Differential Carbon Catabolite Repression and Hemicellulolytic Ability among Pathotypes of Colletotrichum lindemuthianum against Natural Plant Substrates
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Enzyme Assay and Protein Quantitation
2.3. Statistical Analysis
2.4. Secretome Analysis
2.5. Sample Preparation and Processing
2.6. LC-MS/MS Analysis
2.7. Mass Spectrometry
2.8. Data Analysis
3. Results
3.1. Fungal Growth
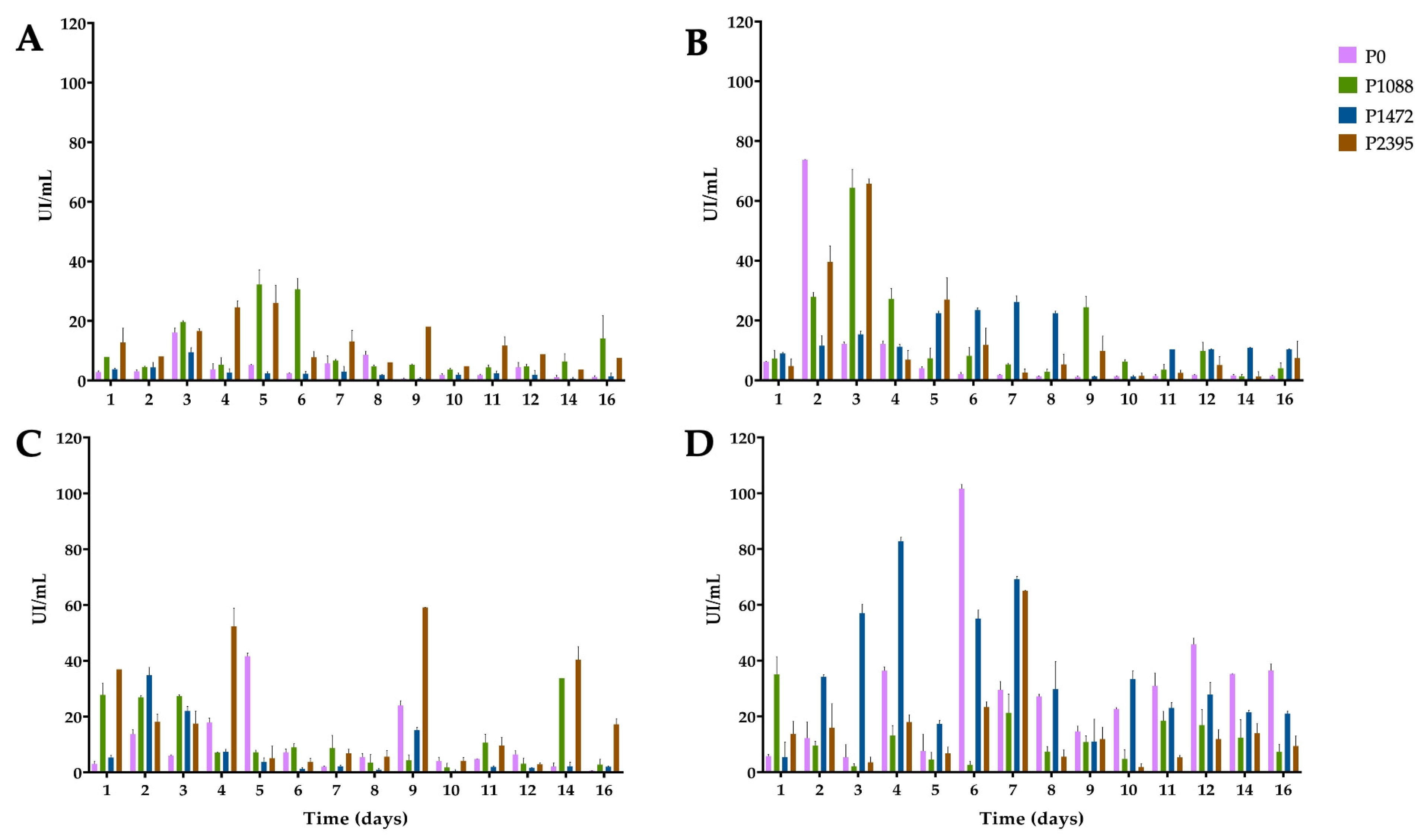
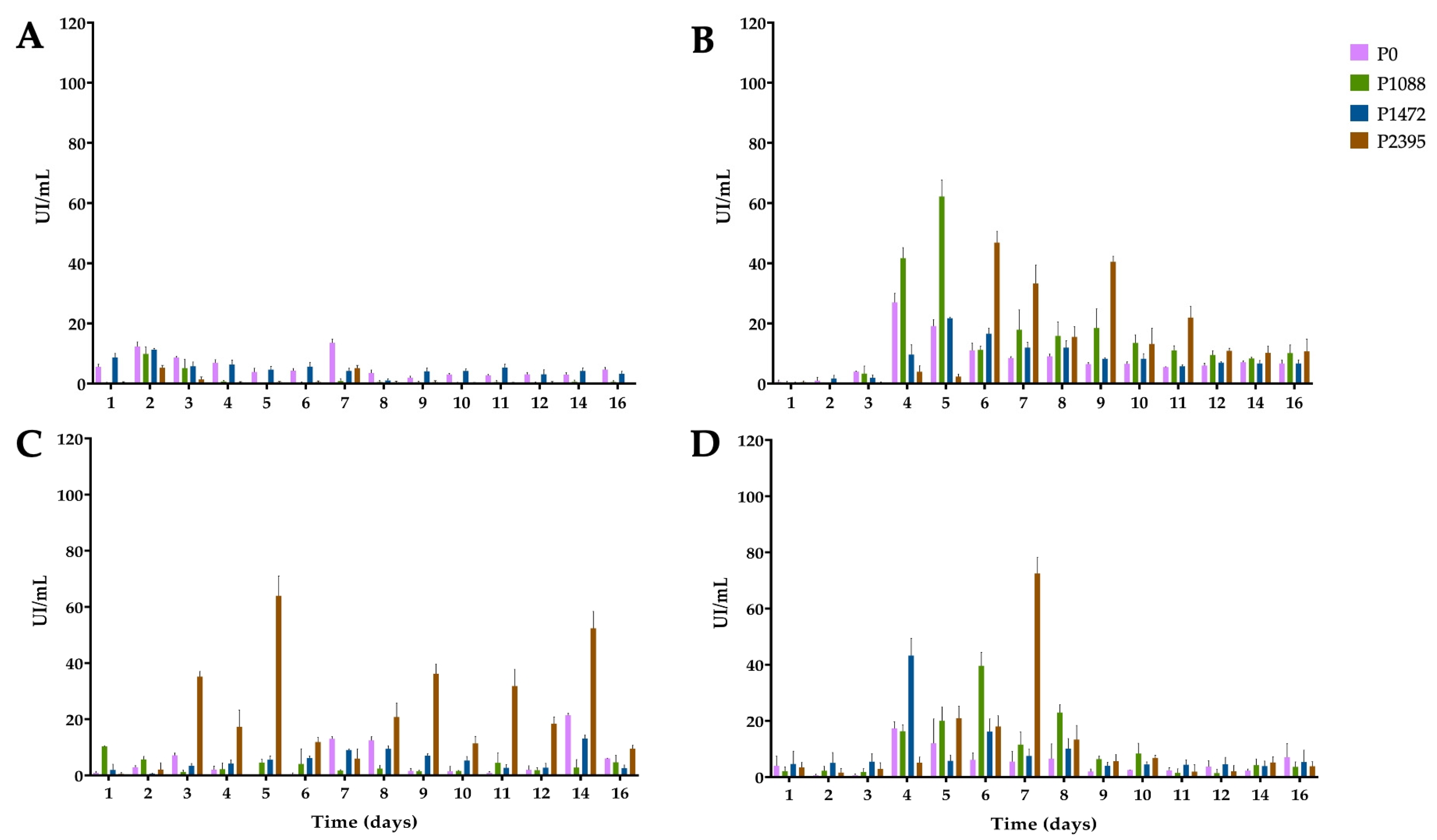
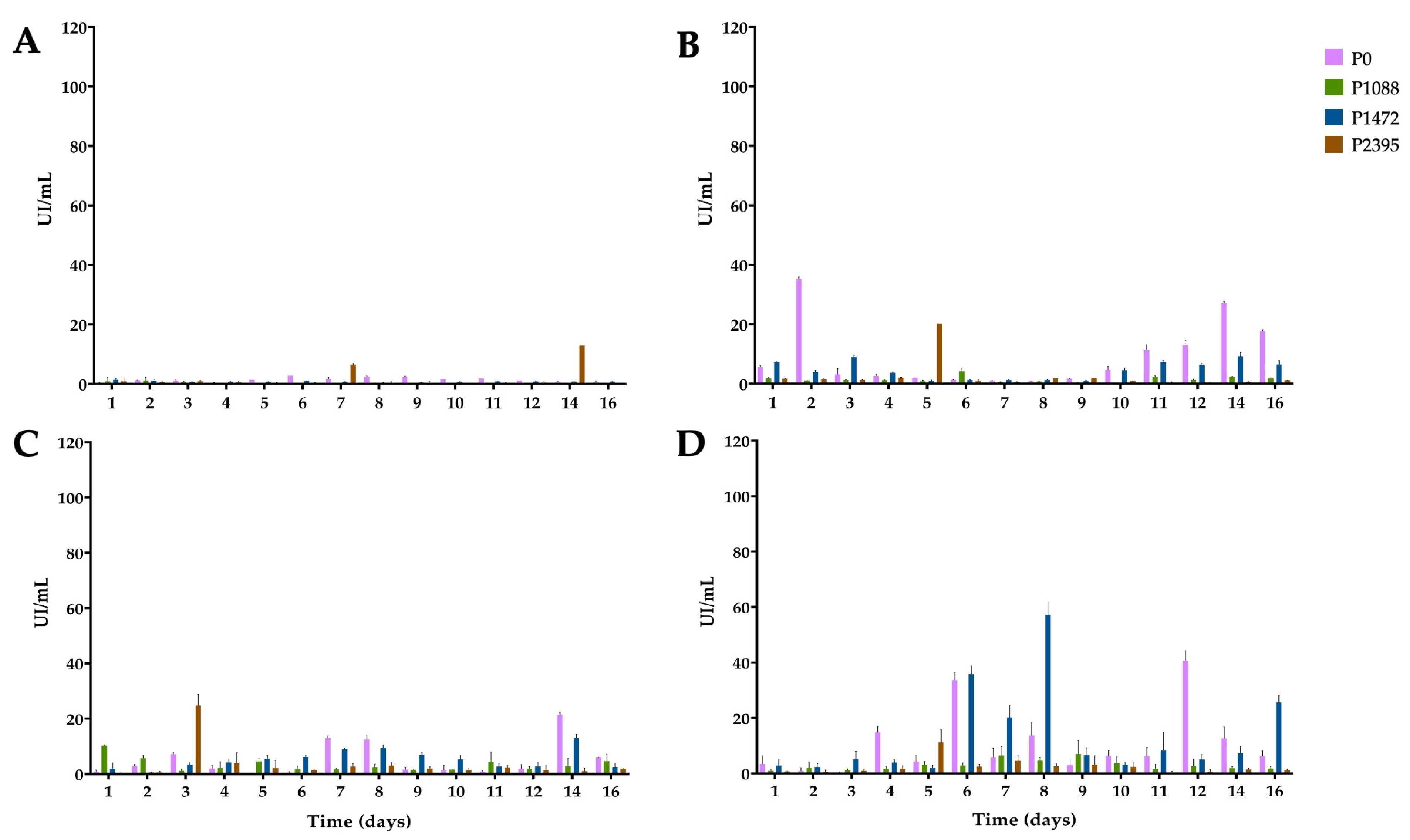
3.2. Endo-β-1,4-xylanase Activity
3.3. α-L-Arabinofuranosidase Activity
3.4. β-xylosidase Activity
3.5. 1,4-β-cellobiohydrolase Activity
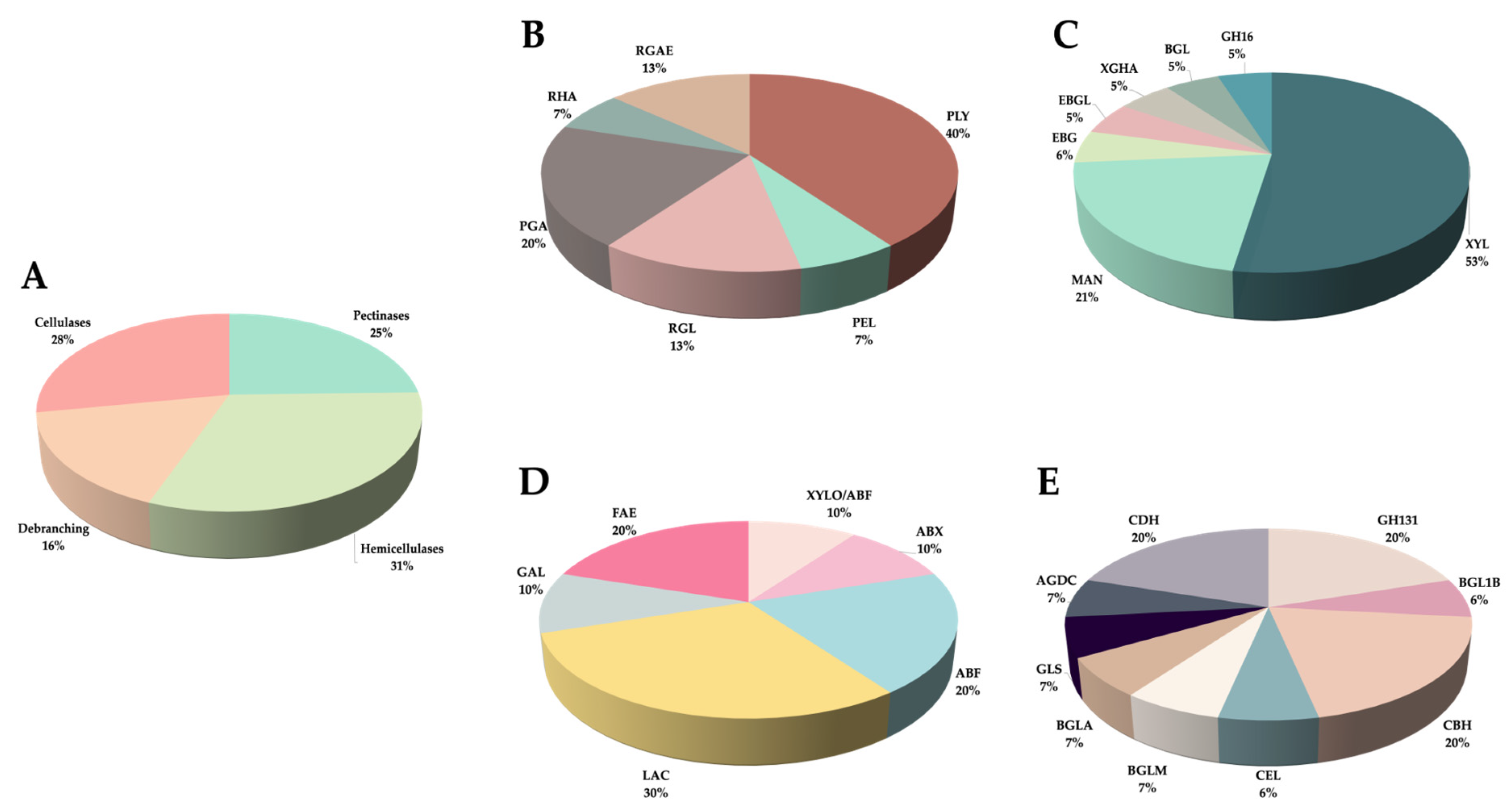
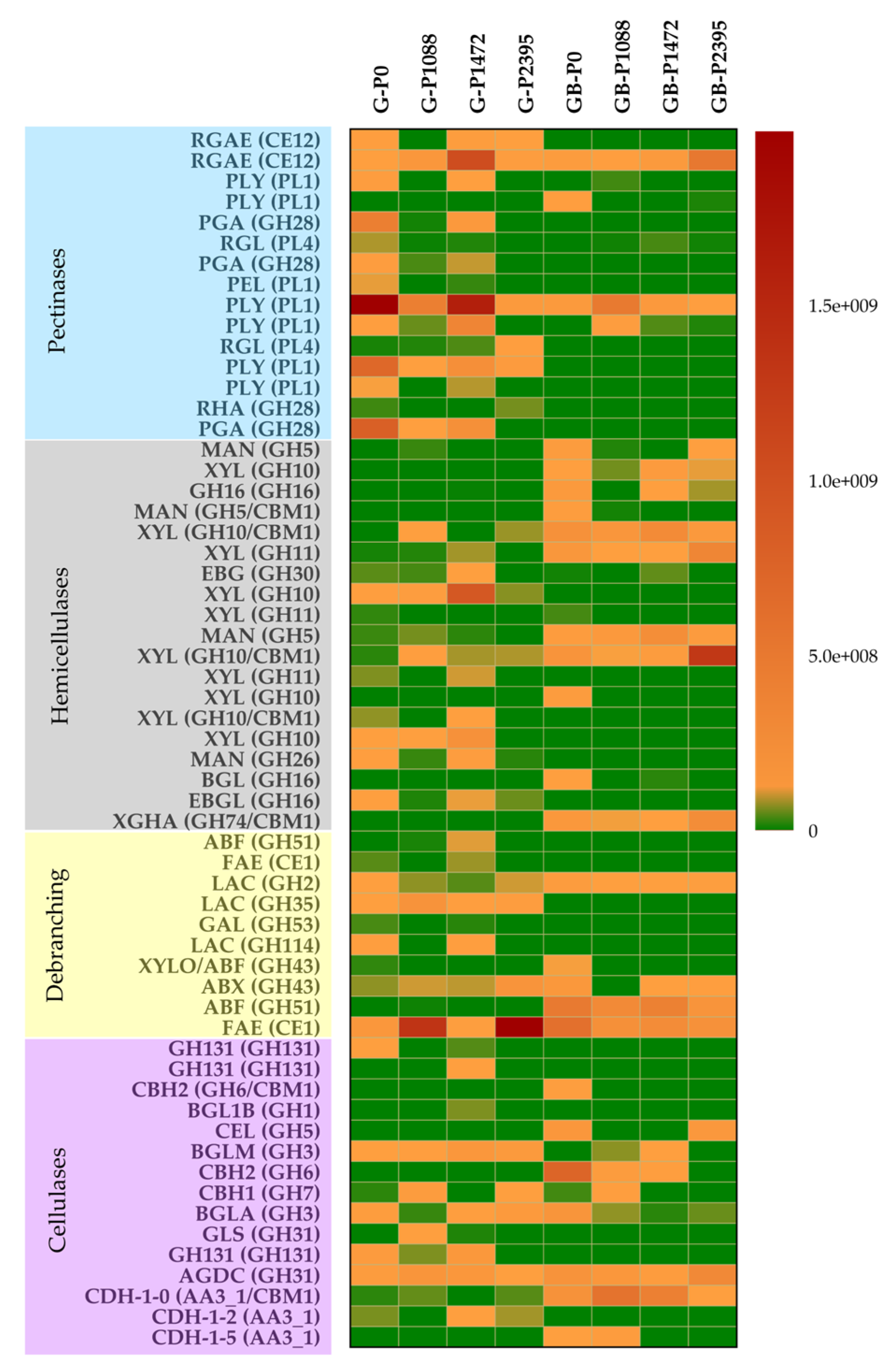
3.6. Secreted PCWDEs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mendgen, K.; Hahn, M. Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 2002, 7, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Münch, S.; Lingner, U.; Floss, D.S.; Ludwig, N.; Sauer, N.; Deising, H.B. The hemibiotrophic lifestyle of Colletotrichum species. J. Plant Physiol. 2008, 165, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.M.; King, B.C.; Hayes, M.L.; Bergstrom, G.C. Plant pathogens as a source of diverse enzymes for lignocellulose digestion. Curr. Opin. Microbiol. 2011, 14, 264–270. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall–degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A. Extracellular enzymes produced by Colletotrichum lindemuthianum and Helminthosporium maydis during growth on isolated bean and corn cell walls. Physiol. Biochem. 1978, 68, 1585–1589. [Google Scholar] [CrossRef]
- Centis, S.; Guillas, I.; Séjalon, N.; Esquerré-Tugayé, M.-T.; Dumas, B. Endopolygalacturonase genes from Colletotrichum lindemuthianum: Cloning of CLPG2 and comparison of its expression to that of CLPG1 during saprophytic and parasitic growth of the fungus. Mol. Plant-Microbe Interact. 1997, 10, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Hugouvieux, V.; Centis, S.; Lafitte, C.; Esquerre-Tugaye, M. Induction by (alpha)-L-arabinose and (alpha)-L-rhamnose of endopolygalacturonase gene expression in Colletotrichum lindemuthianum. Appl. Environ. Microbiol. 1997, 63, 2287–2292. [Google Scholar] [CrossRef] [PubMed]
- Dumas, B.; Centis, S.; Sarrazin, N.; Esquerré-Tugayé, M.-T. Use of Green Fluorescent Protein To Detect Expression of an Endopolygalacturonase Gene of Colletotrichum lindemuthianum during Bean Infection. Appl. Environ. Microbiol. 1999, 65, 1769–1771. [Google Scholar] [CrossRef] [PubMed]
- Herbert, C.; O’Connell, R.; Gaulin, E.; Salesses, V.; Esquerré-Tugayé, M.-T.; Dumas, B. Production of a cell wall-associated endopolygalacturonase by Colletotrichum lindemuthianum and pectin degradation during bean infection. Fungal Genet. Biol. 2004, 41, 140–147. [Google Scholar] [CrossRef]
- Acosta-Rodríguez, I.; Piñón-Escobedo, C.; Zavala-Páramo, M.G.; López-Romero, E.; Cano-Camacho, H. Degradation of cellulose by the bean-pathogenic fungus Colletotrichum lindemuthianum. Production of extracellular cellulolytic enzymes by cellulose induction. Antonie Leeuwenhoek 2005, 87, 301–310. [Google Scholar] [CrossRef]
- Hernández-Silva, L.; Piñón-Escobedo, C.; Cano-Camacho, H.; Zavala-Páramo, M.G.; Acosta-Rodríguez, I.; López-Romero, E. Comparison of fungal growth and production of extracellular pectin lyase activity by pathogenic and non-pathogenic races of Colletotrichum lindemuthianum cultivated under different conditions. Physiol. Mol. Plant Pathol. 2007, 70, 88–95. [Google Scholar] [CrossRef]
- Lara-Márquez, A.; Zavala-Páramo, M.G.; López-Romero, E.; Calderón-Cortés, N.; López-Gómez, R.; Conejo-Saucedo, U.; Cano-Camacho, H. Cloning and characterization of a pectin lyase gene from Colletotrichum lindemuthianum and comparative phylogenetic/structural analyses with genes from phytopathogenic and saprophytic/opportunistic microorganisms. BMC Microbiol. 2011, 11, 260. [Google Scholar] [CrossRef] [PubMed]
- Cnossen-Fassoni, A.; Bazzolli, D.M.S.; Brommonschenkel, S.H.; Fernandes de Araújo, E.; de Queiroz, M.V. The pectate lyase encoded by the pecCl 1 gene is an important determinant for the aggressiveness of Colletotrichum lindemuthianum. J. Microbiol. 2013, 51, 461–470. [Google Scholar] [CrossRef]
- Conejo-Saucedo, U.; Cano-Camacho, H.; López-Romero, E.; Villa-Rivera, M.G.; Lara-Márquez, A.; Zavala-Páramo, M.G. Cloning and characterization of an endo-β-1, 4-xylanase gene from Colletotrichum lindemuthianum and phylogenetic analysis of similar genes from phytopathogenic fungus. Afr. J. Microbiol. Res. 2016, 10, 1292–1305. [Google Scholar] [CrossRef]
- González, M.; Rodríguez, R.; Zavala, M.E.; Jacobo, J.L.; Hernández, F.; Acosta, J.; Martínez, O.; Simpson, J. Characterization of Mexican isolates of Colletotrichum lindemuthianum by using differential cultivars and molecular markers. Phytopathology 1998, 88, 292–299. [Google Scholar] [CrossRef]
- Nunes, M.P.B.A.; Gonçalves-Vidigal, M.C.; Martins, V.S.; Xavier, L.F.; Valentini, G.; Vaz Bisneta, M.; Vidigal Filho, P.S. Relationship of Colletotrichum lindemuthianum races and resistance loci in the Phaseolus vulgaris L. genome. Crop Sci. 2021, 61, 3877–3893. [Google Scholar] [CrossRef]
- Habgood, R. Designation of physiological races of plant pathogens. Nature 1970, 227, 1268–1269. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Corrales, M. Estandarización de variedades diferenciales y de designación de razas de Colletotrichum lindemuthianum. Phytopathology 1991, 81, 694. [Google Scholar]
- Balardin, R.; Jarosz, A.; Kelly, J. Virulence and molecular diversity in Colletotrichum lindemuthianum from South, Central, and North America. Phytopathology 1997, 87, 1184–1191. [Google Scholar] [CrossRef]
- Sicard, D.; Michalakis, Y.; Dron, M.; Neema, C. Genetic diversity and pathogenic variation of Colletotrichum lindemuthianum in the three centers of diversity of its host, Phaseolus vulgaris. Phytopathology 1997, 87, 807–813. [Google Scholar] [CrossRef]
- Calo, S.; Billmyre, R.B.; Heitman, J. Generators of phenotypic diversity in the evolution of pathogenic microorganisms. PLoS Pathog. 2013, 9, e1003181. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Nadales, E.; Nogueira, M.F.A.; Baldin, C.; Castanheira, S.; El Ghalid, M.; Grund, E.; Lengeler, K.; Marchegiani, E.; Mehrotra, P.V.; Moretti, M. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 2014, 70, 42–67. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.-R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.T.; Cherry, J. Progress and challenges in enzyme development for biomass utilization. In Biofuels; Olsson, L., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 108, pp. 95–120. [Google Scholar] [CrossRef]
- King, B.C.; Waxman, K.D.; Nenni, N.V.; Walker, L.P.; Bergstrom, G.C.; Gibson, D.M. Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol. Biofuels 2011, 4, 4. Available online: http://www.biotechnologyforbiofuels.com/content/4/1/4 (accessed on 7 May 2024). [CrossRef] [PubMed]
- Villa-Rivera, M.G.; Zavala-Páramo, M.G.; Conejo-Saucedo, U.; López-Romero, E.; Lara-Márquez, A.; Cano-Camacho, H. Differences in the expression profile of endo-β-(1, 6)-D-galactanase in pathogenic and non-pathogenic races of Colletotrichum lindemuthianum grown in the presence of arabinogalactan, xylan or Phaseolus vulgaris cell walls. Physiol. Mol. Plant Pathol. 2017, 99, 75–86. [Google Scholar] [CrossRef]
- French, E.R.; Hebert, T.T. Metodos de Investigacion Fitopatologica; Catie, B.O.I., Ed.; Instituto Interamericano de Ciencias Agrícolas: San José, Costa Rica, 1982; Volume 43, p. 37. [Google Scholar]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, G.J.; Visser, J. Carbon repression in Aspergilli. FEMS Microbiol. Lett. 1997, 151, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Ries, L.N.; Beattie, S.R.; Espeso, E.A.; Cramer, R.A.; Goldman, G.H. Diverse regulation of the CreA carbon catabolite repressor in Aspergillus nidulans. Genetics 2016, 203, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Stolle-Smits, T.; Beekhuizen, J.G.; Recourt, K.; Voragen, A.G.; Van Dijk, C. Changes in pectic and hemicellulosic polymers of green beans (Phaseolus vulgaris L.) during industrial processing. J. Agric. Food Chem. 1997, 45, 4790–4799. [Google Scholar] [CrossRef]
- González-Rentería, S.; Soto-Cruz, N.; Rutiaga-Quiñones, O.; Medrano-Roldán, H.; Rutiaga-Quinones, J.; López-Miranda, J. Optimization of the enzymatic hydrolysis process of four straw bean varieties (Pinto villa, Pinto saltillo, Pinto mestizo and Flor de mayo). Rev. Mex. Ing. Quim. 2011, 10, 17–28. [Google Scholar]
- Sindhu, R.; Binod, P.; Pandey, A.; Madhavan, A.; Alphonsa, J.A.; Vivek, N.; Gnansounou, E.; Castro, E.; Faraco, V. Water hyacinth a potential source for value addition: An overview. Bioresour. Technol. 2017, 230, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.J.; Wang, H.-T.; York, W.S.; Peña, M.J.; Urbanowicz, B.R. Designer biomass for next-generation biorefineries: Leveraging recent insights into xylan structure and biosynthesis. Biotechnol. Biofuels 2017, 10, 1–14. [Google Scholar]
- Fernandez, J.; Wright, J.D.; Hartline, D.; Quispe, C.F.; Madayiputhiya, N.; Wilson, R.A. Principles of carbon catabolite repression in the rice blast fungus: Tps1, Nmr1-3, and a MATE–family pump regulate glucose metabolism during infection. PLoS Genet. 2012, 8, e1002673. [Google Scholar] [CrossRef] [PubMed]
- Tani, S.; Kawaguchi, T.; Kobayashi, T. Complex regulation of hydrolytic enzyme genes for cellulosic biomass degradation in filamentous fungi. Appl. Microbiol. Biotechnol. 2014, 98, 4829–4837. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Gomi, K. Induction and repression of hydrolase genes in Aspergillus oryzae. Front. Microbiol. 2021, 12, 677603. [Google Scholar] [CrossRef] [PubMed]
- Beldman, G.; Schols, H.; Pitson, S.; Searle-van Leeuwen, M.; Voragen, A. Arabinans and arabinan degrading enzymes. Adv. Macromol. Carbohydr. Res. 1997, 1, 1–64. [Google Scholar]
- Lagaert, S.; Pollet, A.; Courtin, C.M.; Volckaert, G. β-Xylosidases and α-L-arabinofuranosidases: Accessory enzymes for arabinoxylan degradation. Biotechnol. Adv. 2014, 32, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Sakka, M.; Yamada, K.; Kitamura, T.; Kunitake, E.; Kimura, T.; Sakka, K. The modular arabinanolytic enzyme Abf43A-Abf43B-Abf43C from Ruminiclostridium josui consists of three GH43 modules classified in different subfamilies. Enzyme Microb. Technol. 2019, 124, 23–31. [Google Scholar] [CrossRef]
- Biely, P. Microbial xylanolytic systems. Trends Biotechnol. 1985, 3, 286–290. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.M.; Knott, B.C.; Mayes, H.B.; Hansson, H.; Himmel, M.E.; Sandgren, M.; Stahlberg, J.; Beckham, G.T. Fungal cellulases. Chem. Rev. 2015, 115, 1308–1448. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Zheng, W.; Islam, W.; Arif, M.; Abubakar, Y.S.; Wang, Z.; Lu, G. Carbon catabolite repression in filamentous fungi. Int. J. Mol. Sci. 2017, 19, 48. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.A.; Ries, L.N.A.; Goldman, G.H. How nutritional status signalling coordinates metabolism and lignocellulolytic enzyme secretion. Fungal Genet. Biol. 2014, 72, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.; Marroquin-Guzman, M.; Wilson, R.A. Mechanisms of nutrient acquisition and utilization during fungal infections of leaves. Annu. Rev. Phytopathol. 2014, 52, 155–174. [Google Scholar] [CrossRef] [PubMed]
- New, A.M.; Cerulus, B.; Govers, S.K.; Perez-Samper, G.; Zhu, B.; Boogmans, S.; Xavier, J.B.; Verstrepen, K.J. Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol. 2014, 12, e1001764. [Google Scholar] [CrossRef] [PubMed]
- Villamagna, A.; Murphy, B. Ecological and socio-economic impacts of invasive water hyacinth (Eichhornia crassipes): A review. Freshw. Biol. 2010, 55, 282–298. [Google Scholar] [CrossRef]
- Anuja Sharma, A.S.; Aggarwal, N.; Anita Saini, A.S.; Anita Yadav, A.Y. Beyond biocontrol: Water hyacinth-opportunities and challenges. J. Environ. Sci. Technol. 2016, 9, 26–48. [Google Scholar] [CrossRef]
- Ayanda, O.I.; Ajayi, T.; Asuwaju, F.P. Eichhornia crassipes (Mart.) Solms: Uses, challenges, threats, and prospects. Sci. World J. 2020, 2020, 3452172. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, L.; Ghosh, S. Bioconversion of lignocellulosic fraction of water-hyacinth (Eichhornia crassipes) hemicellulose acid hydrolysate to ethanol by Pichia stipitis. Bioresour. Technol. 2009, 100, 3293–3297. [Google Scholar] [CrossRef]
- Okunowo, W.O.; Gbenle, G.O.; Osuntoki, A.A.; Adekunle, A.A.; Ojokuku, S.A. Production of cellulolytic and xylanolytic enzymes by a phytopathogenic Myrothecium roridum and some avirulent fungal isolates from water hyacinth. Afr. J. Biotechnol. 2010, 9, 1074–1078. [Google Scholar] [CrossRef]
- Battaglia, E.; Visser, L.; Nijssen, A.; van Veluw, G.; Wösten, H.; de Vries, R. Analysis of regulation of pentose utilisation in Aspergillus niger reveals evolutionary adaptations in Eurotiales. Stud. Mycol. 2011, 69, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, M.R.; Donofrio, N.; de Vries, R.P. Plant biomass degradation by fungi. Fungal Genet. Biol. 2014, 72, 2–9. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.L.; Morgan, T.; Garcia, E.A.; Rosa, R.O.; Mendes, T.A.d.O.; de Queiroz, M.V. Pectinolytic arsenal of Colletotrichum lindemuthianum and other fungi with different lifestyles. J. Appl. Microbiol. 2022, 133, 1857–1871. [Google Scholar] [CrossRef] [PubMed]
- Arntzen, M.Ø.; Bengtsson, O.; Várnai, A.; Delogu, F.; Mathiesen, G.; Eijsink, V.G. Quantitative comparison of the biomass-degrading enzyme repertoires of five filamentous fungi. Sci. Rep. 2020, 10, 20267. [Google Scholar] [CrossRef]
- Biely, P.; Vršanská, M.; Tenkanen, M.; Kluepfel, D. Endo-β-1, 4-xylanase families: Differences in catalytic properties. J. Biotechnol. 1997, 57, 151–166. [Google Scholar] [CrossRef] [PubMed]
- de Vries, R.P.; Visser, J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 2001, 65, 497–522. [Google Scholar] [CrossRef]
- Herrmann, M.C.; Vrsanska, M.; Jurickova, M.; Hirsch, J.; Biely, P.; Kubicek, C.P. The β-D-xylosidase of Trichoderma reesei is a multifunctional β-D-xylan xylohydrolase. Biochem. J. 1997, 321, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Huy, N.D.; Thayumanavan, P.; Kwon, T.-H.; Park, S.-M. Characterization of a recombinant bifunctional xylosidase/arabinofuranosidase from Phanerochaete chrysosporium. J. Biosci. Bioeng. 2013, 116, 152–159. [Google Scholar] [CrossRef]
- Yamabhai, M.; Sak-Ubol, S.; Srila, W.; Haltrich, D. Mannan biotechnology: From biofuels to health. Crit. Rev. Biotechnol. 2016, 36, 32–42. [Google Scholar] [CrossRef]
- Mafa, M.S.; Malgas, S. Towards an understanding of the enzymatic degradation of complex plant mannan structures. World J. Microbiol. Biotechnol. 2023, 39, 302. [Google Scholar] [CrossRef] [PubMed]
- Lafond, M.; Navarro, D.; Haon, M.; Couturier, M.; Berrin, J.-G. Characterization of a broad-specificity β-glucanase acting on β-(1, 3)-, β-(1, 4)-, and β-(1, 6)-glucans that defines a new glycoside hydrolase family. Appl. Environ. Microbiol. 2012, 78, 8540–8546. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Yoshida, M.; Tamura, M.; Tanaka, Y.; Umezawa, K.; Nishikawa, A.; Tonozuka, T. Crystal structure of the N-terminal domain of a glycoside hydrolase family 131 protein from Coprinopsis cinerea. FEBS Lett. 2013, 587, 2193–2198. [Google Scholar] [CrossRef] [PubMed]
- Scheiblbrandner, S.; Csarman, F.; Ludwig, R. Cellobiose dehydrogenase in biofuel cells. Curr. Opin. Biotechnol. 2022, 73, 205–212. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]


| PCWDEs | Enzyme | # Genes | CAZy Family |
|---|---|---|---|
| Pectinases | |||
| PLY | Pectate lyase | 6 | PL1 |
| PEL | Pectin lyase | 1 | PL1 |
| RGL | Rhamnogalacturonate lyase | 2 | PL4 |
| PGA | Endopolygalacturonase | 3 | GH28 |
| RHA | Alpha-L-rhamnosidase | 1 | GH28 |
| RGAE | Rhamnogalacturonan acetylesterase | 2 | CE12 |
| Hemicellulases | |||
| XYL | Endo-1,4-beta-xylanase | 4 | GH10 |
| XYL | Endo-1,4-beta-xylanase | 3 | GH10/CBM1 |
| XYL | Endo-1,4-beta-xylanase | 3 | GH11 |
| MAN | Mannan endo-1,4-beta-mannosidase | 2 | GH5 |
| MAN | Mannan endo-1,4-beta-mannosidase | 1 | GH5/CBM1 |
| MAN | Mannan endo-1,4-beta-mannosidase | 1 | GH26 |
| EBG | Endo-beta-1,6-galactanase | 1 | GH30 |
| EBGL | Endo-1,3(4)-beta-glucanase | 1 | GH16 |
| XGHA | Xyloglucanase | 1 | GH74/CBM1 |
| BGL | Beta-glucanase | 1 | GH16 |
| GH16 | Glycosyl hydrolase family 16 | 1 | GH16 |
| Debranching enzymes | |||
| XYLO/ABF | β-xylosidase/arabinofuranosidase | 1 | GH43 |
| ABX | Exo-alpha-(1->5)-L-arabinofuranosidase | 1 | GH43 |
| ABF | Alpha-L-arabinofuranosidase | 2 | GH51 |
| LAC | Beta-galactosidase | 1 | GH35 |
| LAC | Beta-galactosidase | 1 | GH2 |
| LAC | Alpha-galactosidase | 1 | GH114 |
| GAL | Arabinogalactan endo-beta-1,4-galactanase | 1 | GH53 |
| FAE | Feruloyl esterase | 2 | CE1 |
| Cellulases | |||
| BGLA | Beta-glucosidase | 1 | GH3 |
| BGL1B | Beta-glucosidase | 1 | GH1 |
| BGLM | Beta-glucosidase | 1 | GH3 |
| CEL | Glycosyl hydrolase 5 family | 1 | GH5 |
| CBH2 | Cellobiohydrolase | 1 | GH6/CBM1 |
| CBH2 | Cellobiohydrolase | 1 | GH6 |
| CBH1 | Cellobiohydrolase | 1 | GH7 |
| GLS | Alpha/beta-glucosidase agdC | 1 | GH31 |
| AGDC | Alpha/beta-glucosidase agdC | 1 | GH31 |
| GH131 | Glycoside hydrolase family 131 | 3 | GH131 |
| Auxiliary Activities | |||
| CDH-1-0 | Cellobiose dehydrogenase | 1 | AA3_1/CBM1 |
| CDH-1-2 | Cellobiose dehydrogenase | 1 | AA3_1 |
| CDH-1-5 | Cellobiose dehydrogenase | 1 | AA3_1 |
| Pathotype | # Genes | Pectinases | Hemicellulases | Debranching | Cellulases | Auxiliar Activities | CAZy Families |
|---|---|---|---|---|---|---|---|
| P0 | 50 | 11 | 19 | 7 | 12 | 3 | 26 |
| P1088 | 33 | 8 | 12 | 5 | 8 | 2 | 24 |
| P1472 | 43 | 11 | 13 | 8 | 11 | 2 | 25 |
| P2395 | 27 | 7 | 10 | 5 | 5 | 2 | 20 |
| Total | 59 | 15 | 19 | 10 | 12 | 3 | 30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Tapia, K.M.; Zavala-Páramo, M.G.; Villa-Rivera, M.G.; Morelos-Martínez, M.I.; López-Romero, E.; Simpson, J.; Bolaños-Rebolledo, J.; Cano-Camacho, H. Differential Carbon Catabolite Repression and Hemicellulolytic Ability among Pathotypes of Colletotrichum lindemuthianum against Natural Plant Substrates. J. Fungi 2024, 10, 406. https://doi.org/10.3390/jof10060406
Díaz-Tapia KM, Zavala-Páramo MG, Villa-Rivera MG, Morelos-Martínez MI, López-Romero E, Simpson J, Bolaños-Rebolledo J, Cano-Camacho H. Differential Carbon Catabolite Repression and Hemicellulolytic Ability among Pathotypes of Colletotrichum lindemuthianum against Natural Plant Substrates. Journal of Fungi. 2024; 10(6):406. https://doi.org/10.3390/jof10060406
Chicago/Turabian StyleDíaz-Tapia, Karla Morelia, María Guadalupe Zavala-Páramo, Maria Guadalupe Villa-Rivera, Ma. Irene Morelos-Martínez, Everardo López-Romero, June Simpson, Jeni Bolaños-Rebolledo, and Horacio Cano-Camacho. 2024. "Differential Carbon Catabolite Repression and Hemicellulolytic Ability among Pathotypes of Colletotrichum lindemuthianum against Natural Plant Substrates" Journal of Fungi 10, no. 6: 406. https://doi.org/10.3390/jof10060406
APA StyleDíaz-Tapia, K. M., Zavala-Páramo, M. G., Villa-Rivera, M. G., Morelos-Martínez, M. I., López-Romero, E., Simpson, J., Bolaños-Rebolledo, J., & Cano-Camacho, H. (2024). Differential Carbon Catabolite Repression and Hemicellulolytic Ability among Pathotypes of Colletotrichum lindemuthianum against Natural Plant Substrates. Journal of Fungi, 10(6), 406. https://doi.org/10.3390/jof10060406








