The Performance and Evolutionary Mechanism of Ganoderma lucidum in Enhancing Selenite Tolerance and Bioaccumulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Laboratory Adaptive Evolution
2.3. Biomass, Extracellular Polysaccharides (EPS) and Intracellular Polysaccharides (IPS)
2.4. Se Accumulation Capacity Determination
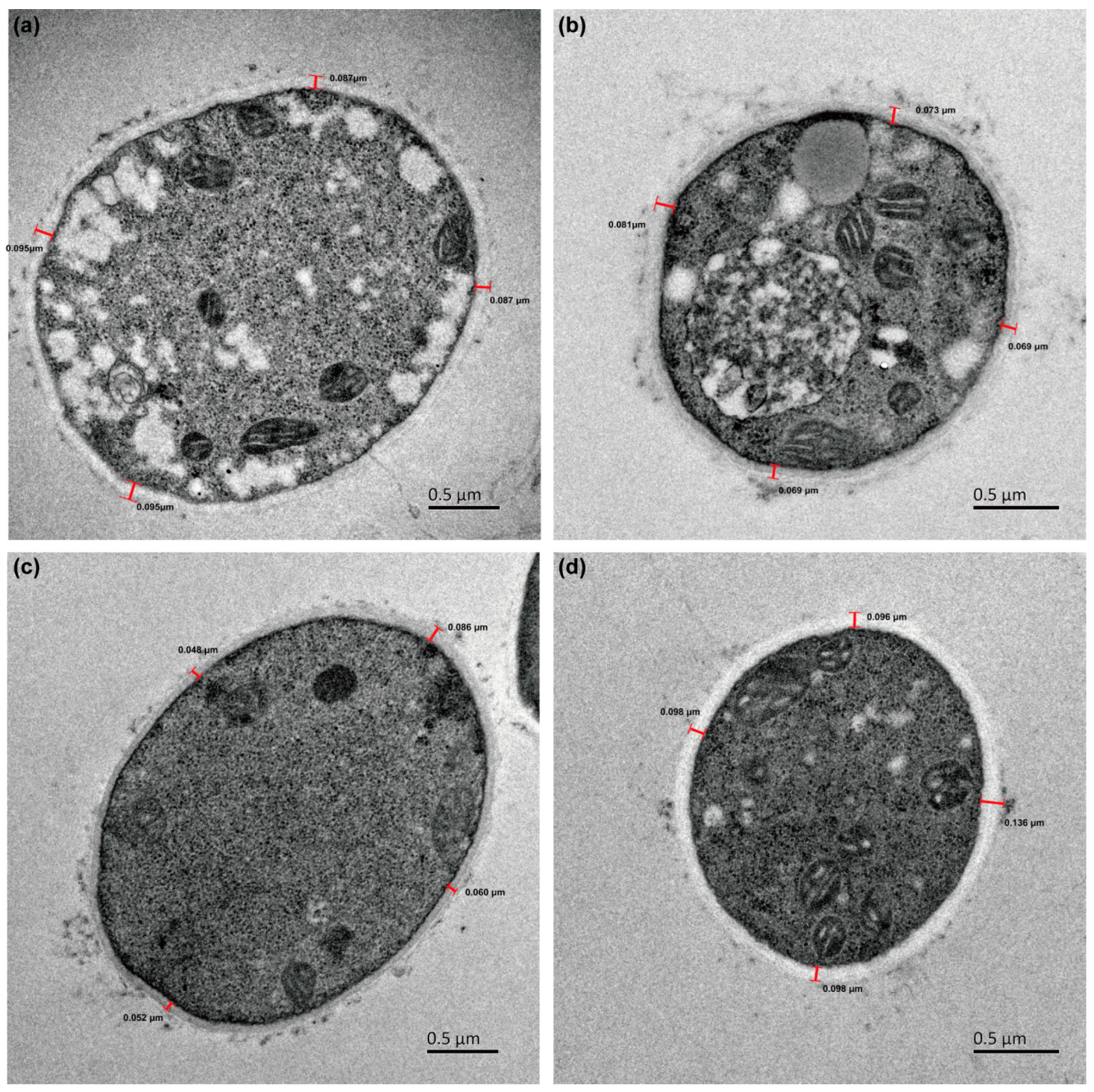
2.5. Macroscopic and Microscopic Morphology
2.6. Selenite Uptake Kinetics
2.7. X-ray Photoelectron Spectroscopy (XPS)
2.8. Transcriptome Profiling Analysis
2.9. Gene Expression Analysis by RT-qPCR
2.10. Statistical Analysis
3. Results and Discussion
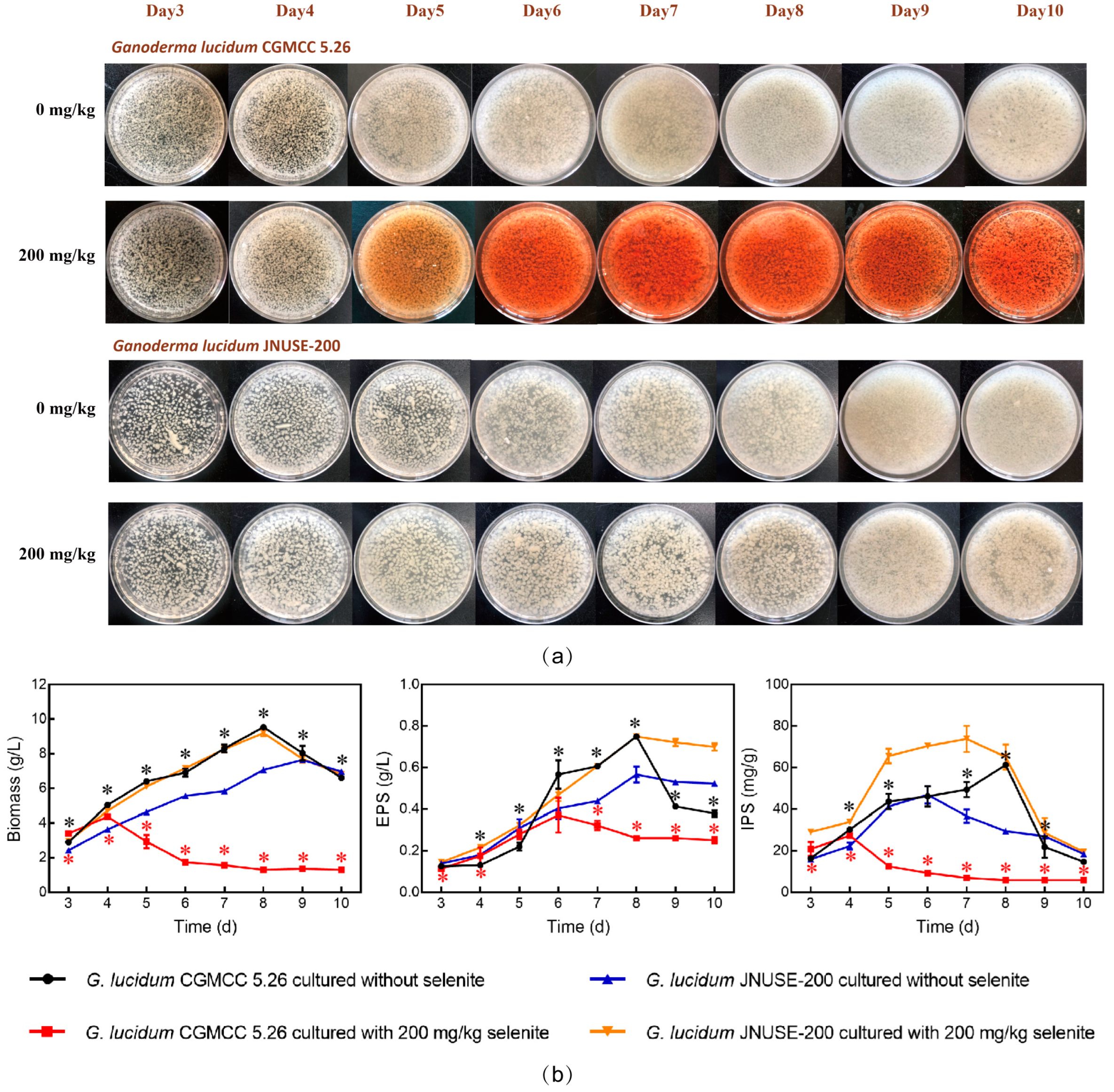
3.1. G. lucidum JNUSE-200 Exhibited Enhanced Selenite Tolerance
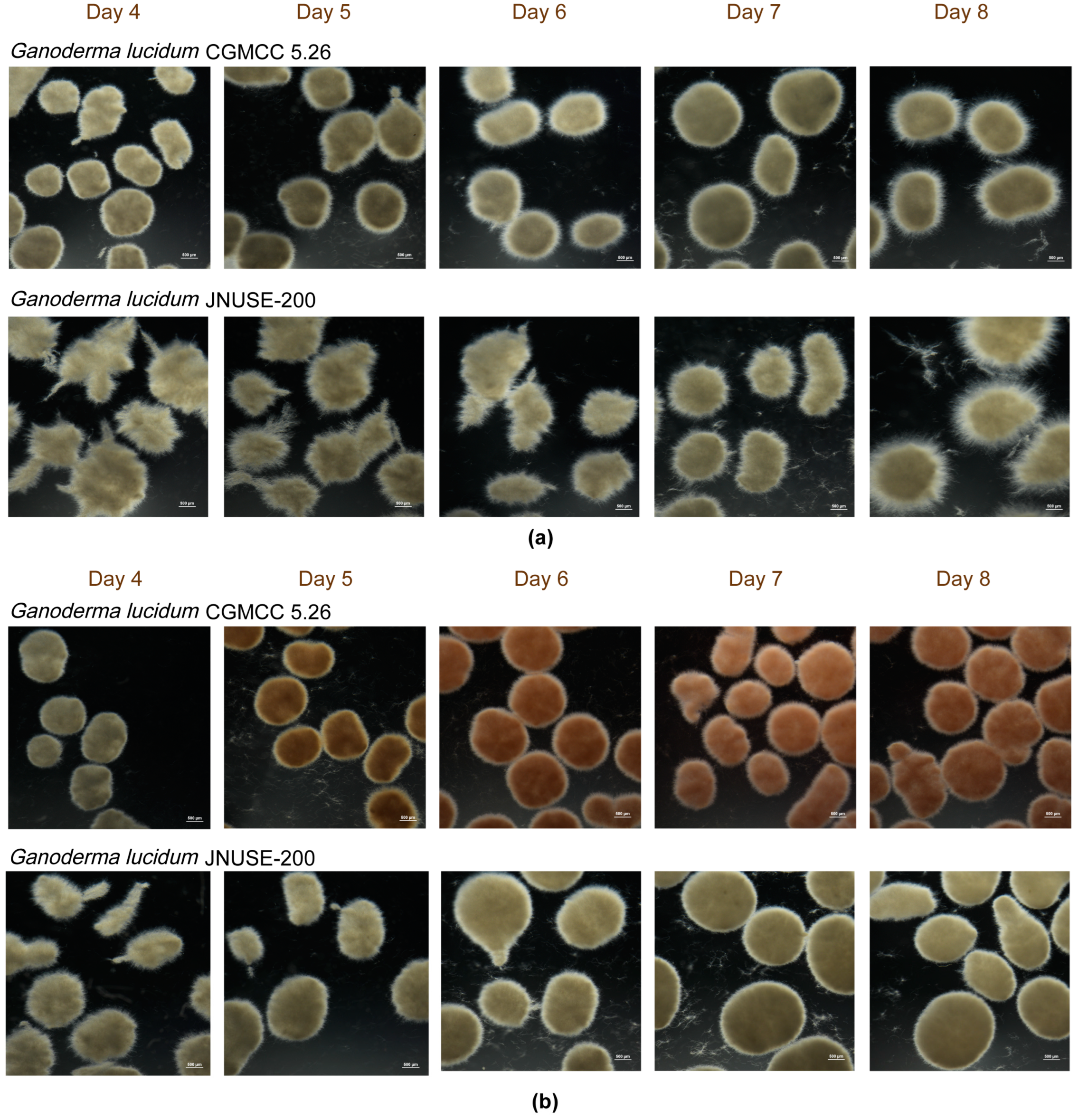
3.2. G. lucidum JNUSE-200 Exhibited Macroscopic and Microscopic Morphological Differences
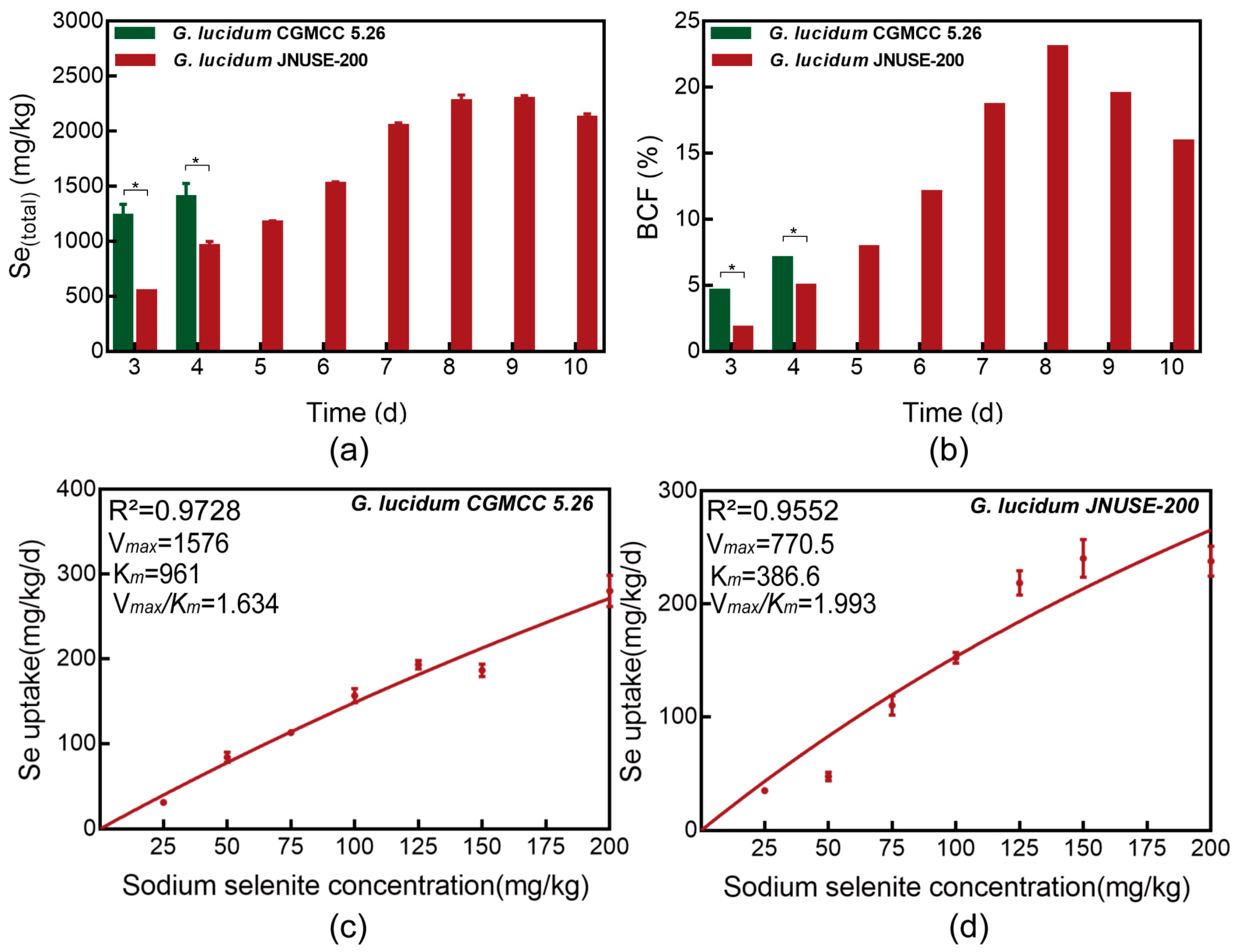
3.3. Total Se Accumulated in G. lucidum CGMCC 5.26 and G. lucidum JNUSE-200
3.4. Uptake Dynamics of Selenite in G. lucidum CGMCC 5.26 and G. lucidum JNUSE-200
3.5. XPS Analysis of Se in G. lucidum JNUSE-200 Mycelia
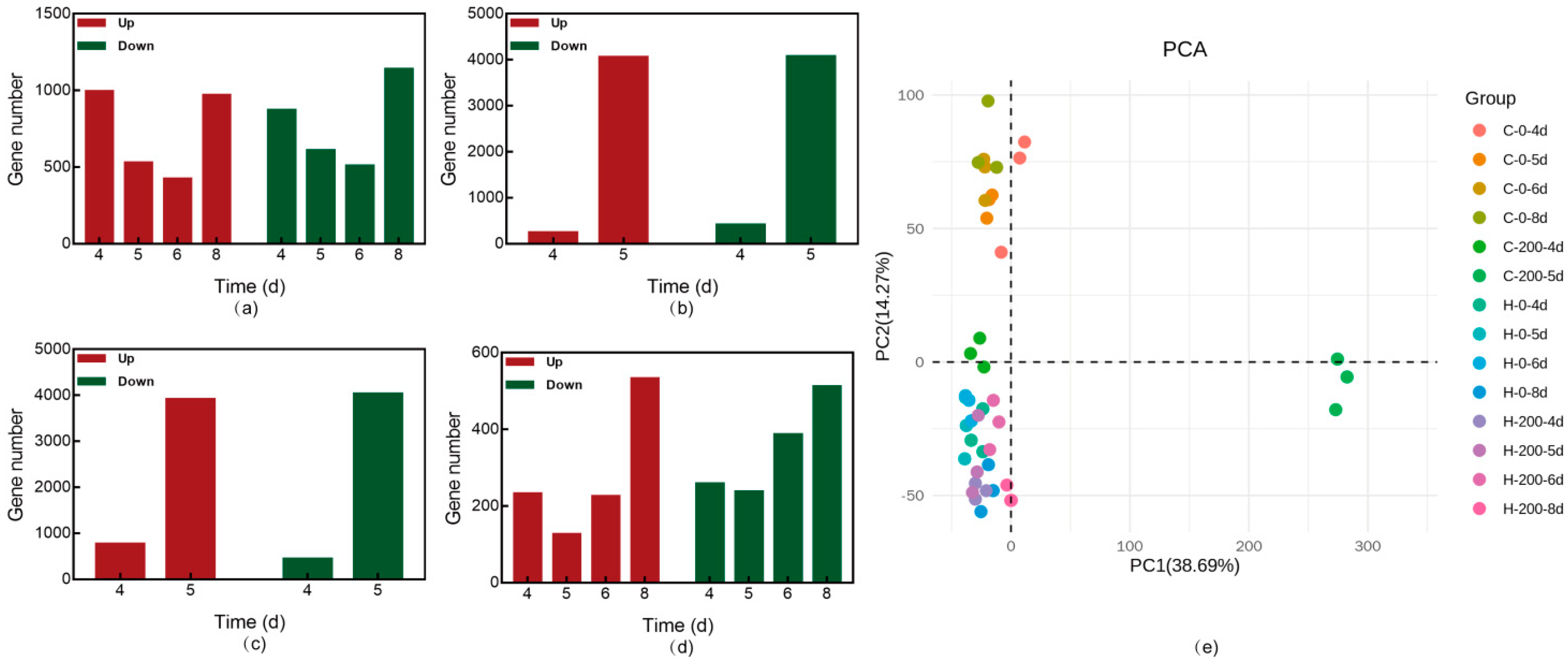
3.6. Transcriptome Analysis of G. lucidum CGMCC 5.26 and G. lucidum JNUSE-200 in Response to Selenite
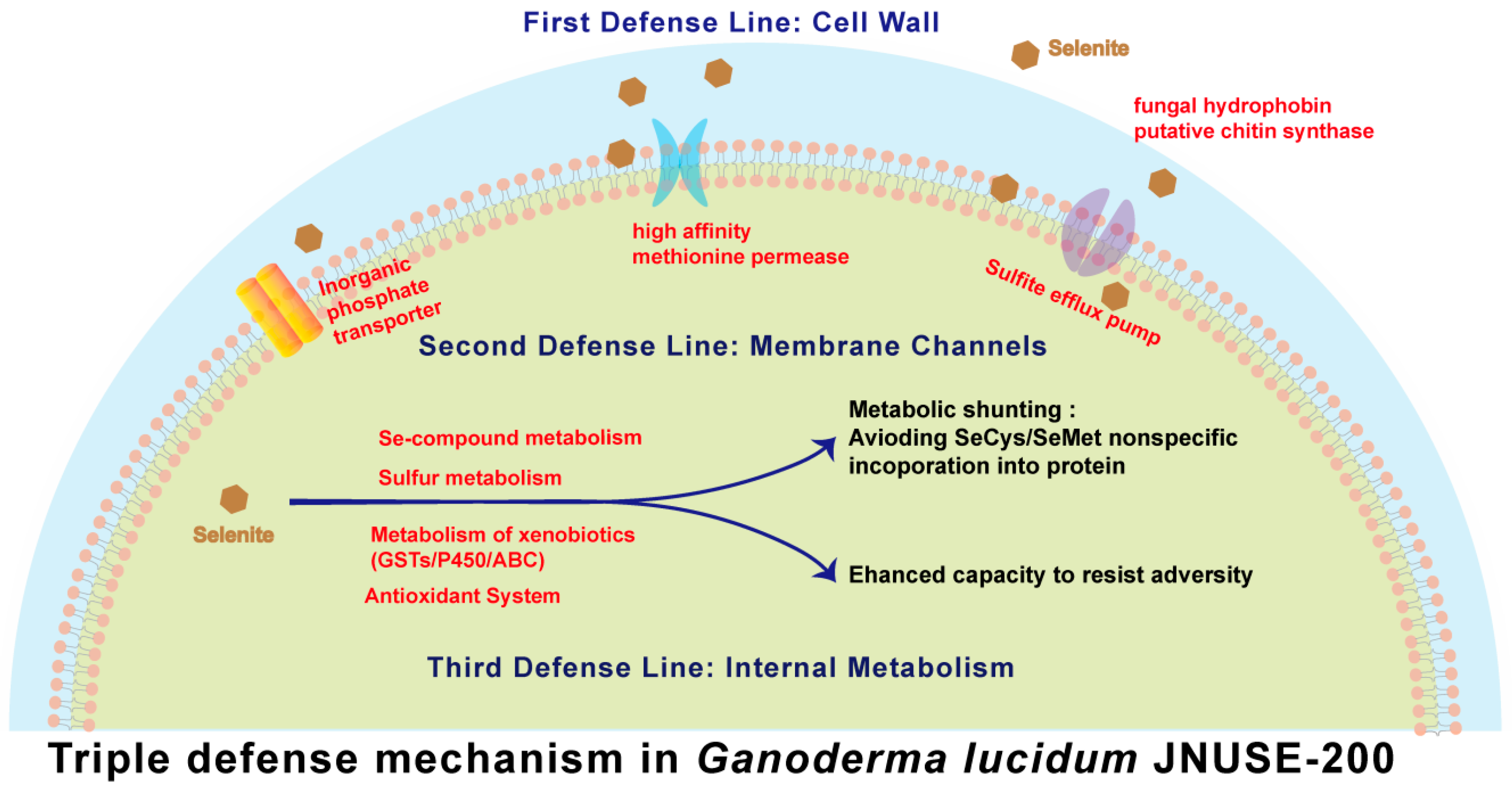
3.6.1. Modulation Selenite Tolerance via the Cell Wall
3.6.2. Modulation Selenite Tolerance via Membrane Channels
3.6.3. Modulation Selenite Tolerance via Internal Metabolic Occurrence
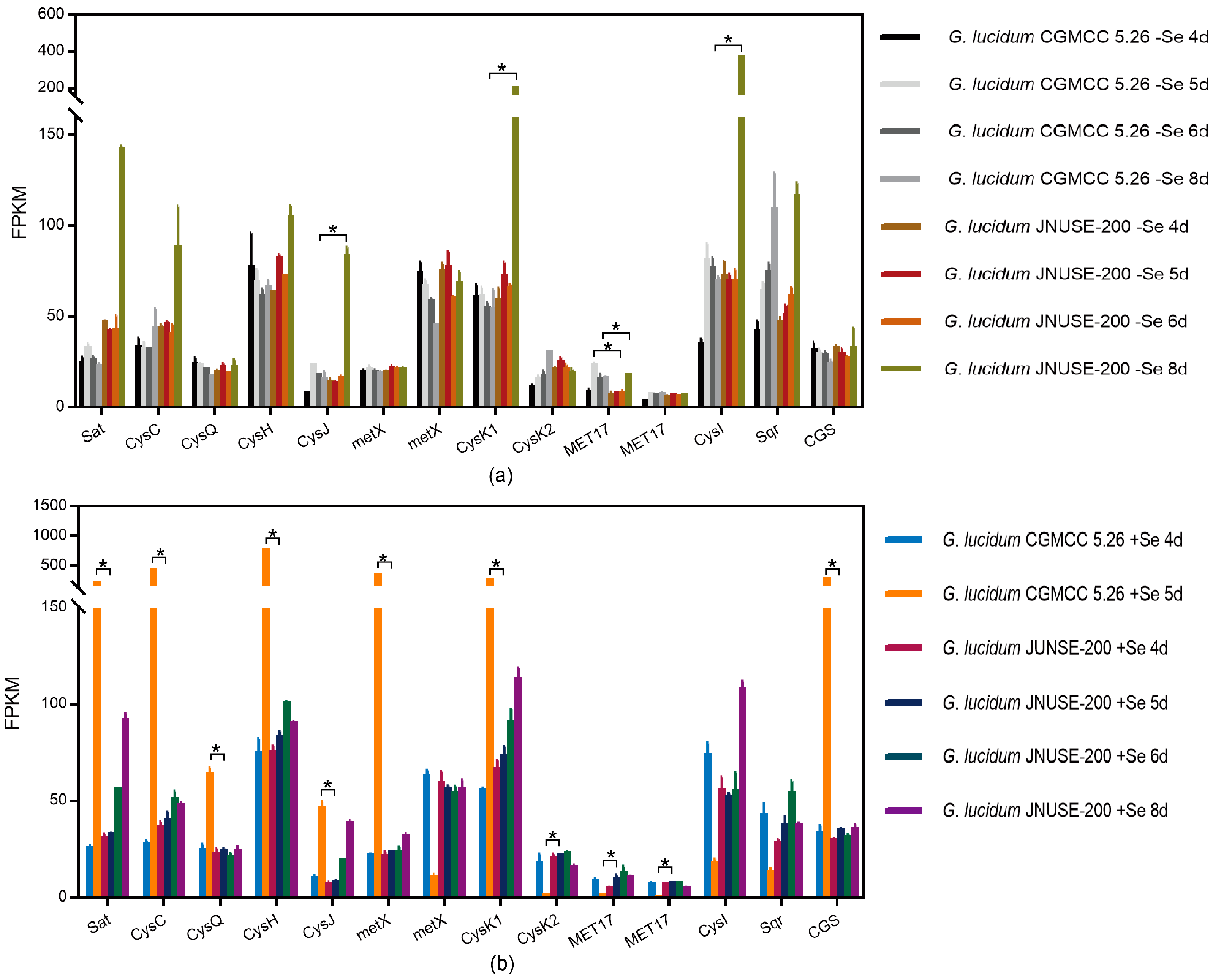
- (1)
- Se compound metabolism
- (2)
- Sulfur metabolism
- (3)
- Metabolism of xenobiotics
- (4)
- Antioxidant defense system
3.6.4. Transcription Factors
3.7. Correlation Coefficients: Transcription Factor–Key Gene–Se Bioaccmulation Capacity
3.8. RT-qPCR Verification of Differential Expression Genes
3.9. Mechanism of Enhanced Selenite Tolerance in G. lucidum JNUSE-200
3.10. Implication
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, H.; Yang, X.; Ning, Z.; Kwon, S.Y.; Li, M.; Tack, F.M.L.; Kwon, E.E.; Rinklebe, J.; Yin, R. The beneficial and hazardous effects of selenium on the health of the soil-plant-human system: An overview. J. Hazard. Mater. 2022, 422, 126876. [Google Scholar] [CrossRef] [PubMed]
- Wadgaonkar, S.L.; Nancharaiah, Y.V.; Esposito, G.; Lens, P.N.L. Environmental impact and bioremediation of seleniferous soils and sediments. Crit. Rev. Biotechnol. 2018, 38, 941–956. [Google Scholar] [CrossRef] [PubMed]
- Eswayah, A.S.; Smith, T.J.; Gardiner, P.H.E. Microbial transformations of selenium species of relevance to bioremediation. Appl. Environ. Microb. 2016, 82, 4848–4859. [Google Scholar] [CrossRef] [PubMed]
- Piacenza, E.; Presentato, A.; Zonaro, E.; Lampis, S.; Vallini, G.; Turner, R.J. Microbial-based bioremediation of selenium and tellurium compounds. Biosorption 2018, 41, 1–8. [Google Scholar] [CrossRef]
- Gan, X.Y.; Huang, J.C.; Zhang, M.P.; Zhou, C.Q.; He, S.B.; Zhou, W.L. Remediation of selenium-contaminated soil through combined use of earthworm Eisenia fetida and organic materials. J. Hazard. Mater. 2021, 405, 124212. [Google Scholar] [CrossRef] [PubMed]
- Pyrzynska, K.; Sentkowska, A. Selenium in plant foods: Speciation analysis, bioavailability, and factors affecting composition. Crit. Rev. Food Sci. 2021, 61, 1340–1352. [Google Scholar] [CrossRef] [PubMed]
- Ruj, B.; Bishayee, B.; Chatterjee, R.P.; Mukherjee, A.; Saha, A.; Nayak, J.; Chakrabortty, S. An economical strategy towards the managing of selenium pollution from contaminated water: A current state-of-the-art review. J. Environ. Manag. 2022, 304, 114143. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.M.; Zhu, S.; Li, Y.R.; Xu, S.; Shi, G.Y.; Ding, Z.Y. Effect of selenium on mushroom growth and metabolism: A review. Trends Food Sci. Tech. 2021, 118, 328–340. [Google Scholar] [CrossRef]
- Sahithya, K.; Mouli, T.; Ankita, B.; Mercy, S.T. Remediation potential of mushrooms and their spent substrate against environmental contaminants: An overview. Biocatal. Agric. Biotechnol. 2022, 42, 102323. [Google Scholar] [CrossRef]
- Irish, K.P.; Harvey, M.A.; Erskine, P.D.; Ent, A. van der. Root foraging and selenium uptake in the Australian hyperaccumulator Neptunia amplexicaulis and non-accumulator Neptunia gracilis. Plant Soil. 2021, 462, 219–233. [Google Scholar] [CrossRef]
- Cappa, J.J.; Yetter, C.; Fakra, S.; Cappa, P.J.; DeTar, R.; Landes, C.; Pilon-Smits, E.A.H.; Simmons, M.P. Evolution of selenium hyperaccumulation in Stanleya (Brassicaceae) as inferred from phylogeny, physiology and X-ray microprobe analysis. New Phytol. 2015, 205, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.L.; Tamaoki, M.; Stushnoff, C.; Quinn, C.F.; Cappa, J.J.; Devonshire, J.; Fakra, S.C.; Marcus, M.A.; McGrath, S.P.; Van Hoewyk, D.; et al. Molecular mechanisms of selenium tolerance and hyperaccumulation in Stanleya pinnata. Plant Physiol. 2010, 153, 1630–1652. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Cappa, J.J.; Harris, J.P.; Edger, P.P.; Zhou, W.; Pires, J.C.; Adair, M.; Unruh, S.A.; Simmons, M.P.; Schiavon, M.; et al. Transcriptome-wide comparison of selenium hyperaccumulator and nonaccumulator Stanleya species provides new insight into key processes mediating the hyperaccumulation syndrome. Plant Biotechnol. J. 2018, 16, 1582–1594. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wu, L.Y.; Zhang, H.; Wu, W.J.; Zhang, M.M.; Li, X.F.; Wu, H. Regulation of the phenylpropanoid pathway: A mechanism of selenium tolerance in peanut (Arachis hypogaea L.) seedlings. J. Agric. Food Chem. 2016, 64, 3626–3635. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Ying, H.Q.; Yang, X.B.; Gao, Y.; Li, T.; Wu, B.; Ren, M.; Zhang, Z.X.; Ding, J.; Gao, J.H.; et al. The Cardamine enshiensis genome reveals whole genome duplication and insight into selenium hyperaccumulation and tolerance. Cell Discov. 2021, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Both, E.B.; Shao, S.X.; Xiang, J.Q.; Jókai, Z.; Yin, H.Q.; Liu, Y.F.; Magyar, A.; Dernovics, M. Selenolanthionine is the major water-soluble selenium compound in the selenium tolerant plant Cardamine violifolia. BBA Gen. Subj. 2018, 1862, 2354–2362. [Google Scholar] [CrossRef] [PubMed]
- Kök, A.B.; Mungan, M.D.; Doganlar, S.; Frary, A. Transcriptomic analysis of selenium accumulation in Puccinellia distans (Jacq.) Parl., a boron hyperaccumulator. Chemosphere 2020, 245, 125665. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Xiao, Y.Q.; Wu, H. Selenium accumulation, speciation, and its effect on nutritive value of Flammulina velutipes (Golden needle mushroom). Food Chem. 2021, 350, 128667. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Li, H.F.; Zhao, G.S.; Guo, Y.B. Selenium enriched Hypsizygus marmoreus, a potential food supplement with improved Se bioavailability. LWT-Food Sci. Technol. 2021, 140, 110819. [Google Scholar] [CrossRef]
- Zhou, F.; Yang, W.X.; Wang, M.K.; Miao, Y.X.; Cui, Z.W.; Li, Z.; Liang, D.L. Effects of selenium application on Se content and speciation in Lentinula edodes. Food Chem. 2018, 265, 182–188. [Google Scholar] [CrossRef]
- Oliveira, A.P.de.; Naozuka, J.; Landero-Figueroa, J.A. Effects of Se(IV) or Se(VI) enrichment on proteins and protein-bound Se distribution and Se bioaccessibility in oyster mushrooms. Food Chem. 2022, 383, 132582. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.M.; Zhu, S.; Wang, L.L.; Wei, Z.Y.; Zhao, L.T.; Shi, G.Y.; Ding, Z.Y. Influence of selenium biofortification on the growth and bioactive metabolites of Ganoderma lucidum. Foods 2021, 10, 1860. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.M.; Zhu, S.; Wang, Q.; Chen, L.; Li, Y.R.; Xu, S.; Gu, Z.H.; Shi, G.Y.; Ding, Z.Y. Pivotal biological processes and proteins for selenite reduction and methylation in Ganoderma lucidum. J. Hazard. Mater. 2023, 444, 130409. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.B.; Xu, M.M.; Wang, Q.; Wang, F.; Zheng, H.H.; Gu, Z.H.; Li, Y.R.; Shi, G.Y.; Ding, Z.Y. Development of an efficient strategy to improve extracellular polysaccharide production of Ganoderma lucidum using L-phenylalanine as an enhancer. Front. Microbiol. 2019, 10, 2306. [Google Scholar] [CrossRef] [PubMed]
- Huo, K.Y.; Zhao, F.J.; Zhang, F.; Liu, R.H.; Yang, C. Morphology engineering: A new strategy to construct microbial cell factories. World J. Microb. Biot. 2020, 36, 127. [Google Scholar] [CrossRef] [PubMed]
- Trippe, R.C.; Pilon-Smits, E.A.H. Selenium transport and metabolism in plants: Phytoremediation and biofortification implications. J. Hazard. Mater. 2021, 404, 124178. [Google Scholar] [CrossRef] [PubMed]
- Mal, J.; Sinharoy, A.; Lens, P.N.L. Simultaneous removal of lead and selenium through biomineralization as lead selenide by anaerobic granular sludge. J. Hazard. Mater. 2021, 420, 126663. [Google Scholar] [CrossRef] [PubMed]
- Brauer, V.S.; Pessoni, A.M.; Freitas, M.S.; Cavalcanti-Neto, M.P.; Ries, L.N.A.; Almeida, F. Chitin Biosynthesis in Aspergillus Species. J. Fungi 2023, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Z.; Li, J.H.; Yang, H.T.; Shin, H.J. Fungal and mushroom hydrophobins: A review. Int. J. Med. Mushrooms 2017, 15, 1–7. [Google Scholar] [CrossRef]
- Alemayehu, H.A.; Dumbuya, G.; Hasan, M.; Tadesse, T.; Nakajyo, S.; Fujioka, T.; Abe, A.; Matsunami, M.; Shimono, H. Genotypic variation in cold tolerance of 18 Ethiopian rice cultivars in relation to their reproductive morphology. Field Crop Res. 2021, 262, 108042. [Google Scholar] [CrossRef]
- Kuang, L.H.; Shen, Q.F.; Chen, L.Y.; Ye, L.Z.; Yan, T.; Chen, Z.H.; Waugh, R.; Li, Q.; Huang, L.; Cai, S.G.; et al. The genome and gene editing system of sea barleygrass provide a novel platform for cereal domestication and stress tolerance studies. Plant Commun. 2022, 3, 100333. [Google Scholar] [CrossRef] [PubMed]
- Léchenne, B.; Reichard, U.; Zaugg, C.; Fratti, M.; Kunert, J.; Boulat, O.; Monod, M. Sulphite efflux pumps in Aspergillus fumigatus and dermatophytes. Microbiology 2007, 153, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Pinson, B.; Sagot, I.; Daignan-Fornier, B. Identification of genes affecting selenite toxicity and resistance in Saccharomyces cerevisiae. Mol. Microbiol. 2000, 36, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Gong, A.; Liu, W.Y.; Lin, Y.L.; Huang, L.L.; Xie, Z.X. Adaptive laboratory evolution reveals the selenium efflux process to improve selenium tolerance mediated by the membrane sulfite pump in Saccharomyces cerevisiae. Microbiol. Spectr. 2023, 11, e01326-23. [Google Scholar] [CrossRef] [PubMed]
- Pitsch, E.; Hughes, A. Elucidating the cellular mechanism of methionine toxicity. FASEB J. 2022, 36. [Google Scholar] [CrossRef]
- Qu, L.L.; Xu, J.Y.; Dai, Z.H.; Elyamine, A.M.; Huang, W.X.; Han, D.; Dang, B.J.; Xu, Z.C.; Jia, W. Selenium in soil-plant system: Transport, detoxification and bioremediation. J. Hazard. Mater. 2023, 452, 131272. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.P.; Shao, H.F.; Huang, H.G.; Shen, Y.; Wang, L.Z.; Wu, F.Y.; Han, D.; Song, J.Y.; Jia, H.F. Overexpression of the phosphate transporter gene OsPT8 improves the Pi and selenium contents in Nicotiana tabacum. Environ. Exp. Bot. 2017, 137, 158–165. [Google Scholar] [CrossRef]
- Jiang, L.; Cao, H.M.; Chen, Z.P.; Liu, C.X.; Cao, S.Q.; Wei, Z.J.; Han, Y.; Gao, Q.C.; Wang, W.Y. Cytokinin is involved in TPS22-mediated selenium tolerance in Arabidopsis thaliana. Ann. Bot. 2018, 122, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Y.; Zhou, Y.F.; Zhou, D.Z.; Hong, J.; Zhao, L.M.; Bu, G.J.; Chen, F.; Tang, L. The ethylene response factor RAP2.6 plays a positive role in the regulation of selenium tolerance in Arabidopsis thaliana. Plant Growth Regul. 2023, 99, 241–250. [Google Scholar] [CrossRef]
- El-Badri, A.M.; Hashem, A.M.; Batool, M.; Sherif, A.; Nishawy, E.; Ayaad, M.; Hassan, H.M.; Elrewainy, I.M.; Wang, J.; Kuai, J.; et al. Comparative efficacy of bio-selenium nanoparticles and sodium selenite on morpho-physiochemical attributes under normal and salt stress conditions, besides selenium detoxification pathways in Brassica napus L. J. Nanobiotechnol. 2022, 20, 163. [Google Scholar] [CrossRef]
- Cui, L.W.; Zhao, J.T.; Chen, J.Y.; Zhang, W.; Gao, Y.X.; Li, B.; Li, Y.F. Translocation and transformation of selenium in hyperaccumulator plant Cardamine enshiensis from Enshi, Hubei, China. Plant Soil. 2018, 425, 577–588. [Google Scholar] [CrossRef]
- Baggio, G.; Groves, R.A.; Chignola, R.; Piacenza, E.; Presentato, A.; Lewis, I.A.; Lampis, S.; Vallini, G.; Turner, R.J. Untargeted metabolomics investigation on selenite reduction to elemental selenium by Bacillus mycoides SeITE01. Front. Microbiol. 2021, 12, 711000. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.Y.; Lin, W.Q.; Jiao, H.P.; Liu, J.G.; Chan, L.; Liu, X.Y.; Wang, R.; Chen, T.F. Uptake, transport, and metabolism of selenium and its protective effects against toxic metals in plants: A review. Metallomics 2021, 13, mfab040. [Google Scholar] [CrossRef] [PubMed]
- Lampis, S.; Vallini, G. Microbial reduction of selenium oxyanions: Energy-yielding and detoxification reactions. In Environmental Technologies to Treat Selenium Pollution: Principles and Engineering; Lens, P.N.L., Pakshirajan, K., Eds.; Chapter 3; IWA Publishing: London, UK, 2021. [Google Scholar] [CrossRef]
- Chauhan, R.; Awasthi, S.; Srivastava, S.; Dwivedi, S.; Pilon-Smits, E.A.H.; Dhankher, O.P.; Tripathi, R.D. Understanding selenium metabolism in plants and its role as a beneficial element. Crit. Rev. Environ. Sci. Technol. 2019, 49, 1937–1958. [Google Scholar] [CrossRef]
- Gupta, M.; Gupta, S. An Overview of Selenium Uptake, Metabolism, and Toxicity in Plants. Front. Plant Sci. 2017, 7, 2074. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.A.D.; Gomes, F.T.D.; Antonio, J.R.R.; Guilherme, L.R.G.; Liu, J.P.; Li, L.; Silva, M.L.D. Sulfate availability and soil selenate adsorption alleviate selenium toxicity in rice plants. Environ. Exp. Bot. 2022, 201, 104971. [Google Scholar] [CrossRef]
- El Mehdawi, A.F.; Jiang, Y.; Guignardi, Z.S.; Esmat, A.; Pilon, M.; Pilon-Smits, E.A.H.; Schiavon, M. Influence of sulfate supply on selenium uptake dynamics and expression of sulfate/selenate transporters in selenium hyperaccumulator and nonhyperaccumulator Brassicaceae. New Phytol. 2018, 217, 194–205. [Google Scholar] [CrossRef]
- Kurmanbayeva, A.; Bekturova, A.; Soltabayeva, A.; Oshanova, D.; Nurbekova, Z.; Srivastava, S.; Tiwari, P.; Dubey, A.K.; Sagi, M. Active O-acetylserine-(thiol) lyase A and B confer improved selenium resistance and degrade l-Cys and l-SeCys in Arabidopsis. J. Exp. Bot. 2022, 73, 2525–2539. [Google Scholar] [CrossRef]
- Van Hoewyk, D.; Garifullina, G.F.; Ackley, A.R.; Abdel-Ghany, S.E.; Marcus, M.A.; Fakra, S.; Ishiyama, K.; Inoue, E.; Pilon, M.; Takahashi, H.; et al. Overexpression of AtCpNifS enhances selenium tolerance and accumulation in Arabidopsis. Plant Physiol. 2005, 139, 1518–1528. [Google Scholar] [CrossRef]
- Tan, L.R.; Lu, Y.C.; Zhang, J.J.; Luo, F.; Yang, H. A collection of cytochrome P450 monooxygenase genes involved in modification and detoxification of herbicide atrazine in rice (Oryza sativa) plants. Ecotoxicol. Environ. Saf. 2015, 119, 25–34. [Google Scholar] [CrossRef]
- Wu, X.; Chen, J.H.; Yue, X.M.; Wei, X.N.; Zou, J.W.; Chen, Y.H.; Su, N.N.; Cui, J. The zinc-regulated protein (ZIP) family genes and glutathione s-transferase (GST) family genes play roles in Cd resistance and accumulation of pak choi (Brassica campestris ssp. chinensis). Ecotoxicol. Environ. Saf. 2019, 183, 109571. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, B.; Hao, X.; Diao, J.; Cao, J.X.; Tan, R.A.; Ma, W.; Ma, L. Functional analysis of 3 genes in xenobiotic detoxification pathway of Bursaphelenchus xylophilus against matrine. Pestic. Biochem. Phys. 2023, 190, 105334. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tao, M.Z.; Meng, Y.; Zhu, X.Y.; Qian, L.W.; Shah, A.; Wang, W.; Cao, S.Q. The role of WRKY47 gene in regulating selenium tolerance in Arabidopsis thaliana. Plant Biotechnol. Rep. 2020, 14, 121–129. [Google Scholar] [CrossRef]
- Jiang, L.; Yang, J.; Liu, C.X.; Chen, Z.P.; Yao, Z.C.; Cao, S.Q. Overexpression of ethylene response factor ERF96 gene enhances selenium tolerance in Arabidopsis. Plant Physiol. Biochem. 2020, 149, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Schiavon, M.; Lima, L.W.; Jones, R.R.; El Mehdawi, A.F.; Royer, S.; Zeng, Z.H.; Hu, Y.G.; Pilon-Smits, E.A.H.; Pilon, M. Comparison of ATP sulfurylase 2 from selenium hyperaccumulator Stanleya pinnata and non-accumulator Stanleya elata reveals differential intracellular localization and enzyme activity levels. BBA Gen. Subj. 2018, 1862, 2363–2371. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Uraji, M.; Shimoishi, Y.; Mori, I.C.; Nakamura, Y.; Murata, Y. Mechanisms of the Selenium Tolerance of the Arabidopsis thaliana Knockout Mutant of Sulfate Transporter SULTR1;2. Biosci. Biotechnol. Biochem. 2012, 76, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Grant, K.; Carey, N.M.; Mendoza, M.; Schulze, J.; Pilon, M.; Pilon-Smits, E.A.H.; Van Hoewyk, D. Adenosine 5’-phosphosulfate reductase (APR2) mutation in Arabidopsis implicates glutathione deficiency in selenate toxicity. Biochem. J. 2011, 438, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Chen, Z.P.; Gan, Q.C.; Ci, L.K.; Cao, S.Y.; Han, Y.; Wang, W.Y. Loss-of-function mutations in the APX1 gene result in enhanced selenium tolerance in Arabidopsis thaliana. Plant Cell Environ. 2016, 39, 2133–2144. [Google Scholar] [CrossRef] [PubMed]
- Tamaoki, M.; Freeman, J.L.; Pilon-Smits, E.A.H. Cooperative ethylene and jasmonic acid signaling regulates selenite resistance in Arabidopsis. Plant Physiol. 2008, 146, 1219–1230. [Google Scholar] [CrossRef]
- Cardoso, A.A.D.; Namorato, F.A.; Guilherme, L.R.G.; Silva, M.L.D.; Liu, J.P.; Li, L. Glutathione is involved in selenium detoxification and suppresses the selenate-induced SULTR1;1 gene expression in plants. Environ. Exp. Bot. 2023, 213, 105424. [Google Scholar] [CrossRef]
- Rao, S.; Yu, T.; Cong, X.; Lai, X.Z.; Xiang, J.Q.; Cao, J.; Liao, X.L.; Gou, Y.Y.; Chao, W.; Xue, H.; et al. Transcriptome, proteome, and metabolome reveal the mechanism of tolerance to selenate toxicity in Cardamine violifolia. J. Hazard. Mater. 2021, 406, 124283. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, H.; Lai, F.R.; Wu, H. Germinating Peanut (Arachis hypogaea L.) Seedlings Attenuated Selenite-Induced Toxicity by Activating the Antioxidant Enzymes and Mediating the Ascorbate-Glutathione Cycle. J. Agric. Food Chem. 2016, 64, 1298–1308. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.J.; Tian, Y.S.; Wang, B.; Peng, R.H.; Xu, J.; Fu, X.Y.; Han, H.J.; Wang, L.J.; Zhang, W.H.; Deng, Y.D.; et al. Enhanced phytoremediation of selenium using genetically engineered rice plants. J. Plant Physiol. 2022, 271, 153665. [Google Scholar] [CrossRef] [PubMed]



| Plant | Target Gene | Mechanism | Reference |
|---|---|---|---|
| Arabidopsis | Transporter gene | Sultr1;2 mutation conferred Se tolerance | [57] |
| Metabolism related gene | O-acetylserine-(thiol) lyase A and B degrade L-Cys and L-SeCys and confer improved resistance | [49] | |
| APR2 mutant exhibited decreased selenate tolerance through disruption glutathione biosynthesis | [58] | ||
| Increased AtCpNifS expression enhanced selenite tolerance | [50] | ||
| Loss of function of terpenoid synthase (TPS22) enhanced Se tolerance | [38] | ||
| Antioxidant gene | Loss of function of ascorbate peroxidase (APX1) enhanced selenite tolerance | [59] | |
| Signaling pathway gene | Ethylene and jasmonic acid signaling regulated selenite resistance | [60] | |
| Transcription factor gene | Gain- and loss-of-function mutations in WRKY47 enhanced the sensitivity to Se stress | [54] | |
| Increased ERF96 expression enhanced Se tolerance | [55] | ||
| Increased RAP2.6 expression improved resistance to Se | [39] | ||
| Metabolites | Glutathione | [61] | |
| Stanleya pinnata | Metabolism related gene | The high ATP sulfurylase 2 activity in the cytosol and concomitant reduced ATPS activity in the plastids diverted Se fluxes | [56] |
| Metabolism related gene | Up-regulated JA, SA and ethylene-mediated defense systems, elevated expression of genes in sulphate/selenate uptake and assimilation or in antioxidant activity | [13] | |
| Antioxidant gene | Increased antioxidants and up-regulated sulfur assimilation | [12] | |
| Cardamine violifolia | Metabolism related gene | The downregulation of cysteine-rich kinases and calcium proteins enhanced Se tolerance | [62] |
| Metabolites | Flavonoid, glutathione, and lignin | [15] | |
| Selenolanthionine | [16] | ||
| Arachis hypogaea L. | Antioxidant gene | Activating the antioxidant enzymes and mediating the ascorbate-glutathione cycle | [63] |
| Nicotiana tabacum | Transporter gene | Increased Pi transporter OsPT8 gene expression improved Se content | [37] |
| Rice ZH11 | Metabolism related gene | Increased selenocysteine lyase and selenocysteine methyltransferase gene expressions enhanced selenate and selenite tolerance | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Meng, Q.; Zhu, S.; Yu, R.; Chen, L.; Shi, G.; Wong, K.-H.; Fan, D.; Ding, Z. The Performance and Evolutionary Mechanism of Ganoderma lucidum in Enhancing Selenite Tolerance and Bioaccumulation. J. Fungi 2024, 10, 415. https://doi.org/10.3390/jof10060415
Xu M, Meng Q, Zhu S, Yu R, Chen L, Shi G, Wong K-H, Fan D, Ding Z. The Performance and Evolutionary Mechanism of Ganoderma lucidum in Enhancing Selenite Tolerance and Bioaccumulation. Journal of Fungi. 2024; 10(6):415. https://doi.org/10.3390/jof10060415
Chicago/Turabian StyleXu, Mengmeng, Qi Meng, Song Zhu, Ruipeng Yu, Lei Chen, Guiyang Shi, Ka-Hing Wong, Daming Fan, and Zhongyang Ding. 2024. "The Performance and Evolutionary Mechanism of Ganoderma lucidum in Enhancing Selenite Tolerance and Bioaccumulation" Journal of Fungi 10, no. 6: 415. https://doi.org/10.3390/jof10060415
APA StyleXu, M., Meng, Q., Zhu, S., Yu, R., Chen, L., Shi, G., Wong, K.-H., Fan, D., & Ding, Z. (2024). The Performance and Evolutionary Mechanism of Ganoderma lucidum in Enhancing Selenite Tolerance and Bioaccumulation. Journal of Fungi, 10(6), 415. https://doi.org/10.3390/jof10060415






