Control of Fusarium Head Blight of Wheat with Bacillus velezensis E2 and Potential Mechanisms of Action
Abstract
:1. Introduction
2. Materials and Methods
2.1. Wheat
2.2. Isolation, Identification and Pathogenicity Verification of the Fungal Pathogen
2.3. Analysis of the Toxigenic Chemotype of F1
2.4. Culturing the Potential Antagonist
2.5. Effect of B. velezensis E2 on F. asiaticum F1 Growth
2.6. SEM Analysis
2.7. Effect of Bacillus velezensis E2 on Toxin Accumulation in Wheat
2.8. Sample Preparation and RNA Extraction
2.9. Transcriptome Sequencing and DEGs Obtention for F1
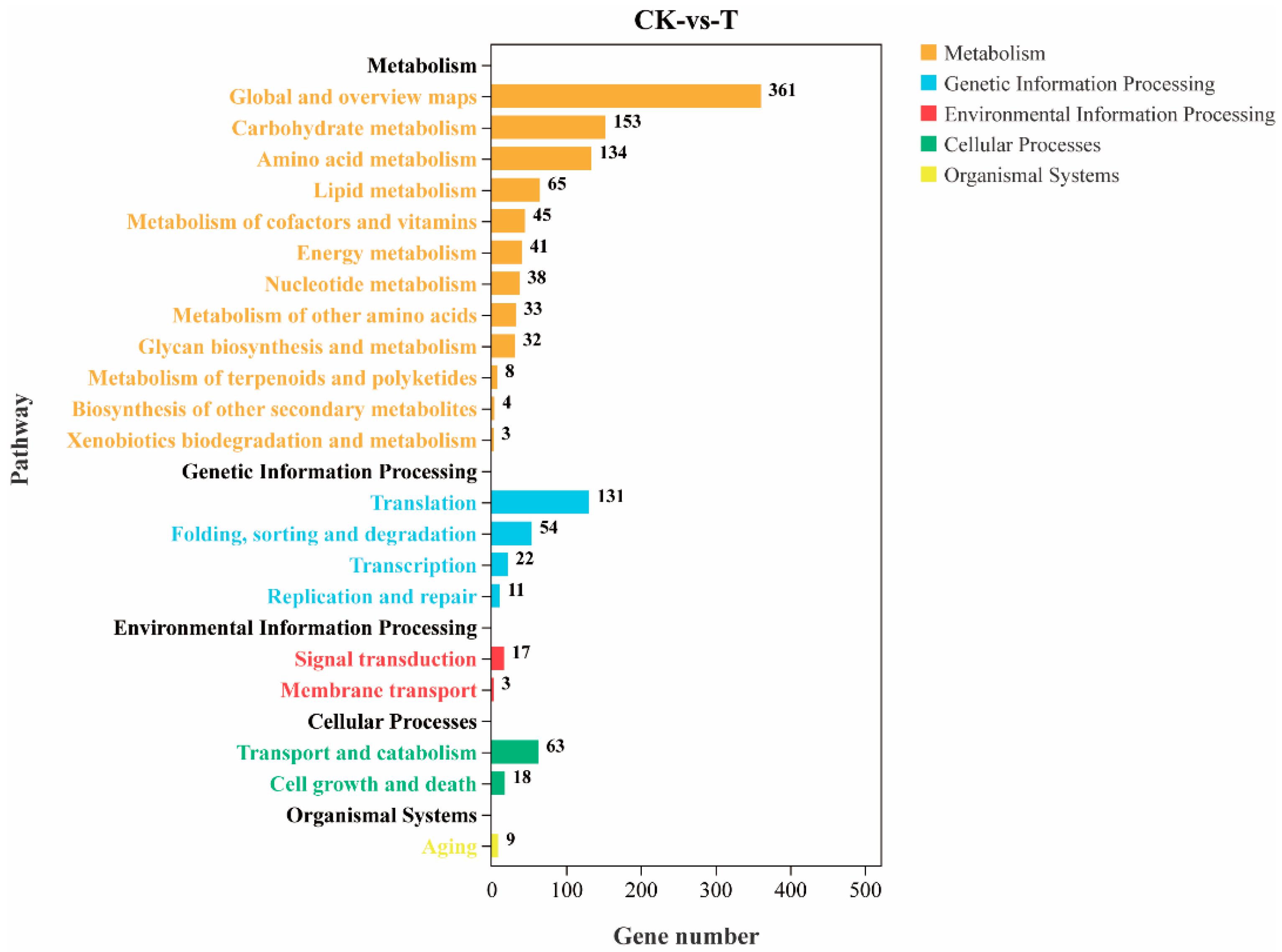
2.10. GO and KEGG Classification
2.11. Gene Expression Validation by RT-qPCR
2.11.1. Primer Design
2.11.2. RT-qPCR Reaction System and Procedure
2.12. Statistical Analyses
3. Results
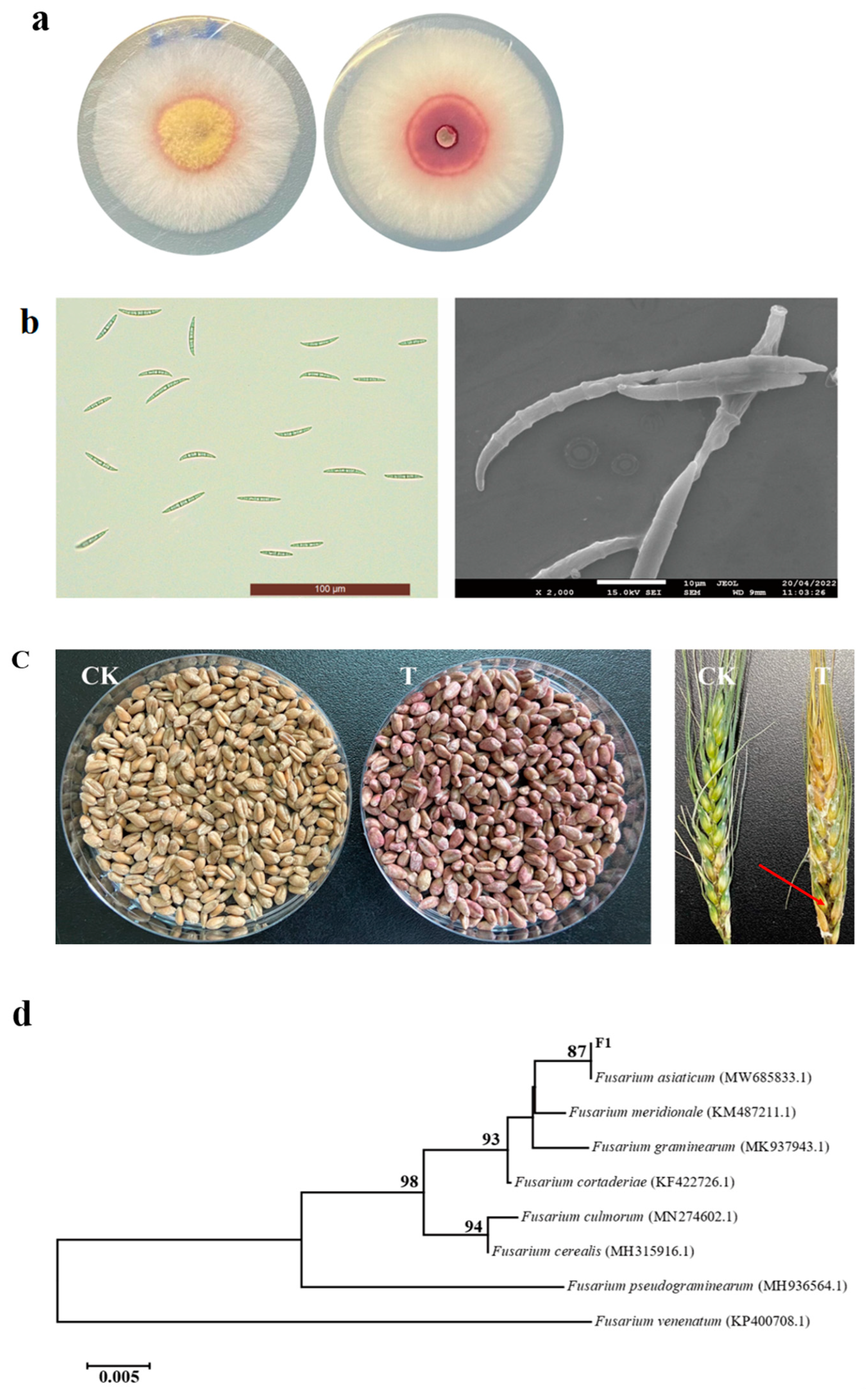
3.1. Isolation, Identification and Pathogenicity Verification of FHB Pathogens
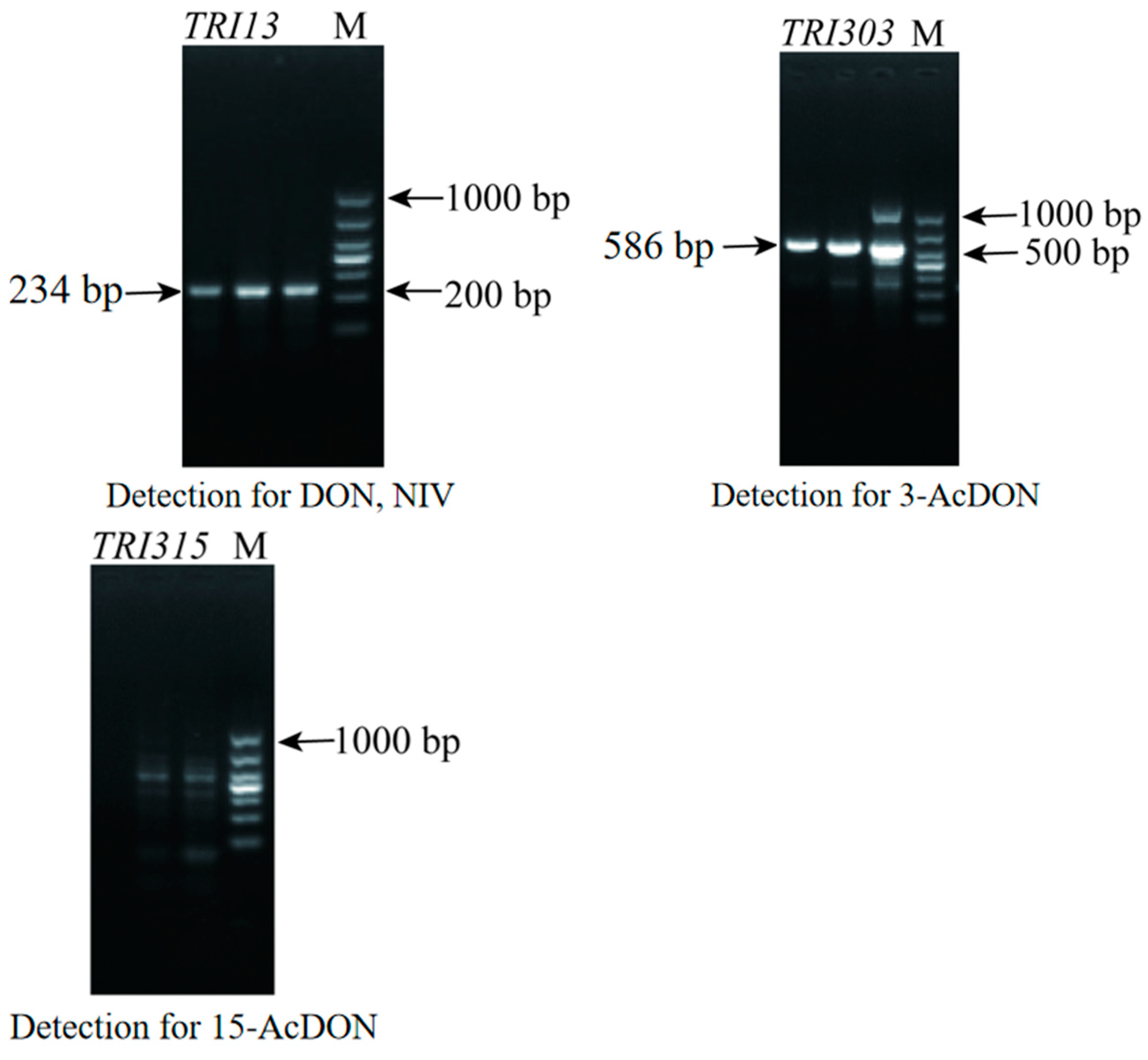
3.2. Analysis of the Toxigenic Chemotype of F1
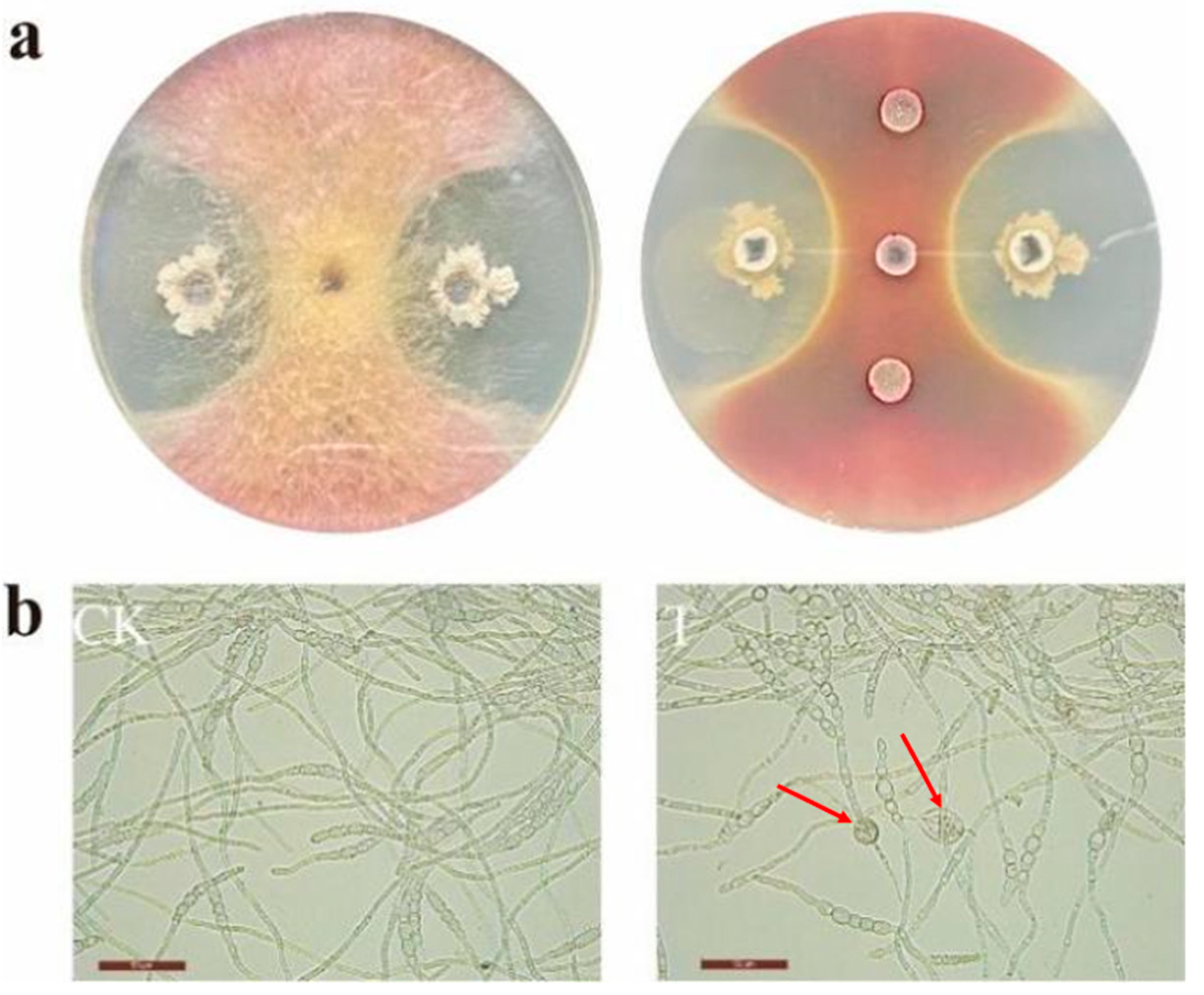
3.3. Antagonistic Activity of Bacillus velezensis
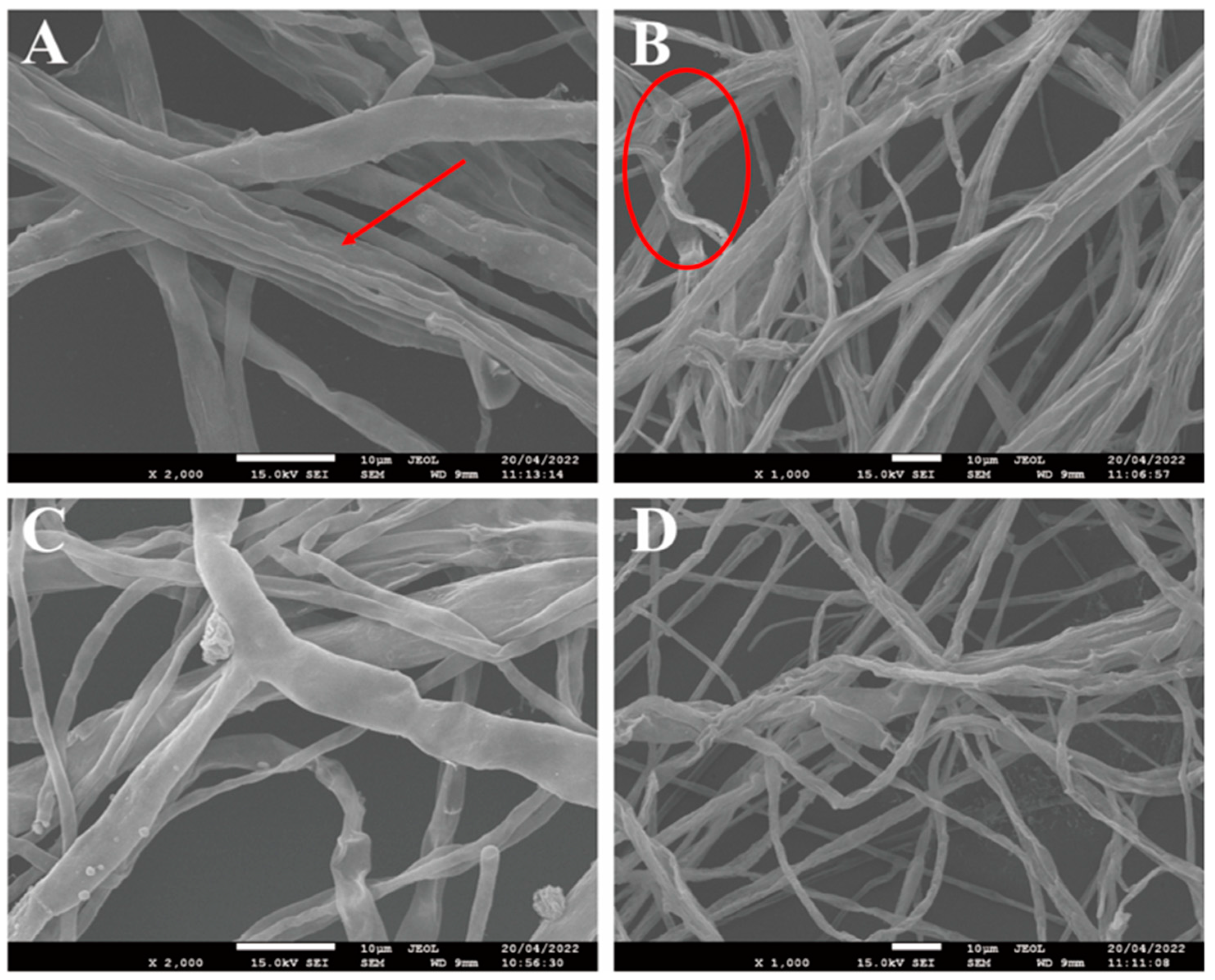
3.4. SEM
3.5. Effect of B. velezensis E2 on DON Accumulation in Storage Wheat
3.6. Sequencing Data and Its Quality Control
3.7. Differential Expression Gene Analysis
3.8. GO Enrichment Analysis of Differentially Expressed Genes
3.9. KEGG Enrichment Analysis of Differentially Expressed Genes
3.9.1. Effects on DON Synthesis Pathways
3.9.2. Effects on Secondary Metabolic Pathways
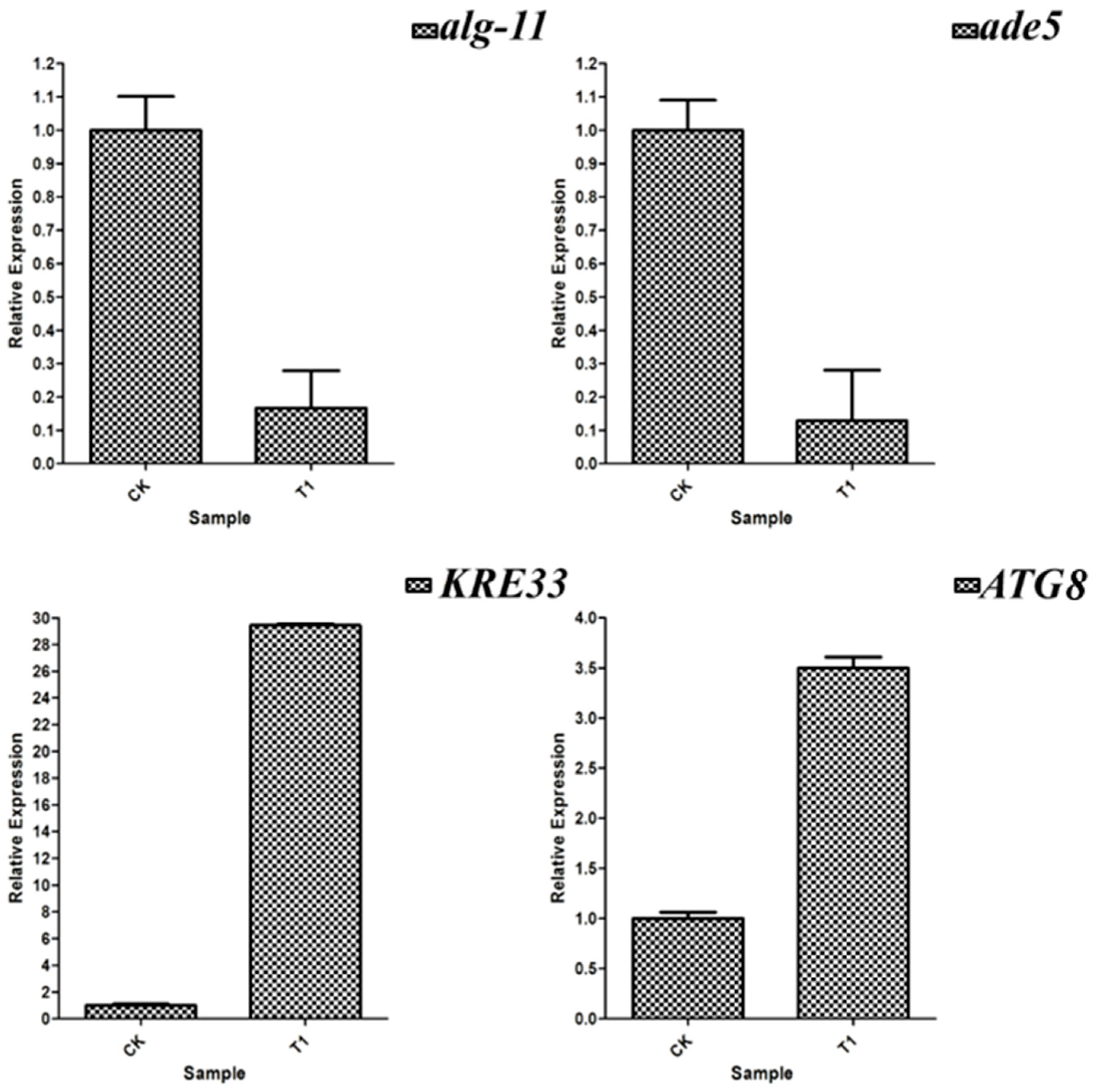
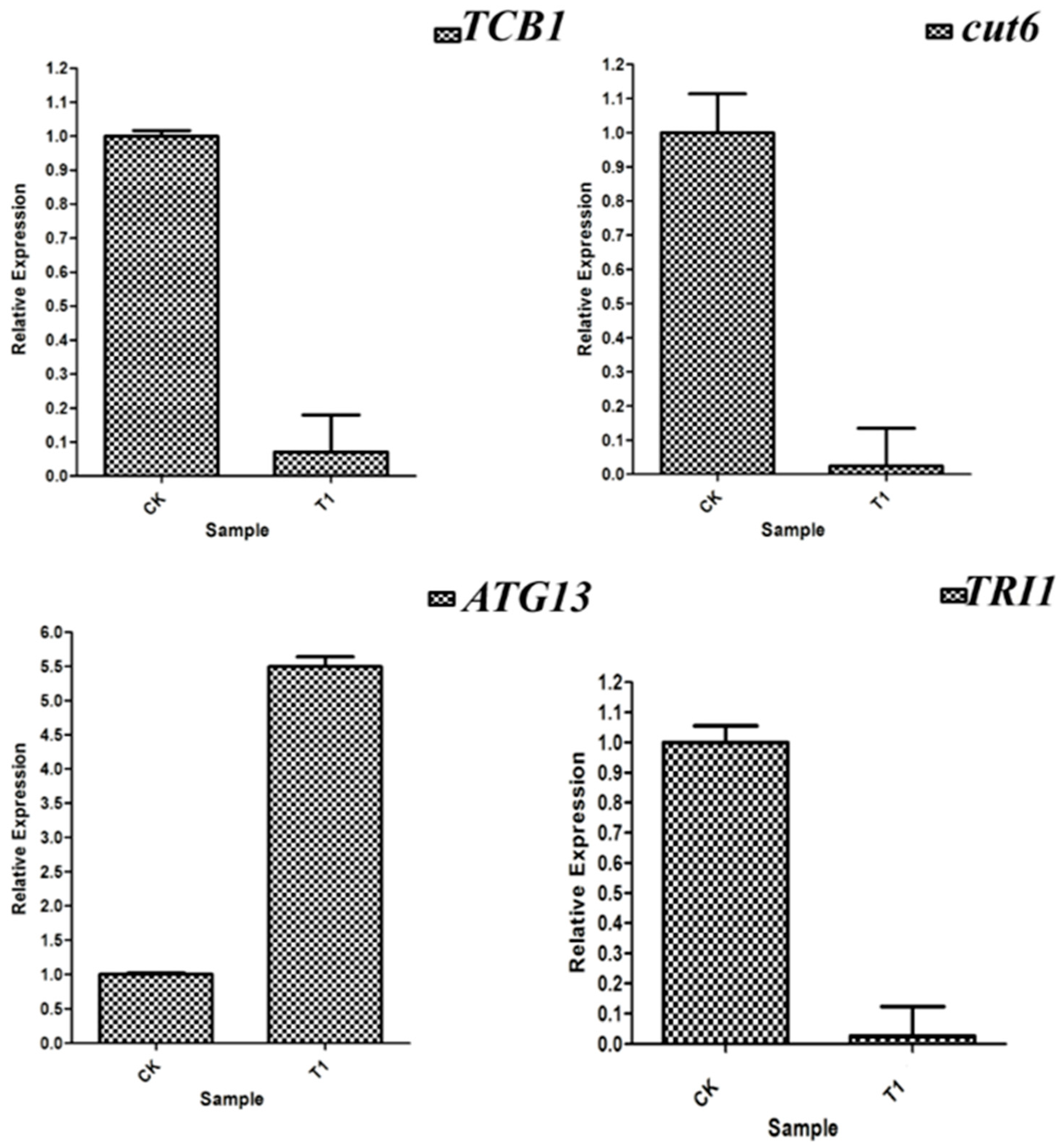
3.10. RT-qPCR Verification of Differentially Expressed Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Niu, H.; Jiang, Y.; Niu, J. Research Advances in the Genetics and Breeding of Wheat (Triticum aestivum) Resistance to Fusarium Head Blight. J. Agric. Biotechnol. 2020, 28, 530–542. [Google Scholar]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases-a field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Yli-Mattila, T.; Gagkaeva, T.; Ward, T.J.; Aoki, T.; Kistler, H.C.; O‘Donnell, K. A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian Far East. Mycologia 2009, 101, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Xing, Y.J.; Lee, Y.W.; Mokoena, M.P.; Olaniran, A.O.; Xu, J.H.; Shi, J.R. Occurrence of Fusarium mycotoxins and toxigenic Fusarium species in freshly harvested rice in Jiangsu, China. World Mycotoxin J. 2020, 13, 201–211. [Google Scholar] [CrossRef]
- Lee, T.; Lee, S.H.; Shin, J.Y.; Kim, H.K.; Yun, S.H.; Kim, H.Y.; Lee, S.; Ryu, J.G. Comparison of Trichothecene Biosynthetic Gene Expression between Fusarium graminearum and Fusarium asiaticum. Plant Pathol. J. 2014, 30, 33–42. [Google Scholar] [CrossRef]
- Wegulo, S.N. Factors influencing deoxynivalenol accumulation in small grain cereals. Toxins 2012, 4, 1157–1180. [Google Scholar] [CrossRef]
- Liu, N.; Yang, Y.; Chen, J.; Jia, H.; Zhang, Y.; Jiang, D.; Wu, G.; Wu, Z. 3-Acetyldeoxynivalenol induces lysosomal membrane permeabilization-mediated apoptosis and inhibits autophagic flux in macrophages. Env. Pollut. 2020, 265, 114697. [Google Scholar] [CrossRef]
- Choi, J.-H.; Nah, J.-Y.; Lee, M.-J.; Jang, J.-y.; Lee, T.; Kim, J. Fusarium diversity and mycotoxin occurrence in proso millet in Korea. LWT 2021, 14, 110964. [Google Scholar] [CrossRef]
- Rotter, B.A.; Prelusky, D.B.; Pestka, J.J. Toxicology of deoxynivalenol (vomitoxin). J. Toxicol. Env. Health 1996, 48, 1–34. [Google Scholar] [CrossRef]
- Xu, C.; Li, M.; Zhou, Z.; Li, J.; Chen, D.; Duan, Y.; Zhou, M. Impact of Five Succinate Dehydrogenase Inhibitors on DON Biosynthesis of Fusarium asiaticum, Causing Fusarium Head Blight in Wheat. Toxins 2019, 11, 272. [Google Scholar] [CrossRef]
- Wang, H.W.; Sun, S.L.; Ge, W.Y.; Zhao, L.F.; Hou, B.Q.; Wang, K.; Lyu, Z.F.; Chen, L.Y.; Xu, S.S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, 844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.G.; Zhu, Y.X.; Ma, D.F.; Xu, W.J.; Zhou, J.J.; Yan, H.W.; Yang, L.; Yin, J.L. Screening, Identification, and Optimization of Fermentation Conditions of an Antagonistic Endophyte to Wheat Head Blight. Agronomy 2019, 9, 476. [Google Scholar] [CrossRef]
- Stumbriene, K.; Gudiukaite, R.; Semaskiene, R.; Svegzda, P.; Jonaviciene, A.; Suproniene, S. Screening of new bacterial isolates with antifungal activity and application of selected Bacillus sp. cultures for biocontrol of Fusarium graminearum under field conditions. Crop Prot. 2018, 113, 22–28. [Google Scholar] [CrossRef]
- Siahmoshteh, F.; Hamidi-Esfahani, Z.; Spadaro, D.; Shams-Ghahfarokhi, M.; Razzaghi-Abyaneh, M. Unraveling the mode of antifungal action of Bacillus subtilis and Bacillus amyloliquefaciens as potential biocontrol agents against aflatoxigenic Aspergillus parasiticus. Food Control 2018, 89, 300–307. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, J.; Sun, J.; Lu, F.; Bie, X.; Lu, Z. Membrane disruption and DNA binding of Fusarium graminearum cell induced by C16-Fengycin A produced by Bacillus amyloliquefaciens. Food Control 2019, 102, 206–213. [Google Scholar] [CrossRef]
- Marin-Bruzos, M.; Grayston, S.J.; Forge, T.; Nelson, L.M. Isolation and characterization of streptomycetes and pseudomonad strains with antagonistic activity against the plant parasitic nematode Pratylenchus penetrans and fungi associated with replant disease. Biol. Control 2021, 158, 104599. [Google Scholar] [CrossRef]
- Cai, X.C.; Liu, C.H.; Wang, B.T.; Xue, Y.R. Genomic and metabolic traits endow Bacillus velezensis CC09 with a potential biocontrol agent in control of wheat powdery mildew disease. Microbiol. Res. 2017, 196, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Zalila-Kolsi, I.; Ben Mahmoud, A.; Ali, H.; Sellami, S.; Nasfi, Z.; Tounsi, S.; Jamoussi, K. Antagonist effects of Bacillus spp. strains against Fusarium graminearum for protection of durum wheat (Triticum turgidum L. subsp. durum). Microbiol. Res. 2016, 192, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Teng, K.; Wang, T.; Dong, E.; Zhang, M.; Tao, Y.; Zhong, J. Antimicrobial Bacillus velezensis HC6: Production of three kinds of lipopeptides and biocontrol potential in maize. J. Appl. Microbiol. 2020, 128, 242–254. [Google Scholar] [CrossRef]
- Chen, K.; Tian, Z.; He, H.; Long, C.-A.; Jiang, F. Bacillus species as potential biocontrol agents against citrus diseases. Biol. Control 2020, 151, 104419. [Google Scholar] [CrossRef]
- Ye, W.-Q.; Sun, Y.-F.; Tang, Y.-J.; Zhou, W.-W. Biocontrol potential of a broad-spectrum antifungal strain Bacillus amyloliquefaciens B4 for postharvest loquat fruit storage. Postharvest Biol. Technol. 2021, 174, 111439. [Google Scholar] [CrossRef]
- Srikhong, P.; Lertmongkonthum, K.; Sowanpreecha, R.; Rerngsamran, P. Bacillus sp. strain M10 as a potential biocontrol agent protecting chili pepper and tomato fruits from anthracnose disease caused by Colletotrichum capsici. BioControl 2018, 63, 833–842. [Google Scholar] [CrossRef]
- Platel, R.; Sawicki, M.; Esmaeel, Q.; Randoux, B.; Trapet, P.; El Guilli, M.; Chtaina, N.; Arnauld, S.; Bricout, A.; Rochex, A.; et al. Isolation and Identification of Lipopeptide-Producing Bacillus velezensis Strains from Wheat Phyllosphere with Antifungal Activity against the Wheat Pathogen Zymoseptoria tritici. Agronomy 2022, 12, 95. [Google Scholar] [CrossRef]
- Platel, R.; Lucau-Danila, A.; Baltenweck, R.; Maia-Grondard, A.; Trapet, P.; Magnin-Robert, M.; Randoux, B.; Duret, M.; Halama, P.; Hilbert, J.L.; et al. Deciphering immune responses primed by a bacterial lipopeptide in wheat towards Zymoseptoria tritici. Front. Plant Sci. 2023, 13, 1074447. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Hu, D.; Tian, S.; Peng, J.; Zhang, C.; Jia, N.; Wang, Z. Mechanism Analysis of Fusarium oxysporum HG-11 Responses to Biocontrol Agent Bacillus amyloliquefaciens B501. Acat Agric. Boreali-Occident. Sin. 2021, 30, 1695–1707. [Google Scholar]
- Yang, H.; Wang, L.; Li, S.; Gao, X.; Wu, N.; Zhao, Y.; Sun, W. Control of postharvest grey spot rot of loquat fruit with Metschnikowia pulcherrima E1 and potential mechanisms of action. Biol. Control 2021, 152, 104406. [Google Scholar] [CrossRef]
- Wang, Q.; Song, R.; Fan, S.; Coleman, J.J.; Xu, X.; Hu, X. Diversity of Fusarium community assembly shapes mycotoxin accumulation of diseased wheat heads. Mol. Ecol. 2022, 32, 2504–2518. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Zhang, J.B.; Chen, F.F.; Li, H.P.; Ndoye, M.; Liao, Y.C. A multiplex PCR assay for genetic chemotyping of toxigenic Fusarium graminearum and wheat grains for 3-acetyldeoxynivalenol, 15-acetyldeoxynivalenol and nivalenol mycotoxin. J. Food Agric. Environ. 2012, 10, 505–511. [Google Scholar]
- Yan, F.; Zhang, D.; Wang, X.; Liu, C.; Zhang, F. Reduction of postharvest diseases of loquat fruit by serine protease and possible mechanisms involved. Sci. Hortic. 2022, 304, 111246. [Google Scholar] [CrossRef]
- Ren, Y.; Yao, M.; Chang, P.; Sun, Y.; Li, R.; Meng, D.; Xia, X.; Wang, Y. Isolation and characterization of a Pseudomonas poae JSU-Y1 with patulin degradation ability and biocontrol potential against Penicillium expansum. Toxicon 2021, 195, 1–6. [Google Scholar] [CrossRef]
- Meng, D.; Garba, B.; Ren, Y.; Yao, M.; Xia, X.; Li, M.; Wang, Y. Antifungal activity of chitosan against Aspergillus ochraceus and its possible mechanisms of action. Int. J. Biol. Macromol. 2020, 158, 1063–1070. [Google Scholar] [CrossRef]
- GB5009.111-2016; Determination of Deoxynivalenol in Food—High Performance Liquid Chromatographic Method with Immunoaffinity Column Clean-Up. National Health and Family Planning Commission of the People’s Republic of China, State Food and Drug Administration: Beijing, China, 2016.
- Hsu, S.C.; Tsen, H.Y. PCR primers designed from malic acid dehydrogenase gene and their use for detection of Escherichia coli in water and milk samples. Int. J. Food Microbiol. 2001, 64, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yang, Q.; Serwah Boateng, N.A.; Ahima, J.; Dou, Y.; Zhang, H. Ultrastructure observation and transcriptome analysis of Penicillium expansum invasion in postharvest pears. Postharvest Biol. Technol. 2020, 165, 111198. [Google Scholar] [CrossRef]
- Shao, J.; Pei, Z.; Jing, H.; Wang, L.; Jiang, C.; Du, X.; Jiang, C.; Lou, Z.; Wang, H. Antifungal activity of myriocin against Fusarium graminearum and its inhibitory effect on deoxynivalenol production in wheat grains. Physiol. Mol. Plant Pathol. 2021, 114, 101635. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, J.; Huang, W.; Guo, X.; Li, W.; Zhang, Z. Research Progress in Metabolites Produced by Bacillus Against Three Common Plant Pathogenic Fungi. Biotechnol. Bull. 2023, 39, 59–68. [Google Scholar]
- Adedire, D.E.; Owoeye, T.E.; Farinu, O.M.; Ogundipe, W.F.; Adedire, O.M. Management of Fusarium Wilt Disease of Maize (Zea mays L.) with Selected Antimycotic Plant Extracts and Inhibitory Bacillus Strains. Curr. Microbiol. 2023, 80, 204. [Google Scholar] [CrossRef]
- Wang, J.; Geng, K.; Liu, Y.; Shi, Z.; Gong, Y.; Tian, Y.; Kang, X.; Wang, Y.; Sun, X. Screening and Identification of Strains Degrading Deoxynivalenol. Chin. J. Anim. Nutr. 2023, 35, 5430–5440. [Google Scholar]
- Su, P.; Ge, W.; Wang, H.; Kong, L. Advances in understanding the mechanisms of wheat-Fusarium graminearum interactions. Sci. Sin. Vitae 2021, 51, 1493–1507. [Google Scholar] [CrossRef]
- Ganesan, A.R.; Mohan, K.; Rajan, D.K.; Pillay, A.A.; Palanisami, T.; Sathishkumar, P.; Conterno, L. Distribution, toxicity, interactive effects, and detection of ochratoxin and deoxynivalenol in food: A review. Food Chem. 2022, 378, 131978. [Google Scholar] [CrossRef]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, S.; Safaie, N.; Sadeghi, A.; Shamsbakhsh, M. Streptomyces Strains Induce Resistance to Fusarium oxysporum f. sp. lycopersici Race 3 in Tomato Through Different Molecular Mechanisms. Front. Microbiol. 2019, 10, 1505. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V. What we have learned from ribosome structures. Biochem. Soc. Trans. 2008, 36, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Myong, K.; Shim, W.B.; Yun, S.H.; Lee, Y.W. Functional characterization of acetylglutamate synthase and phosphoribosylamine-glycine ligase genes in Gibberella zeae. Curr. Genet. 2007, 51, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Li, F.Q.; Vierstra, R.D. Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy 2014, 10, 1466–1467. [Google Scholar] [CrossRef]
- Wang, J.; Xu, C.; Sun, Q.; Xu, J.; Chai, Y.; Berg, G.; Cernava, T.; Ma, Z.; Chen, Y. Post-translational regulation of autophagy is involved in intra-microbiome suppression of fungal pathogens. Microbiome 2021, 9, 131. [Google Scholar] [CrossRef]


| Gene Name | Accession Number | Primer | Primer Sequence (5′→3′) | Product Length |
|---|---|---|---|---|
| ITS | NR_121320.1 | ITS1 | TCCGTAGGTGAACCTGCGG | 559 |
| ITS4 | TCCTCCGCTTATTGATATGC | |||
| TEF-α | NW_022157945.1 | TEF-F | ATGGGTAAGGAGGAGAAGAC | 700 |
| TEF-R | GGAAGTACCAGTGATCATGTT |
| Gene Name | Accession Number | Primer | Primer Sequence (5′→3′) | Toxigenic Chemotype | Product Length |
|---|---|---|---|---|---|
| Tri13 | NC_058400.1 | Tri13-F | TACGTGAAACATTGTTGGC | deoxynivalenol (DON), nivalenol (NIV) | 234, 415 |
| Tri13-R | GGTGTCCCAGGATCTGCG | ||||
| Tri3 | NC_058400.1 | Tri303-F | GATGGCCGCAAGTGGA | 3-AcDON (3-acetyldeoxynivalenol) | 586 |
| Tri303-R | GCCGGACTGCCCTATTG | ||||
| Tri315-F | CTCGCTGAAGTTGGACGTAA | 15-AcDON (15-acetyldeoxynivalenol) | 864 | ||
| Tri315-R | GTCTATGCTCTCAACGGACAAC |
| Gene ID | Gene Name | Accession Number | Revers Primer (5′ to 3′) |
|---|---|---|---|
| FGSG_00732 | KRE33 | NC_026474.1 | F: CGCAAAGCGGTGGACT R: GAATCTTGTCGGTCTCCTTGT |
| FGSG_06580 | cut6 | NC_026477.1 | F: AGCGAGCCATTCACTTCACT R: TCCAGGGGGTCCGATAAAGA |
| FGSG_07896 | TRI101 | NC_026477.1 | F: GTTCTGCCGTGCTGTTGATG R: GTCTCACAGTCTCGGGCTTAC |
| FGSG_08429 | ade5 | NC_026475.1 | F: GGGCAGACAAAGCAGGCA R: TAGGCTCGCTCAATGGCAC |
| FGSG_08491 | ATG13 | NC_026475.1 | F: AACGACCCAGCCGAATCTAC R: ACTGCTCCATCACATCCCAC |
| FGSG_10740 | ATG8 | NC_026476.1 | F: GAGGTTCTACCCCCGACAG R: CGCCAAAAGTGTTCTCGCC |
| FGSG_10858 | ALG-11 | NC_026476.1 | F: CCGACCCGAGAAGAACCATC R: CTCAGCCAGTCCAGAACCTC |
| FGSG_06885 | TCB1 | NC_026477.1 | F: TCAAGGGCGAGGATGGAC R: GGCAGGTCGGAACAGAAGTC |
| FGSG_09286 | cpc-1 | NC_026477.1 | F: GCCTTTTCCTCACCTGCTGT R: CCGACTTGCGACGGTTCA |
| FGSG_04400 | rhoA | NC_026475.1 | F: GGCGATGGTGCTTGTGGTAA R: GAGGGAGTCGGGAGAGTCAA |
| Treatment | CK | B. velezensis E2 |
|---|---|---|
| Spore germination rate | 93.33 ± 0.76 | 25.17 ± 3.32 ** |
| Experiment | Control Group | Treatment Group |
|---|---|---|
| DON content (μg/kg) | 46.02 ± 1.19 | 36.72 ± 0.83 ** |
| Samples | Raw Data (bp) | Clean Data (bp) | Clean Reads | GC (%) | Q20 (%) | Unique-Mapped (%) |
|---|---|---|---|---|---|---|
| CK1 | 6,112,122,000 | 6,075,824,999 | 40,647,974 | 52.91 | 98.70 | 35,427,322 (87.32%) |
| CK2 | 6,529,115,400 | 6,479,907,696 | 43,433,248 | 52.96 | 98.69 | 38,186,463 (88.06%) |
| CK3 | 6,884,337,000 | 6,836,297,207 | 45,781,106 | 53.10 | 98.65 | 40,361,865 (88.30%) |
| T1 | 5,622,680,700 | 5,585,596,453 | 37,401,952 | 52.36 | 98.72 | 32,604,351 (87.36%) |
| T2 | 5,586,865,500 | 5,542,412,171 | 37,159,506 | 52.26 | 98.76 | 32,255,302 (86.95%) |
| T3 | 5,876,491,800 | 5,830,758,067 | 39,079,466 | 52.58 | 98.64 | 33,887,979 (86.88%) |
| Gene ID | Gene Name | log2FC | Definition |
|---|---|---|---|
| FGSG_07896 | TRI101 | −5.02 | richothecene 3-O-acetyltransferase |
| FGSG_03538 | TRI10 | −10.75 | TRI10 [Fusarium graminearum] |
| FGSG_00071 | TRI1 | −6.28 | cytochrome P450 monooxygenase |
| FGSG_06580 | cut6 | −4.3 | acetyl-CoA carboxylase |
| FGSG_05243 | ALG9 | −3.13 | alpha-1,2-mannosyltransferase |
| FGSG_10858 | alg-11 | −2.24 | alpha-1,2-mannosyltransferase |
| FGSG_01233 | ALG12 | −3.25 | alpha-1,6-mannosyltransferase |
| FGSG_04044 | dpm1 | −2.37 | dolichol-phosphate mannosyltransferase |
| FGSG_05973 | gls2 | −3.28 | alpha-glucosidase |
| FGSG_06885 | TCB1 | −2.62 | transmembrane protein |
| FGSG_08429 | ade5 | −2.64 | phosphoribosylglycinamide formyltransferase |
| FGSG_10740 | ATG8 | 4.44 | GABA(A) receptor-associated protein |
| FGSG_08491 | ATG13 | 3.34 | autophagy-related protein 13 |
| FGSG_10033 | mpp10 | 3.56 | U3 small nucleolar RNA-associated protein |
| FGSG_08759 | UTP14 | 4.90 | U3 small nucleolar RNA-associated protein 14 |
| FGSG_10060 | UTP15 | 3.70 | U3 small nucleolar RNA-associated protein 15 |
| FGSG_00270 | UTP4 | 4.69 | U3 small nucleolar RNA-associated protein 4 |
| FGSG_06165 | NOG1 | 3.57 | nucleolar GTP-binding protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Gao, C.; Lin, M.; Sun, Z.; Zhao, Y.; Li, X.; Zhao, T.; Xu, X.; Sun, W. Control of Fusarium Head Blight of Wheat with Bacillus velezensis E2 and Potential Mechanisms of Action. J. Fungi 2024, 10, 390. https://doi.org/10.3390/jof10060390
Ma J, Gao C, Lin M, Sun Z, Zhao Y, Li X, Zhao T, Xu X, Sun W. Control of Fusarium Head Blight of Wheat with Bacillus velezensis E2 and Potential Mechanisms of Action. Journal of Fungi. 2024; 10(6):390. https://doi.org/10.3390/jof10060390
Chicago/Turabian StyleMa, Jianing, Chen Gao, Meiwei Lin, Zhenzhong Sun, Yuhao Zhao, Xin Li, Tianyuan Zhao, Xingang Xu, and Weihong Sun. 2024. "Control of Fusarium Head Blight of Wheat with Bacillus velezensis E2 and Potential Mechanisms of Action" Journal of Fungi 10, no. 6: 390. https://doi.org/10.3390/jof10060390
APA StyleMa, J., Gao, C., Lin, M., Sun, Z., Zhao, Y., Li, X., Zhao, T., Xu, X., & Sun, W. (2024). Control of Fusarium Head Blight of Wheat with Bacillus velezensis E2 and Potential Mechanisms of Action. Journal of Fungi, 10(6), 390. https://doi.org/10.3390/jof10060390




