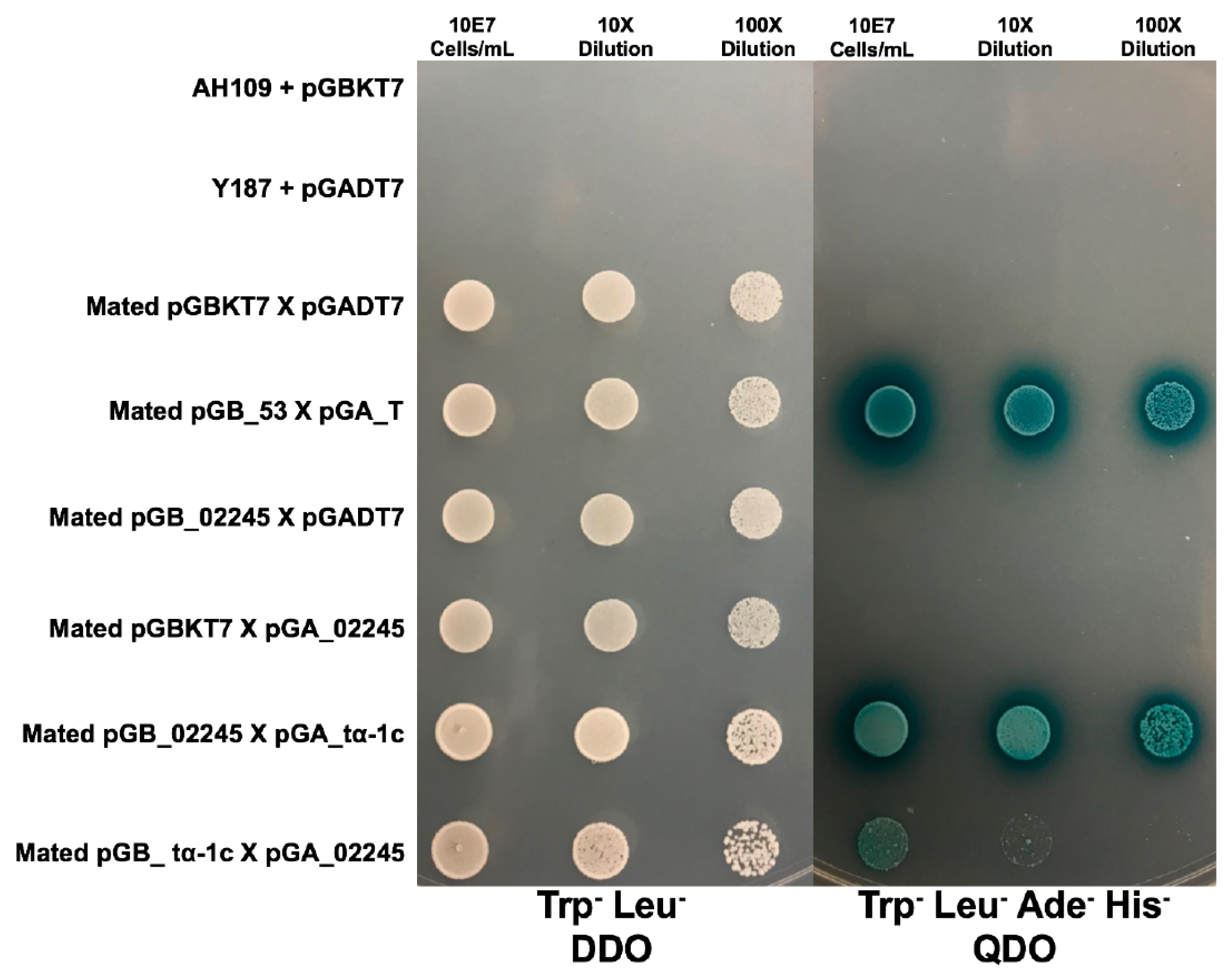
3.3.3. MVLG_02245 Interacts with Four Plant Host Proteins
3.4. Potential Locations of Protein-Protein Interactions in the Plant Host
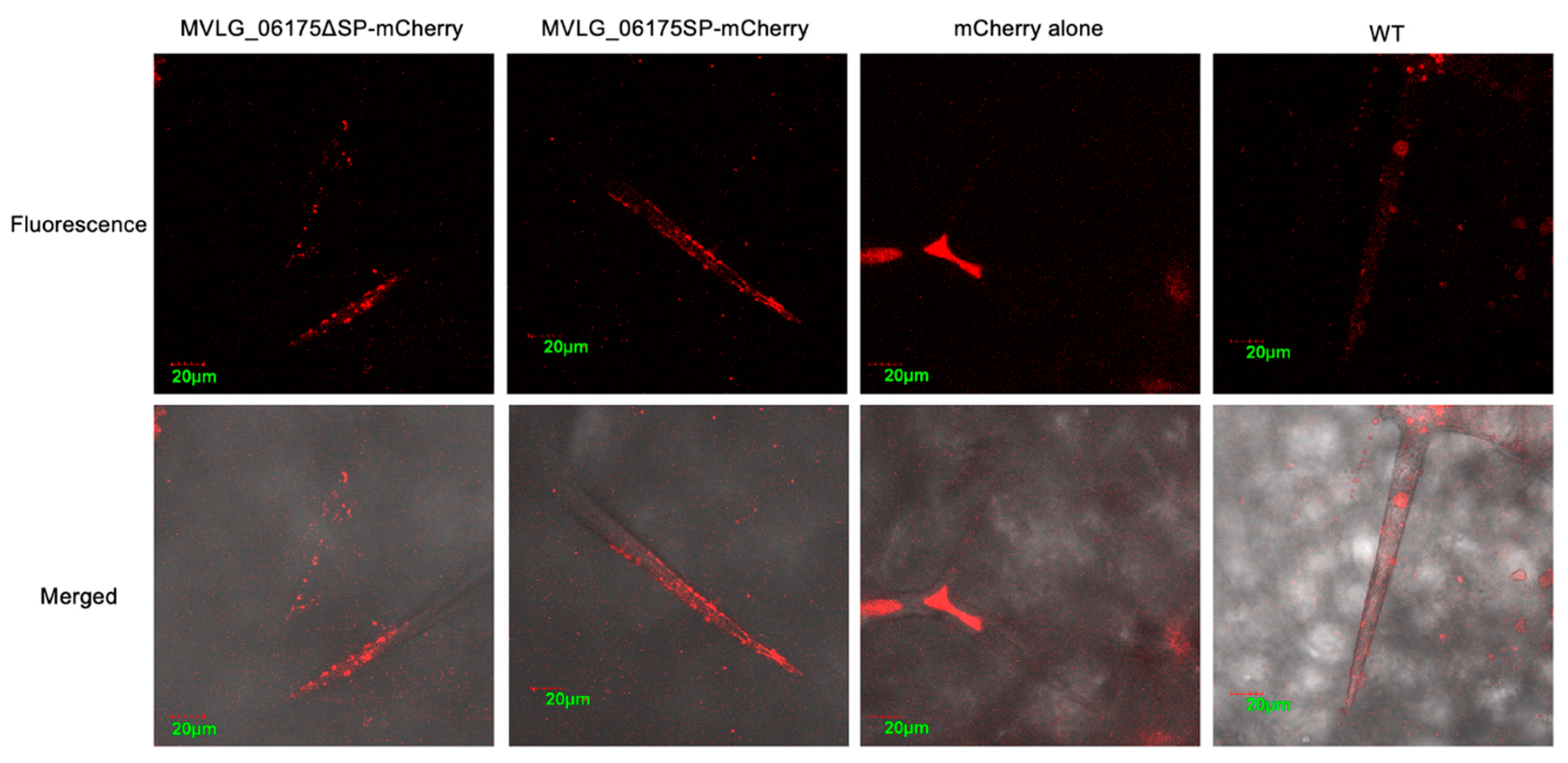
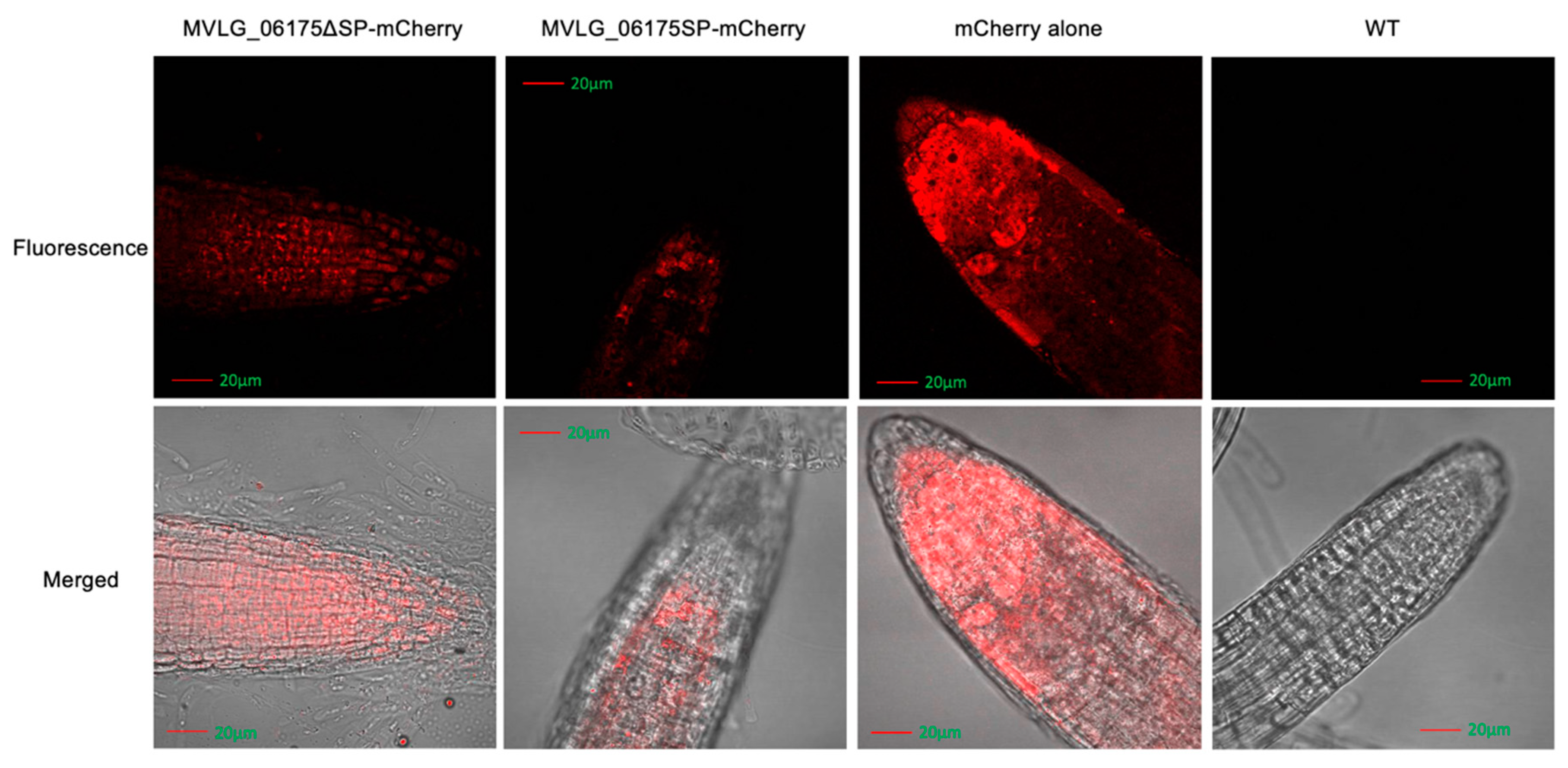
3.4.1. Localization of Effector Protein MVLG_06175 in Transgenic Plant Tissues
3.4.2. Localization of Effector Protein MVLG_05122 in Transgenic Plant Tissues
3.5. Determination of Phenotype Changes in Transgenic A. thaliana
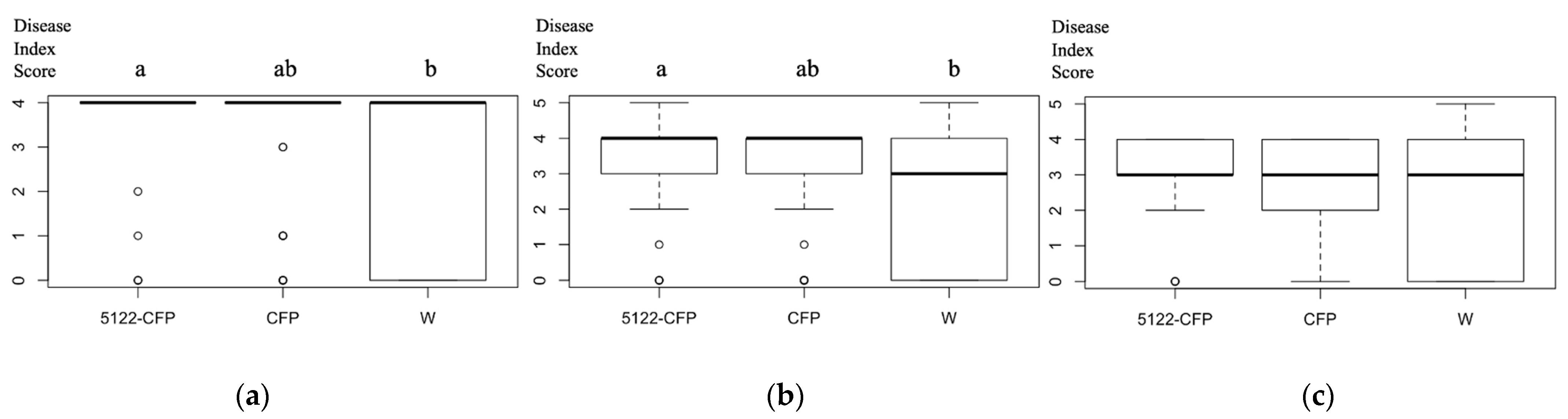
3.6. Fusarium Infection Assays on Transgenic A. thaliana
4. Discussion
4.1. MVLG_06175 Could Affect the Formation of the Casparian Strip and Growth of Trichome by Interacting with CASPL2C1
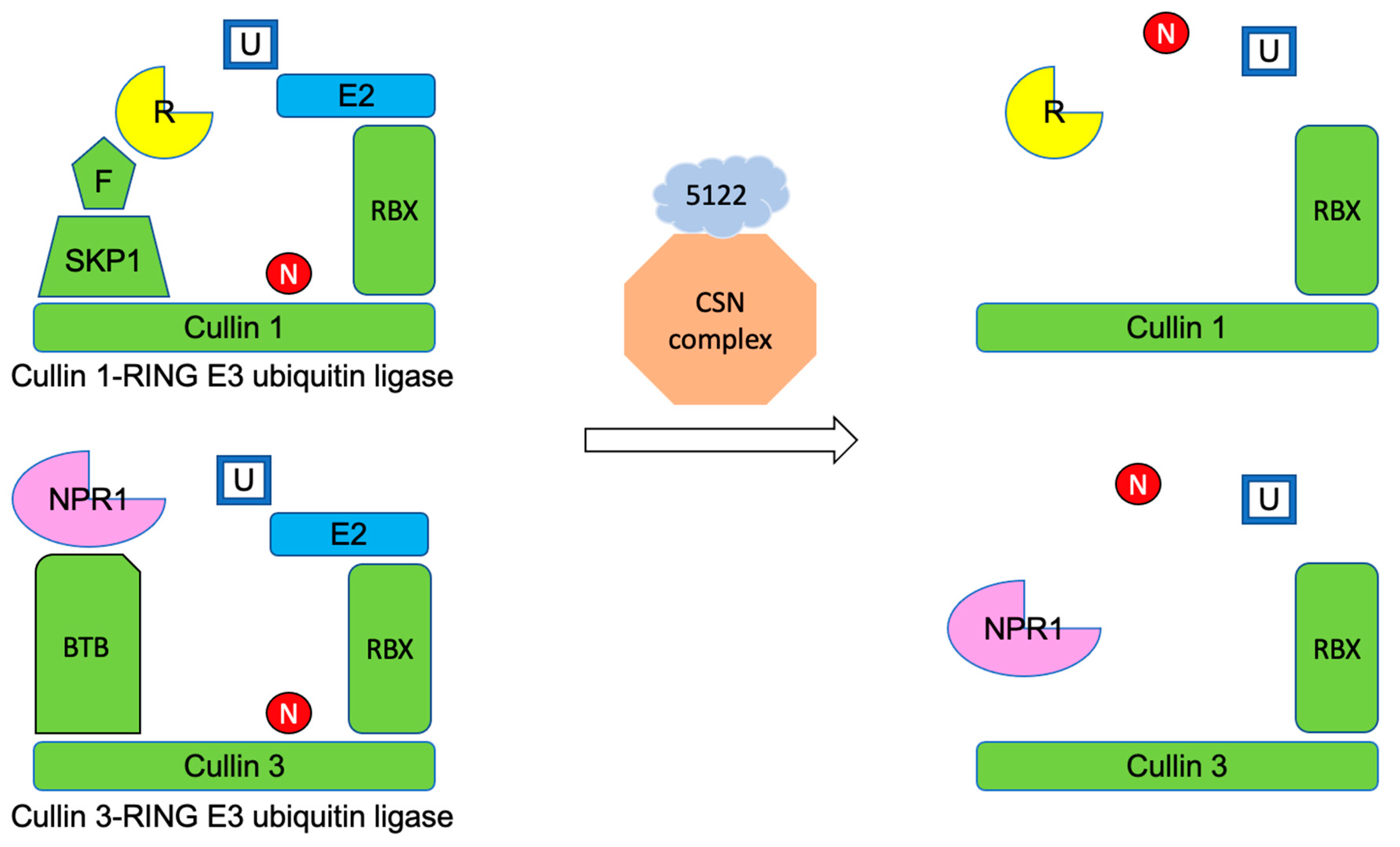
4.2. MVLG_05122 Might Alter the Protein Turnover Rate Associated with the Plant Defense and Trichome Growth by Interacting with the Plant Proteins CSN5a/5b
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kemler, M.; Goker, M.; Oberwinkler, F.; Begerow, D. Implications of Molecular Characters for the Phylogeny of the Microbotryaceae (Basidiomycota: Urediniomycetes). BMC Evol. Biol. 2006, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Giraud, T.; Yockteng, R.; Lopez-Villavicencio, M.; Refregier, G.; Hood, M.E. Mating System of the Anther Smut Fungus Microbotryum violaceum: Selfing under Heterothallism. Eukaryot. Cell 2008, 7, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Uchida, W.; Matsunaga, S.; Kawano, S. Ultrastructural Analysis of the Behavior of the Dimorphic Fungus Microbotryum violaceum in Fungus-Induced Anthers of Female Silene latifolia flowers. Protoplasma 2005, 226, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Win, J.; Chaparro-Garcia, A.; Belhaj, K.; Saunders, D.G.; Yoshida, K.; Dong, S.; Schornack, S.; Zipfel, C.; Robatzek, S.; Hogenhout, S.A.; et al. Effector Biology of Plant-Associated Organisms: Concepts and Perspectives. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Rouxel, T.; Grandaubert, J.; Hane, J.K.; Hoede, C.; van de Wouw, A.P.; Couloux, A.; Dominguez, V.; Anthouard, V.; Bally, P.; Bourras, S.; et al. Effector Diversification within Compartments of the Leptosphaeria maculans Genome Affected by Repeat-Induced Point Mutations. Nat. Commun. 2011, 2, 202. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Gardiner, D.M.; Dodds, P.N.; Tini, F.; Covarelli, L.; Singh, K.B.; Manners, J.M.; Taylor, J.M. EffectorP: Predicting Fungal Effector Proteins from Secretomes Using Machine Learning. New Phytol. 2016, 210, 743–761. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; de Wit, P.J. Fungal Effector Proteins. Annu. Rev. Phytopathol. 2009, 47, 233–263. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Fawke, S.; Doumane, M.; Schornack, S. Oomycete Interactions with Plants: Infection Strategies and Resistance Principles. Microbiol. Mol. Biol. Rev. 2015, 79, 263–280. [Google Scholar] [CrossRef]
- Kuppireddy, V.S.; Uversky, V.N.; Toh, S.S.; Tsai, M.; Beckerson, W.C.; Cahill, C.; Carman, B.; Perlin, M.H. Identification and Initial Characterization of the Effectors of an Anther Smut Fungus and Potential Host Target Proteins. Int. J. Mol. Sci. 2017, 18, 2489. [Google Scholar] [CrossRef]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014, 42, D26–D31. [Google Scholar] [CrossRef]
- Perlin, M.H.; Amselem, J.; Fontanillas, E.; Toh, S.S.; Chen, Z.; Goldberg, J.; Duplessis, S.; Henrissat, B.; Young, S.; Zeng, Q.; et al. Sex and parasites: Genomic and transcriptomic analysis of Microbotryum lychnidis-dioicae, the biotrophic and plant-castrating anther smut fungus. BMC Genom. 2015, 16, 461. [Google Scholar] [CrossRef]
- Toh, S.S.; Chen, Z.; Rouchka, E.C.; Schultz, D.J.; Cuomo, C.A.; Perlin, M.H. Pas de deux: An Intricate Dance of Anther Smut and Its Host. G3 (Bethesda) 2018, 8, 505–518. [Google Scholar] [CrossRef]
- Lee, S.; Kelly, B.S.; Damasceno, C.M.; St. John, B.; Kim, B.; Kim, B.D.; Rose, J.K. A Functional Screen to Characterize the Secretomes of Eukaryotic Pathogens and their Hosts in Planta. MPMI 2006, 19, 1368–1377. [Google Scholar] [CrossRef] [PubMed]
- Fields, S.; Song, O. A Novel Genetic System to Detect Protein-Protein Interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- McAlister-Henn, L.; Gibson, N.; Panisko, E. Applications of the Yeast Two-Hybrid System. Methods 1999, 19, 330–337. [Google Scholar] [CrossRef]
- Greener, M.; Perkel, J.M. The Yeast Two-Hybrid Assay. Scientist 2005, 19, 32. [Google Scholar]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Mattanovich, D.D.; Ruker, F.; Machado, A.D.; Laimer, M.; Regner, F. Efficient Transformation of Agrobacterium spp. by Electroporation. Nucleic Acids Res. 1989, 17, 6747. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-Mediated Transformation of Arabidopsis thaliana Using the Floral Dip Method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Diener, A.C.; Ausubel, F.M. RESISTANCE TO FUSARIUM OXYSPORUM 1, a Dominant Arabidopsis Disease-Resistance Gene, is not Race Specific. Genetics 2005, 171, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Marín, M.; Uversky, V.N.; Ott, T. Intrinsic disorder in pathogen effectors: Protein flexibility as an evolutionary hallmark in a molecular arms race. Plant Cell 2013, 25, 3153–3157. [Google Scholar] [CrossRef] [PubMed]
- Roppolo, D.; De Rybel, B.; Denervaud Tendon, V.; Pfister, A.; Alassimone, J.; Vermeer, J.E.; Yamazaki, M.; Stierhof, Y.; Beeckman, T.; Geldner, N. A Novel Protein Family Mediates Casparian Strip Formation in the Endodermis. Nature 2011, 473, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Beckerson, W.C.; Rodríguez de la Vega, R.C.; Hartmann, F.E.; Duhamel, M.; Giraud, T.; Perlin, M.H. Cause and Effectors: Whole-Genome Comparisons Reveal Shared but Rapidly Evolving Effector Sets among Host-Specific Plant-Castrating Fungi. mBio 2019, 10, e02391-19. [Google Scholar] [CrossRef]
- Naseer, S.; Lee, Y.; Lapierre, C.; Franke, R.; Nawrath, C.; Geldner, N. Casparian Strip Diffusion Barrier in Arabidopsis Is Made of Lignin Polymer without Suberin. Proc. Natl. Acad. Sci. USA 2012, 109, 10101–10106. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.E., 2nd; Trontin, C.; Duan, L.; Dinneny, J.R. Beyond the Barrier: Communication in the Root through the Endodermis. Plant Physiol. 2014, 166, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Steudle, E. Water Uptake by Plant Roots: An Integration of Views. Plant Soil 2000, 226, 45–56. [Google Scholar] [CrossRef]
- Karahara, I.; Takaya, E.; Fujibayashi, S.; Inoue, H.; Weller, J.L.; Reid, J.B.; Sugai, M. Development of the Casparian Strip is Delayed by Blue Light in Pea Stems. Planta 2011, 234, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lin, J.; Zhu, J.; Hu, Y.; Hartmann, K.; Schreiber, L. Casparian Strips in Needles of Pinus bungeana: Isolation and Chemical Characterization. Physiol. Plant 2003, 117, 421–424. [Google Scholar] [CrossRef]
- Romeo, D. Casparian Strips in the Leaf Intrastelar Canals of Isoetes duriei Bory, a Mediterranean Terrestrial Species. Ann. Bot. 2000, 86, 1051–1054. [Google Scholar] [CrossRef][Green Version]
- Machado, A.; Pereira, H.; Teixeira, R.T. Anatomy and Development of the Endodermis and Phellem of Quercus suber L. Roots. Microsc. Microanal. 2013, 19, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pulido, L.; Martin-Belmonte, F.; Valencia, A.; Alonso, M.A. MARVEL: A Conserved Domain Involved in Membrane Apposition Events. Trends Biochem. Sci. 2002, 27, 599–601. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.M.; Kemler, M.; Bauer, R.; Begerow, D. The Illustrated Life Cycle of Microbotryum on the Host Plant Silene latifolia. Botany 2010, 88, 875–885. [Google Scholar] [CrossRef]
- Roppolo, D.; Boeckmann, B.; Pfister, A.; Boutet, E.; Rubio, M.C.; Denervaud-Tendon, V.; Vermeer, J.E.; Gheyselinck, J.; Xenarios, I.; Geldner, N. Functional and Evolutionary Analysis of the CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN Family. Plant Physiol. 2014, 165, 1709–1722. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Jeon, H.S.; Kim, S.H.; Chung, J.H.; Roppolo, D.; Lee, H.J.; Cho, J.H.; Tobimatsu, Y.; Ralph, J.; Park, O.K. Lignin-Based Barrier Restricts Pathogens to the Infection Site and Confers Resistance in Plants. EMBO J. 2019, 38, e101948. [Google Scholar] [CrossRef] [PubMed]
- Karabourniotis, G.; Liakopoulos, G.; Nikolopoulos, D.; Bresta, P. Protective and Defensive Roles of Non-Glandular Trichomes against Multiple Stresses: Structure–Function Coordination. J. For. Res. 2019, 31, 1–12. [Google Scholar] [CrossRef]
- Wagner, G.J.; Wang, E.; Shepherd, R.W. New Approaches for Studying and Exploiting an Old Protuberance, the Plant Trichome. Ann. Bot. 2004, 93, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Gusmaroli, G.; Feng, S.; Deng, X.W. The Arabidopsis CSN5A and CSN5B Subunits are Present in Distinct COP9 Signalosome Complexes, and Mutations in their JAMM Domains Exhibit Differential Dominant Negative Effects on Development. Plant Cell 2004, 16, 2984–3001. [Google Scholar] [CrossRef] [PubMed]
- Dohmann, E.M.; Kuhnle, C.; Schwechheimer, C. Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 Signalosome Subunit 5 is Sufficient to Cause the cop/det/fus Mutant Phenotype in Arabidopsis. Plant Cell 2005, 17, 1967–1978. [Google Scholar] [CrossRef]
- Gusmaroli, G.; Figueroa, P.; Serino, G.; Deng, X.W. Role of the MPN Subunits in COP9 Signalosome Assembly and Activity, and their Regulatory Interaction with Arabidopsis Cullin3-Based E3 Ligases. Plant Cell 2007, 19, 564–581. [Google Scholar] [CrossRef]
- Choi, C.M.; Gray, W.M.; Mooney, S.; Hellmann, H. Composition, Roles, and Regulation of Cullin-Based Ubiquitin E3 Ligases. Arab. Book 2014, 12, e0175. [Google Scholar] [CrossRef] [PubMed]
- Wei, N.; Serino, G.; Deng, X.W. The COP9 Signalosome: More than a Protease. Trends Biochem. Sci. 2008, 33, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, M.; Shirasu, K. Ubiquitination in Plant immunity. Curr. Opin. Plant Biol. 2010, 13, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Seo, K.-I.; Song, E.; Chung, S.; Lee, J.-H. Roles of Various Cullin-RING E3 Ligases Involved in Hormonal and Stress Responses in Plants. J. Plant Biol. 2013, 55, 421–428. [Google Scholar] [CrossRef]
- Vierstra, R.D. The Ubiquitin-26S Proteasome System at the Nexus of Plant Biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.I.; Armstrong, M.R.; Gilroy, E.M.; Boevink, P.C.; Hein, I.; Taylor, R.M.; Zhendong, T.; Engelhardt, S.; Vetukuri, R.R.; Harrower, B.; et al. Phytophthora Infestans Effector AVR3a is Essential for Virulence and Manipulates Plant Immunity by Stabilizing Host E3 Ligase CMPG1. Proc. Natl. Acad. Sci. USA 2010, 107, 9909–9914. [Google Scholar] [CrossRef] [PubMed]
- Rosebrock, T.R.; Zeng, L.; Brady, J.J.; Abramovitch, R.B.; Xiao, F.; Martin, G.B. A Bacterial E3 Ubiquitin Ligase Targets a Host Protein Kinase to Disrupt Plant Immunity. Nature 2007, 448, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Schellenberg, B.; Bachmann, A.S.; Archer, C.R.; Huber, R.; Powell, T.K.; Lindow, S.; Kaiser, M.; Dudler, R. A Plant Pathogen Virulence Factor Inhibits the Eukaryotic Proteasome by a Novel Mechanism. Nature 2008, 452, 755–758. [Google Scholar] [CrossRef]
- Misas-Villamil, J.C.; Kolodziejek, I.; Crabill, E.; Kaschani, F.; Niessen, S.; Shindo, T.; Kaiser, M.; Alfano, J.R.; van der Hoorn, R.A. Pseudomonas syringae pv. syringae Uses Proteasome Inhibitor Syringolin A to Colonize from Wound Infection Sites. PLoS Pathog. 2013, 9, e1003281. [Google Scholar] [CrossRef]
- Yan, S.; Dong, X. Perception of the Plant Immune Signal Salicylic Acid. Curr. Opin. Plant Biol. 2014, 20, 64–68. [Google Scholar] [CrossRef]
- Furniss, J.J.; Spoel, S.H. Cullin-RING Ubiquitin Ligases in Salicylic Acid-Mediated Plant Immune Signaling. Front. Plant Sci. 2015, 6, 154. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Calabria, J.; Chen, H.W.; Somssich, M. The Arabidopsis thaliana-Fusarium oxysporum strain 5176 pathosystem: An overview. J. Exp. Bot. 2022, 73, 6052–6067. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Amornsiripanitch, N.; Dong, X. A Genomic Approach to Identify Regulatory Nodes in the Transcriptional Network of Systemic Acquired Resistance in Plants. PLoS Pathog. 2006, 2, e123. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Li, X.; Gruber, M.Y.; Feyissa, B.A.; Amyot, L.; Hannoufa, A. COP9 Signalosome Subunit 5A Affects Phenylpropanoid Metabolism, Trichome Formation and Transcription of Key Genes of a Regulatory Tri-Protein Complex in Arabidopsis. BMC Plant Biol. 2018, 18, 134. [Google Scholar] [CrossRef] [PubMed]
- Traw, M.B.; Bergelson, J. Interactive Effects of Jasmonic Acid, Salicylic Acid, and Gibberellin on Induction of Trichomes in Arabidopsis. Plant Physiol. 2003, 133, 1367–1375. [Google Scholar] [CrossRef]
- Hind, S.R.; Pulliam, S.E.; Veronese, P.; Shantharaj, D.; Nazir, A.; Jacobs, N.S.; Stratmann, J.W. The COP9 Signalosome Controls Jasmonic Acid Synthesis and Plant Responses to Herbivory and Pathogens. Plant J. 2011, 65, 480–491. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, M.-C.; Barati, M.T.; Kuppireddy, V.S.; Beckerson, W.C.; Long, G.; Perlin, M.H. Characterization of Microbotryum lychnidis-dioicae Secreted Effector Proteins, Their Potential Host Targets, and Localization in a Heterologous Host Plant. J. Fungi 2024, 10, 262. https://doi.org/10.3390/jof10040262
Tsai M-C, Barati MT, Kuppireddy VS, Beckerson WC, Long G, Perlin MH. Characterization of Microbotryum lychnidis-dioicae Secreted Effector Proteins, Their Potential Host Targets, and Localization in a Heterologous Host Plant. Journal of Fungi. 2024; 10(4):262. https://doi.org/10.3390/jof10040262
Chicago/Turabian StyleTsai, Ming-Chang, Michelle T. Barati, Venkata S. Kuppireddy, William C. Beckerson, Grace Long, and Michael H. Perlin. 2024. "Characterization of Microbotryum lychnidis-dioicae Secreted Effector Proteins, Their Potential Host Targets, and Localization in a Heterologous Host Plant" Journal of Fungi 10, no. 4: 262. https://doi.org/10.3390/jof10040262
APA StyleTsai, M.-C., Barati, M. T., Kuppireddy, V. S., Beckerson, W. C., Long, G., & Perlin, M. H. (2024). Characterization of Microbotryum lychnidis-dioicae Secreted Effector Proteins, Their Potential Host Targets, and Localization in a Heterologous Host Plant. Journal of Fungi, 10(4), 262. https://doi.org/10.3390/jof10040262






