The Autophagy-Related Protein ATG8 Orchestrates Asexual Development and AFB1 Biosynthesis in Aspergillus flavus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Bioinformatic Identification and Analysis of ATG8
2.3. Strain Construction
2.4. Phenotype Analysis
2.5. Aflatoxin B1 Extraction and Determination
2.6. Stress Analysis
2.7. ROS Measurement
2.8. Seed Infections
2.9. Real-Time Quantitative Reverse Transcription PCR
2.10. Statistical Analysis
3. Results
3.1. Identification of ATG8 Protein in A. flavus and Prediction of the Tertiary Structure of ATG8 Protein
3.2. Deletion and Complementation of atg8 in A. flavus
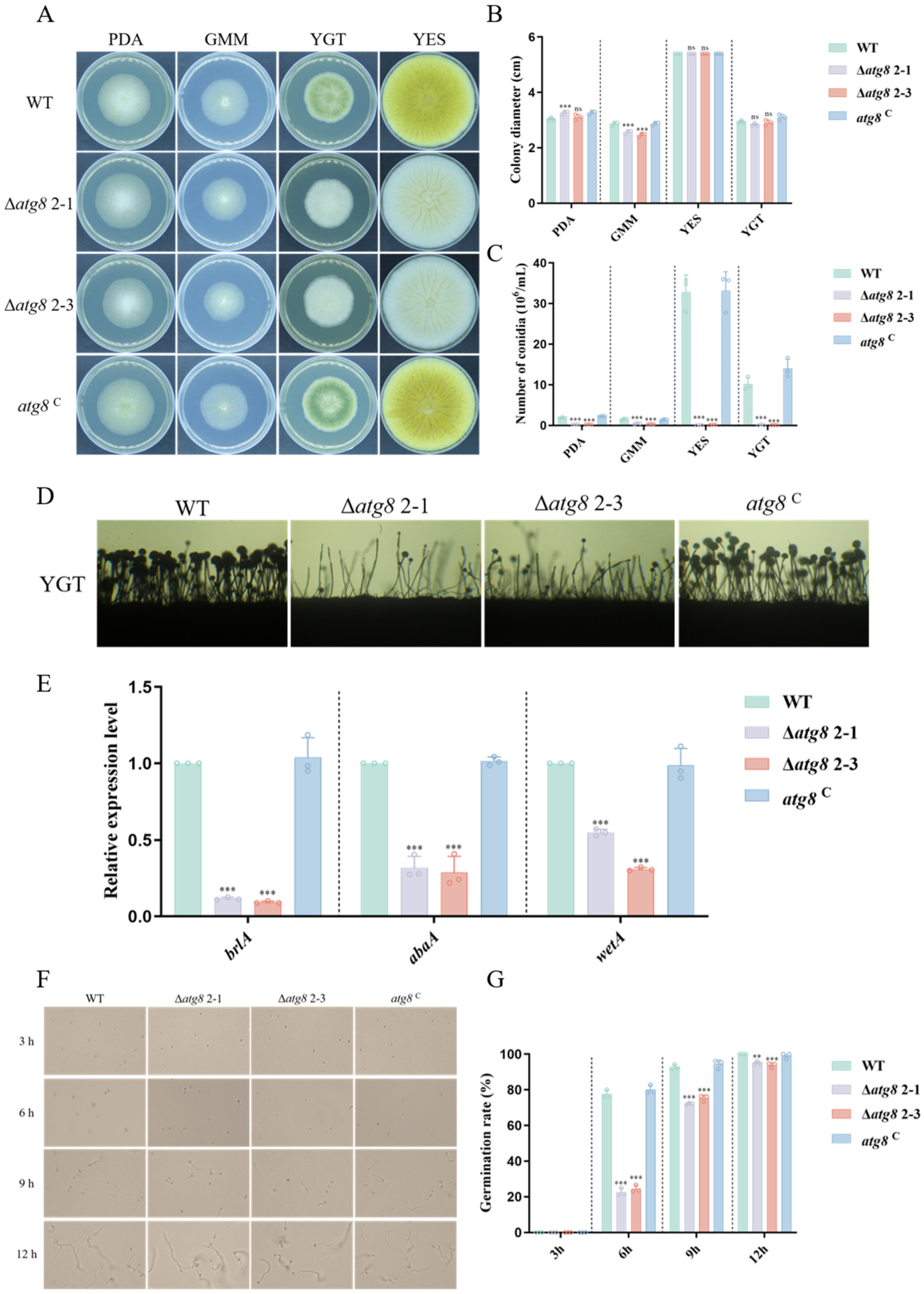
3.3. ATG8 Is Crucial for Hyphal Growth and Conidiation in A. flavus
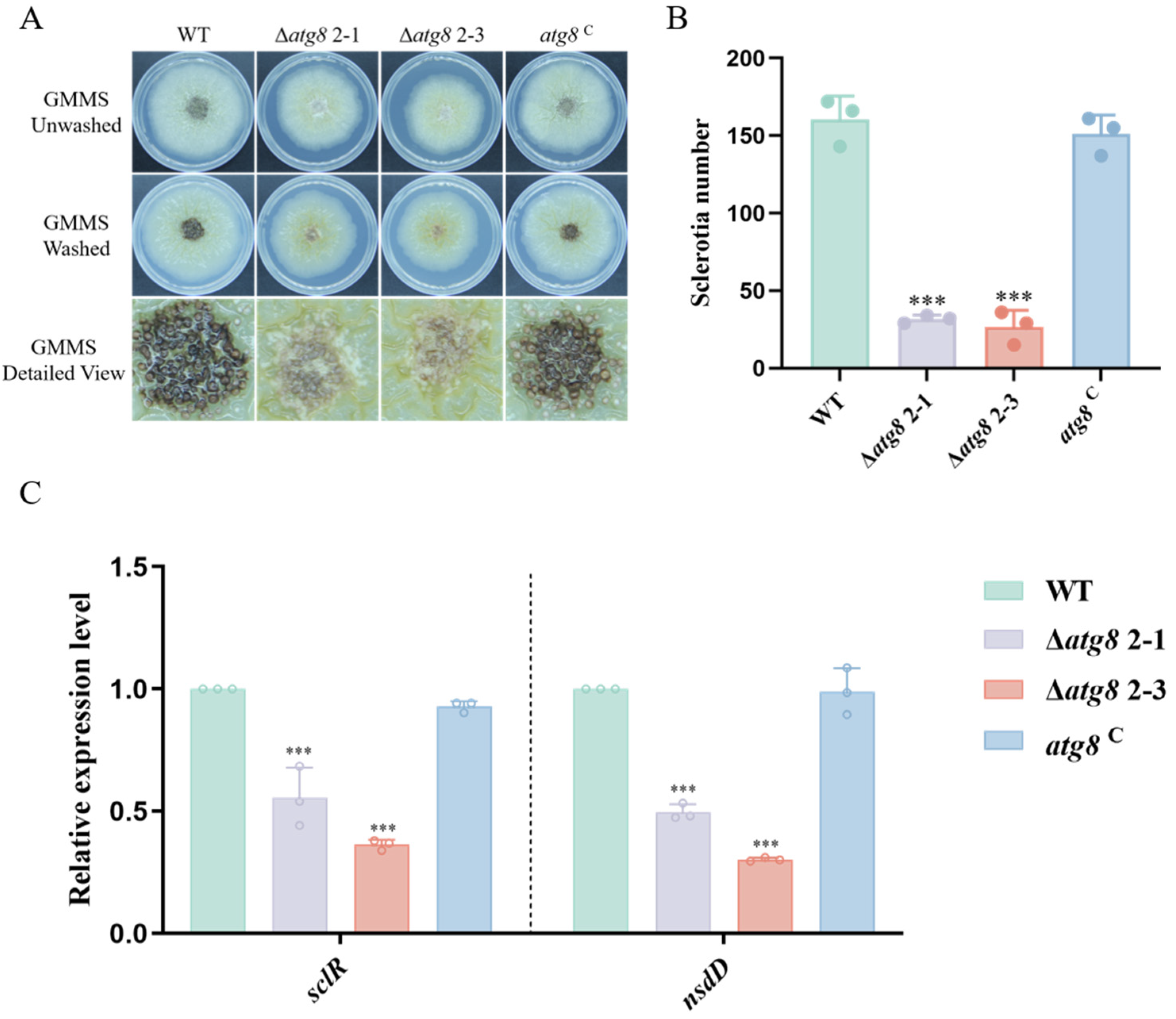
3.4. ATG8 Contributes to Sclerotia Formation
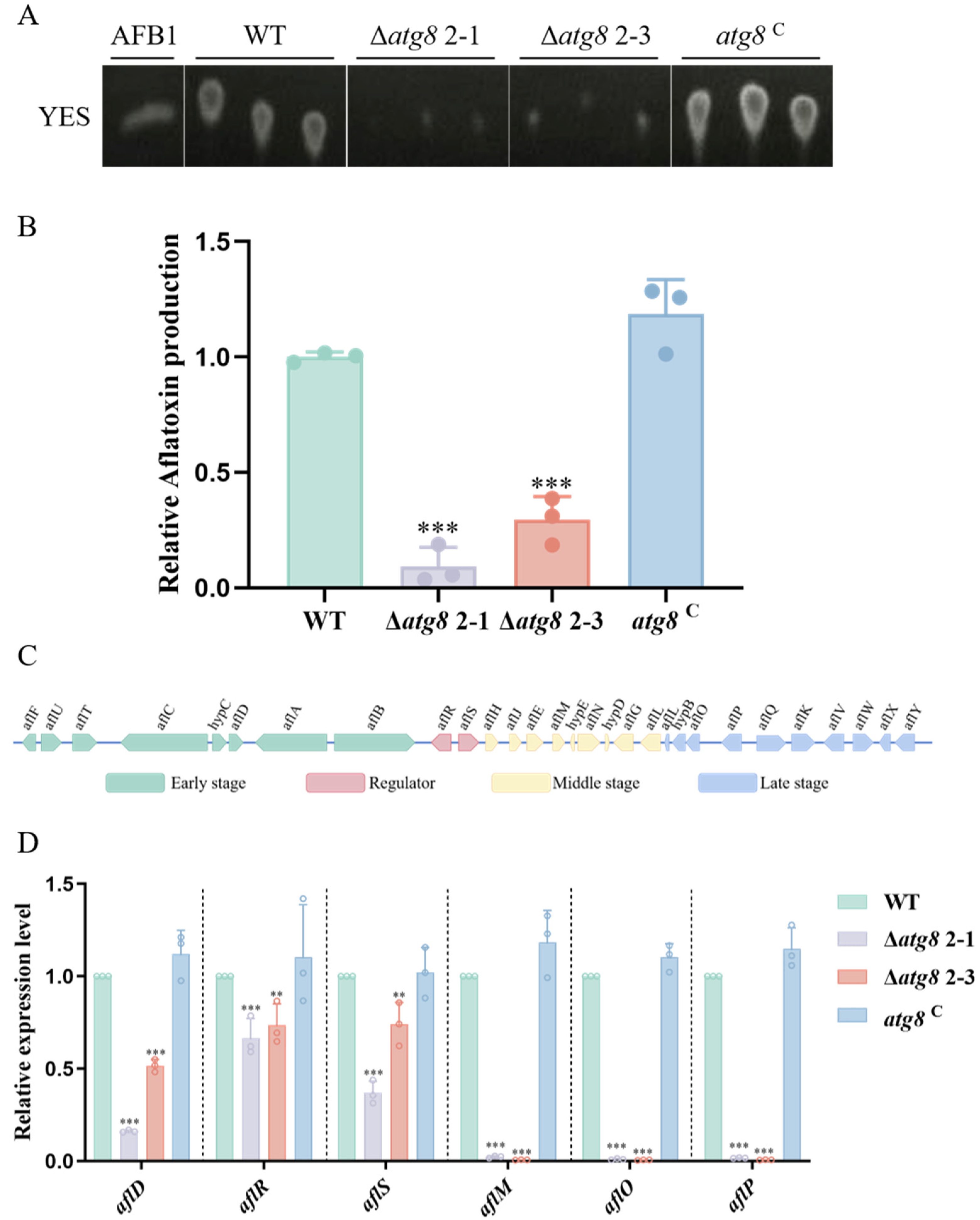
3.5. ATG8 Regulates Aflatoxin Biosynthesis in A. flavus
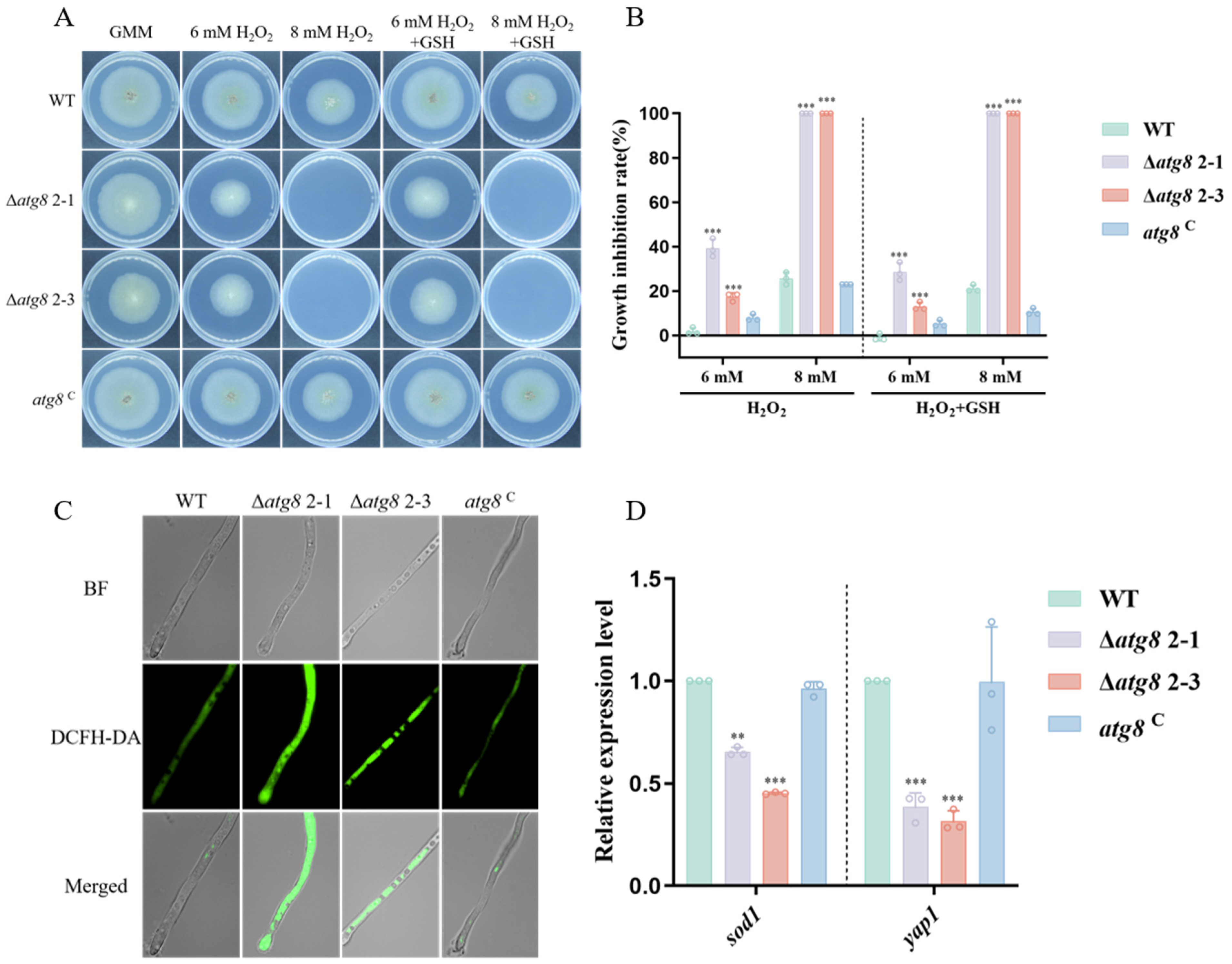
3.6. Response of ATG8 to Multiple Stresses in A. flavus
3.7. Effect of ATG8 Mutants on Pathogenicity to Crop Seeds
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amaike, S.; Keller, N.P. Aspergillus flavus. Annu. Rev. Phytopathol. 2011, 49, 107–133. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Tian, J.; Keller, N.P. Post-translational modifications drive secondary metabolite biosynthesis in Aspergillus: A review. Environ. Microbiol. 2022, 24, 2857–2881. [Google Scholar] [CrossRef]
- Hedayati, M.T.; Pasqualotto, A.C.; Warn, P.A.; Bowyer, P.; Denning, D.W. Aspergillus flavus: Human pathogen, allergen and mycotoxin producer. Microbiology 2007, 153, 1677–1692. [Google Scholar] [CrossRef]
- Ding, C.; Tian, T.; Liu, Q.; Zhao, S.; Tao, T.; Wu, H.; Guo, L. Transcriptomics and metabolomic profiling identify molecular mechanism for Aspergillus flavus infection in grain. Food Front. 2023, 4, 1845–1858. [Google Scholar] [CrossRef]
- Lohmar, J.M.; Puel, O.; Cary, J.W.; Calvo, A.M. The Aspergillus flavus rtfA gene regulates plant and animal pathogenesis and secondary metabolism. Appl. Environ. Microbiol. 2019, 85, e02446-18. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Gu, M.; Yan, H.; Zhang, M.; Xie, H.; Yue, X.; Zhang, Q.; Li, P. Protein biomarker for early diagnosis of microbial toxin contamination: Using Aspergillus flavus as an example. Food Front. 2023, 4, 2013–2023. [Google Scholar] [CrossRef]
- Yang, K.; Geng, Q.; Luo, Y.; Xie, R.; Sun, T.; Wang, Z.; Qin, L.; Zhao, W.; Liu, M.; Li, Y.; et al. Dysfunction of FadA-cAMP signalling decreases Aspergillus flavus resistance to antimicrobial natural preservative Perillaldehyde and AFB1 biosynthesis. Environ. Microbiol. 2022, 24, 1590–1607. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Zhang, Y.; Ti, Y.; Qiao, H.; Niu, C.; Ban, Y.; Wang, X.; Hou, Y.; Lu, R.; Song, Z. A magnetic nanocomposite combined with cinnamic acid for capture–inhibition–separation of Aspergillus fumigatus. Food Front. 2023, 4, 867–882. [Google Scholar] [CrossRef]
- Shankar, J.; Thakur, R.; Clemons, K.V.; Stevens, D.A. Interplay of cytokines and chemokines in aspergillosis. J. Fungi 2024, 10, 251. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Singh, R. The emerging epidemiology of mould infections in developing countries. Curr. Opin. Infect. Dis. 2011, 24, 521–526. [Google Scholar] [CrossRef]
- Krishnan, S.; Manavathu, E.K.; Chandrasekar, P.H. Aspergillus flavus: An emerging non-fumigatus Aspergillus species of significance. Mycoses 2009, 52, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Mellon, J.E.; Cotty, P.J.; Dowd, M.K. Aspergillus flavus hydrolases: Their roles in pathogenesis and substrate utilization. Appl. Microbiol. Biotechnol. 2007, 77, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Luo, Y.; Sun, T.; Qiu, H.; Geng, Q.; Li, Y.; Liu, M.; Keller, N.P.; Song, F.; Tian, J. Nitric oxide-mediated regulation of Aspergillus flavus asexual development by targeting TCA cycle and mitochondrial function. J. Hazard. Mater. 2024, 471, 134385. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, C.; Müller, G.; Mourabit, S.; Carnal, S.; Bandara, K. Occurrence of fungi and fungal toxins in fish feed during storage. Toxins 2020, 12, 171. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Li, Z.Y.; Wang, Y.D.; Wang, J.Q.; Yang, P.L. Quercetin inhibits the proliferation and aflatoxins biosynthesis of Aspergillus flavus. Toxins 2019, 11, 154. [Google Scholar] [CrossRef]
- Sun, D.; Mao, J.; Wang, Z.; Li, H.; Zhang, L.; Zhang, W.; Zhang, Q.; Li, P. Inhibition of Aspergillus flavus growth and aflatoxins production on peanuts over α-Fe2O3 nanorods under sunlight irradiation. Int. J. Food Microbiol. 2021, 353, 109296. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, N.; Luo, Y.; Qiu, H.; Geng, Q.; Li, Y.; Peng, X.; Song, F.; Tian, J.; Yang, K. p-Anisaldehyde suppresses Aspergillus flavus infection and aflatoxin biosynthesis via targeting cell structure and Ap1-regulatory antioxidant system. Food Front. 2024, 5, 1302–1319. [Google Scholar] [CrossRef]
- Ren, X.; Branà, M.T.; Haidukowski, M.; Gallo, A.; Zhang, Q.; Logrieco, A.F.; Li, P.; Zhao, S.; Altomare, C. Potential of Trichoderma spp. for biocontrol of aflatoxin-producing Aspergillus flavus. Toxins 2022, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Klionsky, D.J. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 2009, 335, 1–32. [Google Scholar]
- Xu, D.D.; Du, L.L. Fission yeast autophagy machinery. Cells 2022, 11, 1086. [Google Scholar] [CrossRef]
- Wang, J.; Xu, C.; Sun, Q.; Xu, J.; Chai, Y.; Berg, G.; Cernava, T.; Ma, Z.; Chen, Y. Post-translational regulation of autophagy is involved in intra-microbiome suppression of fungal pathogens. Microbiome 2021, 9, 131. [Google Scholar] [CrossRef]
- Ivory, B.J.; Smith, H.M.; Cabrera, E.; Robinson, M.R.; Sparks, J.T.; Solem, A.; Ishihara, J.I.; Takahashi, H.; Tsuji, M.; Segarra, V.A. ATG8 is conserved between Saccharomyces cerevisiae and psychrophilic, polar-collected fungi. MicroPubl Biol. 2021, 2021. [Google Scholar] [CrossRef]
- Schaaf, M.B.; Keulers, T.G.; Vooijs, M.A.; Rouschop, K.M. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 2016, 30, 3961–3978. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Wang, Y.; Xu, J.; Yang, Z.; Zhang, Z.; Qian, J.; Hu, J.; Yin, Z.; Yang, L.; Liu, M.; et al. Autophagy and cell wall integrity pathways coordinately regulate the development and pathogenicity through MoAtg4 phosphorylation in Magnaporthe oryzae. PLoS Pathog. 2024, 20, e1011988. [Google Scholar] [CrossRef]
- Xu, F.; Liu, X.H.; Zhuang, F.L.; Zhu, J.; Lin, F.C. Analyzing autophagy in Magnaporthe oryzae. Autophagy 2011, 7, 525–530. [Google Scholar] [CrossRef]
- Deng, Y.Z.; Ramos-Pamplona, M.; Naqvi, N.I. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy 2009, 5, 33–43. [Google Scholar] [CrossRef]
- Josefsen, L.; Droce, A.; Sondergaard, T.E.; Sørensen, J.L.; Bormann, J.; Schäfer, W.; Giese, H.; Olsson, S. Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium graminearum. Autophagy 2012, 8, 326–337. [Google Scholar] [CrossRef]
- Khalid, A.R.; Lv, X.; Naeem, M.; Mehmood, K.; Shaheen, H.; Dong, P.; Qiu, D.; Ren, M. Autophagy related gene (ATG3) is a key regulator for cell growth, development, and virulence of Fusarium oxysporum. Genes 2019, 10, 658. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, S.; Ju, Z.; Zhang, C.; Tang, G.; Wang, J.; Wen, Z.; Chen, W.; Ma, Z. Contribution of peroxisomal docking machinery to mycotoxin biosynthesis, pathogenicity and pexophagy in the plant pathogenic fungus Fusarium graminearum. Environ. Microbiol. 2018, 20, 3224–3245. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Wang, C.; Yang, N.; Que, Y.; Talbot, N.J.; Wang, Z. Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci. Rep. 2017, 7, 11062. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Shadkchan, Y.; Tannous, J.; Landero Figueroa, J.A.; Wiemann, P.; Osherov, N.; Wang, S.; Keller, N.P. Contribution of ATPase copper transporters in animal but not plant virulence of the crossover pathogen Aspergillus flavus. Virulence 2018, 9, 1273–1286. [Google Scholar] [CrossRef]
- Yang, K.; Liang, L.; Ran, F.; Liu, Y.; Li, Z.; Lan, H.; Gao, P.; Zhuang, Z.; Zhang, F.; Nie, X.; et al. The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 2016, 6, 23259. [Google Scholar] [CrossRef]
- Luis, J.M.; Carbone, I.; Payne, G.A.; Bhatnagar, D.; Cary, J.W.; Moore, G.G.; Lebar, M.D.; Wei, Q.; Mack, B.; Ojiambo, P.S. Characterization of morphological changes within stromata during sexual reproduction in Aspergillus flavus. Mycologia 2020, 112, 908–920. [Google Scholar] [CrossRef]
- Shabeer, S.; Asad, S.; Jamal, A.; Ali, A. Aflatoxin contamination, its impact and management strategies: An updated review. Toxins 2022, 14, 307. [Google Scholar] [CrossRef]
- Fasoyin, O.E.; Yang, K.; Qiu, M.; Wang, B.; Wang, S.; Wang, S. Regulation of morphology, aflatoxin production, and virulence of Aspergillus flavus by the major nitrogen regulatory gene areA. Toxins 2019, 11, 718. [Google Scholar] [CrossRef]
- Wu, P.C.; Choo, C.Y.L.; Lu, H.Y.; Wei, X.Y.; Chen, Y.K.; Yago, J.I.; Chung, K.R. Pexophagy is critical for fungal development, stress response, and virulence in Alternaria alternata. Mol. Plant Pathol. 2022, 23, 1538–1554. [Google Scholar] [CrossRef]
- Bhatnagar, D.; Cary, J.W.; Ehrlich, K.; Yu, J.; Cleveland, T.E. Understanding the genetics of regulation of aflatoxin production and Aspergillus flavus development. Mycopathologia 2006, 162, 155–166. [Google Scholar] [CrossRef]
- Jayaprakash, A.; Roy, A.; Thanmalagan, R.R.; Arunachalam, A.; Ptv, L. Immune response gene coexpression network analysis of Arachis hypogaea infected with Aspergillus flavus. Genomics 2021, 113, 2977–2988. [Google Scholar] [CrossRef]
- Yin, Z.; Chen, C.; Yang, J.; Feng, W.; Liu, X.; Zuo, R.; Wang, J.; Yang, L.; Zhong, K.; Gao, C.; et al. Histone acetyltransferase MoHat1 acetylates autophagy-related proteins MoAtg3 and MoAtg9 to orchestrate functional appressorium formation and pathogenicity in Magnaporthe oryzae. Autophagy 2019, 15, 1234–1257. [Google Scholar] [CrossRef]
- Zhu, X.M.; Li, L.; Wu, M.; Liang, S.; Shi, H.B.; Liu, X.H.; Lin, F.C. Current opinions on autophagy in pathogenicity of fungi. Virulence 2019, 10, 481–489. [Google Scholar] [CrossRef]
- Pollack, J.K.; Harris, S.D.; Marten, M.R. Autophagy in filamentous fungi. Fungal Genet. Biol. 2009, 46, 1–8. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Xu, Y.; Gu, Y.; Zhang, X.; Zhang, M.; Wen, W.; Lee, Y.W.; Shi, J.; Mohamed, S.R.; et al. The autophagy-related proteins FvAtg4 and FvAtg8 are involved in virulence and fumonisin biosynthesis in Fusarium verticillioides. Virulence 2022, 13, 764–780. [Google Scholar] [CrossRef]
- Asif, N.; Lin, F.; Li, L.; Zhu, X.; Nawaz, S. Regulation of autophagy machinery in Magnaporthe oryzae. Int. J. Mol. Sci. 2022, 23, 8366. [Google Scholar] [CrossRef]
- Liu, X.H.; Gao, H.M.; Xu, F.; Lu, J.P.; Devenish, R.J.; Lin, F.C. Autophagy vitalizes the pathogenicity of pathogenic fungi. Autophagy 2012, 8, 1415–1425. [Google Scholar] [CrossRef]
- Chai, X.; Liu, Y.; Ma, H.; Wang, S.; Niyitanga, E.; He, C. Effects of macroautophagy and mitophagy on the pathogenicity of Fusarium graminearum. Phytopathology 2022, 112, 1928–1935. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, Y.; Li, S.; Li, H. Histone acetyltransferase CfGcn5-mediated autophagy governs the pathogenicity of Colletotrichum fructicola. mBio 2022, 13, e0195622. [Google Scholar] [CrossRef]
- Pickova, D.; Ostry, V.; Toman, J.; Malir, F. Aflatoxins: History, significant milestones, recent data on their toxicity and ways to mitigation. Toxins 2021, 13, 399. [Google Scholar] [CrossRef]
- Liang, L.; Liu, Y.; Yang, K.; Lin, G.; Xu, Z.; Lan, H.; Wang, X.; Wang, S. The putative histone methyltransferase DOT1 regulates aflatoxin and pathogenicity attributes in Aspergillus flavus. Toxins 2017, 9, 232. [Google Scholar] [CrossRef]
- Huang, Z.; Wu, D.; Yang, S.; Fu, W.; Ma, D.; Yao, Y.; Lin, H.; Yuan, J.; Yang, Y.; Zhuang, Z. Regulation of fungal morphogenesis and pathogenicity of Aspergillus flavus by hexokinase AfHxk1 through its domain hexokinase_2. J. Fungi 2023, 9, 1077. [Google Scholar] [CrossRef]



| Name of Strain | Genotype | Background | Source |
|---|---|---|---|
| NRRL 3357 | A. flavus Wild-type | NRRL 3357 | [31] |
| TJES20.1 | pyrG1, Δku70, ΔargB::AfpyrG | NRRL 3357 | [31] |
| TXZ 21.3 | pyrG1, Δku70, ΔargB | NRRL 3357 | [31] |
| Δatg8 | pyrG1, Δku70, ΔargB::AfpyrG, Δatg8::argB | NRRL 3357 | This study |
| Δatg8ΔpyrG | pyrG1, Δku70, Δatg8::argB | NRRL 3357 | This study |
| atg8C | pyrG1, Δku70, Δatg8::argB, atg8::AfpyrG | NRRL 3357 | This study |
| Primers | Sequence (5′-3′) | Application |
|---|---|---|
| atg8/P1 | TGAGAAGTGGCAGAGTGAC | atg8 deletion |
| atg8/P2 | TGTCCCAGTTCCTGCTTAGT | |
| atg8/P3 | TTCTACCGAACTCATCACCACCGGGAAACAGTAGGAAGAAGGTGGAT | |
| atg8/P4 | TGGTGCCCGCATTCACATGTCACGGGCTTCGCCGTCAGTATGTGTT | |
| atg8/P5 | AGTCAGTCATCGCCTGTTTT | |
| atg8/P6 | GCGGTTCCTGGTGGGTTAT | |
| argB/F | TCCCGGTGGTGATGAGTTC | A. flavus argB |
| argB/R | CCCGTGACATGTGAATGCG | |
| pyrG/F | GCCTCAAACAATGCTCTTCACCC | A. fumigatus pyrG |
| pyrG/R | GTCTGAGAGGAGGCACTGATGC | |
| atg8-orf/F | CAACTCTATCTGATCCGTAC | atg8 mutant screen |
| atg8-orf/R | GAGACTATGTCAATATGTGCC | |
| argB-170/R | TGTCCAGTTCGGGTTAGCG | |
| argB-1205/F | ACGGTGTCTCAAAGCCAGG | |
| pyrG-801/R | CAGGAGTTCTCGGGTTGTCG | |
| pyrG-351/F | CAGAGTATGCGGCAAGTCA | |
| c-atg8/p1 | GTCTGCGCTGAGAAGTGG | atg8 complementation |
| c-atg8/P2 | GAAATAGAGGTAGCCTAATCG | |
| c-atg8/P3 | GGGTGAAGAGCATTGTTTGAGGCTTAAAGATCGCCGAAGGTG | |
| c-atg8/P4 | GCATCATGCCTCCTCTCAGACTCCCGGTGGTGATGAGTTC | |
| c-atg8/P5 | GACCCAAACTGTCAGAGC | |
| c-atg8/P6 | ACCCAGGCAATCTTGAGGC |
| Primers | Sequence (5′-3′) | Application |
|---|---|---|
| atg8/QF | GACATCGCCACTATTGATAAG | atg8 RT-qPCR |
| atg8/QR | GTTCTTCGTAGATGCTGCTC | |
| abaA/QF | ACGGAAATCGCCAAAGACA | abaA RT-qPCR |
| abaA/QR | CCGGAATTGCCAAAGTAGG | |
| brlA/QF | CTCCAGCGTCAACCTTCA | brlA RT-qPCR |
| brlA/QR | TCAAATGCTCTTGCCTCTTA | |
| wetA/QF | GGCGTCTAGTTGTCAGGAG | wetA RT-qPCR |
| wetA/QR | ACATTCATTGAGTTGGAGGA | |
| sclR/QF | CAATGAGCCTATGGGAGTGG | sclR RT-qPCR |
| sclR/QR | ATCTTCGCCCGAGTGGTT | |
| nsdD/QF | GGACTTGCGGGTCGTGCTA | nsdD RT-qPCR |
| nsdD/QR | AGAACGCTGGGTCTGGTGC | |
| aflD/QF | GCTCCCGTCCTACTGTTTC | aflD RT-qPCR |
| aflD/QR | CATGTTGGTGATGGTGCTG | |
| aflR/QF | AAAGCACCCTGTCTTCCCTAAC | aflR RT-qPCR |
| aflR/QR | GAAGAGGTGGGTCAGTGTTTGTAG | |
| aflS/QF | CGAGTCGCTCAGGCGCTCAA | aflS RT-qPCR |
| aflS/QR | GCTCAGACTGACCGCCGCTC | |
| aflM/QF | CCCCAGAAGAATTTGACCG | aflM RT-qPCR |
| aflM/QR | ACGCAAGCAGTGTTAGAGC | |
| aflO/QF | GATTGGGATGTGGTCATGCGATT | aflO RT-qPCR |
| aflO/QR | GCCTGGGTCCGAAGAATGC | |
| aflP/QF | ACGAAGCCACTGGTAGAGGAGATG | aflP RT-qPCR |
| aflP/QR | GTGAATGACGGCAGGCAGGT | |
| sod1/QF | ATGGTCAAGGCTGGTAGG | sod1 RT-qPCR |
| sod1/QR | CAGTGATAGGCTGGGAGG | |
| yap1/QF | CTTCTTCTTGCCGCTCTT | yap1 RT-qPCR |
| yap1/QR | TCCGTAACCCAATCCACC | |
| actin/QF | ACGGTGTCGTCACAAACTGG | actin RT-qPCR |
| actin/QR | CGGTTGGACTTAGGGTTGATAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, Q.; Hu, J.; Xu, P.; Sun, T.; Qiu, H.; Wang, S.; Song, F.; Shen, L.; Li, Y.; Liu, M.; et al. The Autophagy-Related Protein ATG8 Orchestrates Asexual Development and AFB1 Biosynthesis in Aspergillus flavus. J. Fungi 2024, 10, 349. https://doi.org/10.3390/jof10050349
Geng Q, Hu J, Xu P, Sun T, Qiu H, Wang S, Song F, Shen L, Li Y, Liu M, et al. The Autophagy-Related Protein ATG8 Orchestrates Asexual Development and AFB1 Biosynthesis in Aspergillus flavus. Journal of Fungi. 2024; 10(5):349. https://doi.org/10.3390/jof10050349
Chicago/Turabian StyleGeng, Qingru, Jixiang Hu, Pingzhi Xu, Tongzheng Sun, Han Qiu, Shan Wang, Fengqin Song, Ling Shen, Yongxin Li, Man Liu, and et al. 2024. "The Autophagy-Related Protein ATG8 Orchestrates Asexual Development and AFB1 Biosynthesis in Aspergillus flavus" Journal of Fungi 10, no. 5: 349. https://doi.org/10.3390/jof10050349
APA StyleGeng, Q., Hu, J., Xu, P., Sun, T., Qiu, H., Wang, S., Song, F., Shen, L., Li, Y., Liu, M., Peng, X., Tian, J., & Yang, K. (2024). The Autophagy-Related Protein ATG8 Orchestrates Asexual Development and AFB1 Biosynthesis in Aspergillus flavus. Journal of Fungi, 10(5), 349. https://doi.org/10.3390/jof10050349






