Comparative Bioremediation of Tetradecane, Cyclohexanone and Cyclohexane by Filamentous Fungi from Polluted Habitats in Kazakhstan
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Fungal Cultures and Their Identification
2.2. Growth Experiment on the Fungal Isolation Substrates
2.3. Biodegradation
2.4. Determination of the Biomass
2.5. Extraction and Identification of the Biodegradation Products
3. Results
3.1. Identification of the Fungal Strains
3.2. Growth of the Fungal Strains on Hydrocarbons
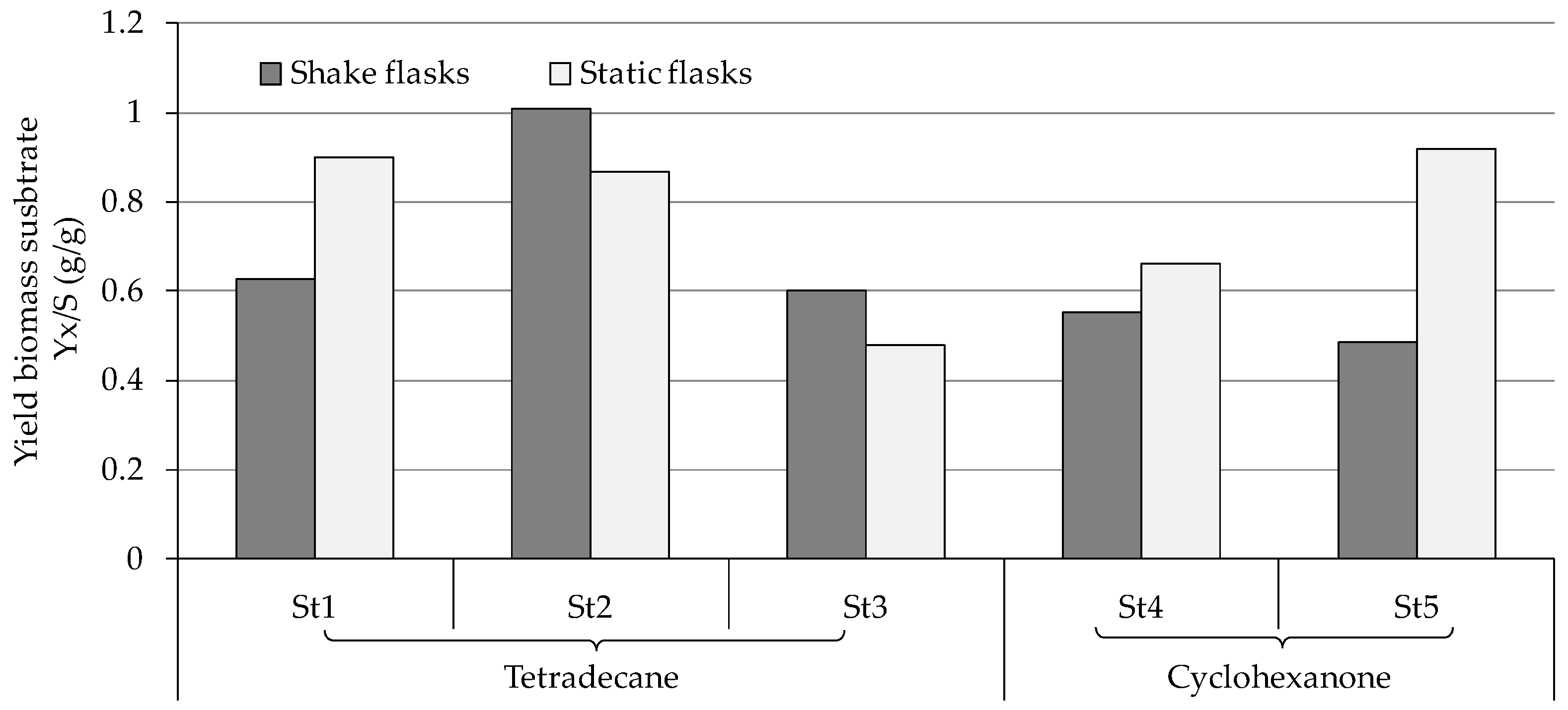
3.3. Ability of the Fungal Strains to Biodegrade Hydrocarbons
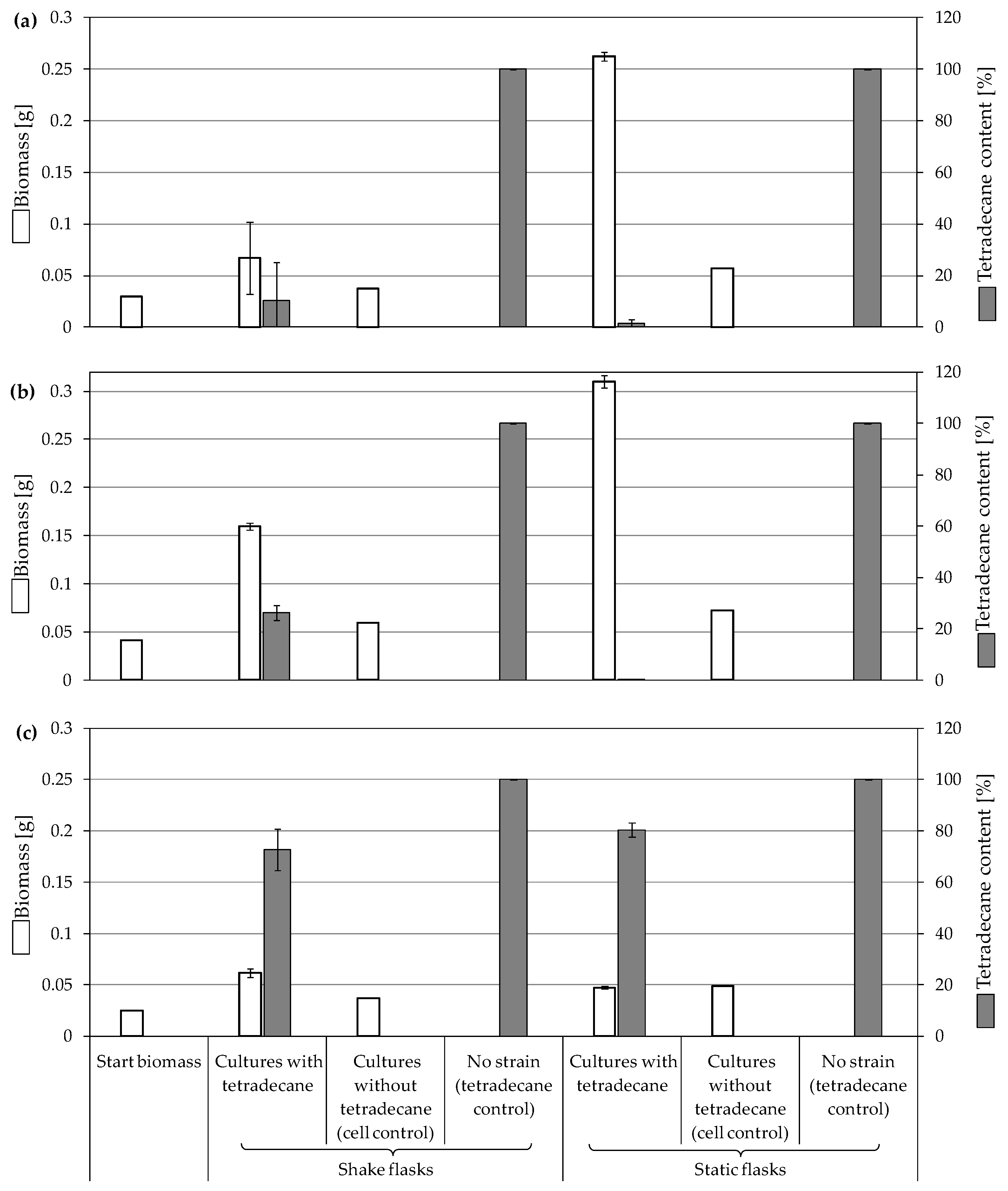
3.3.1. Biodegradation of Tetradecane
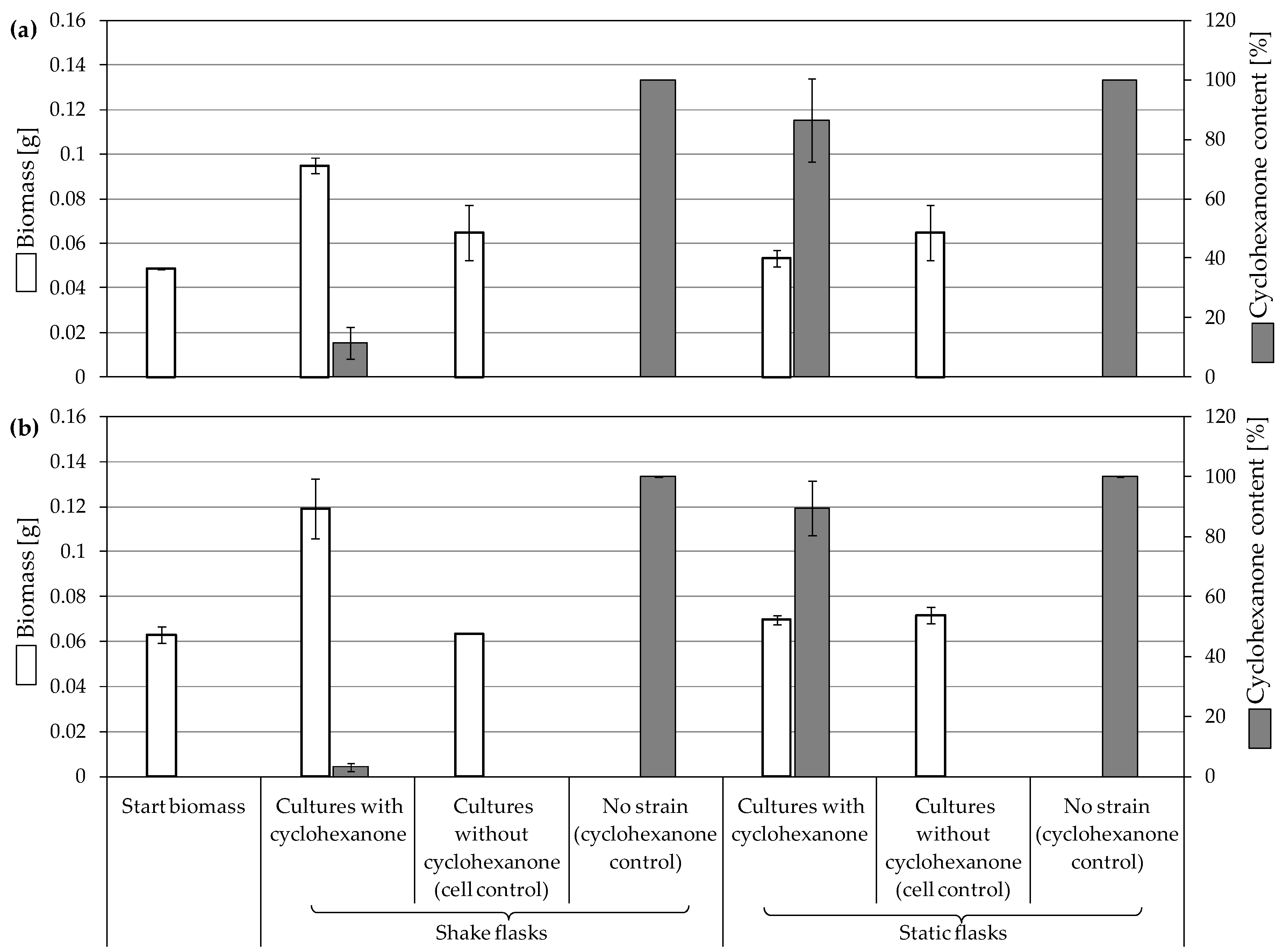
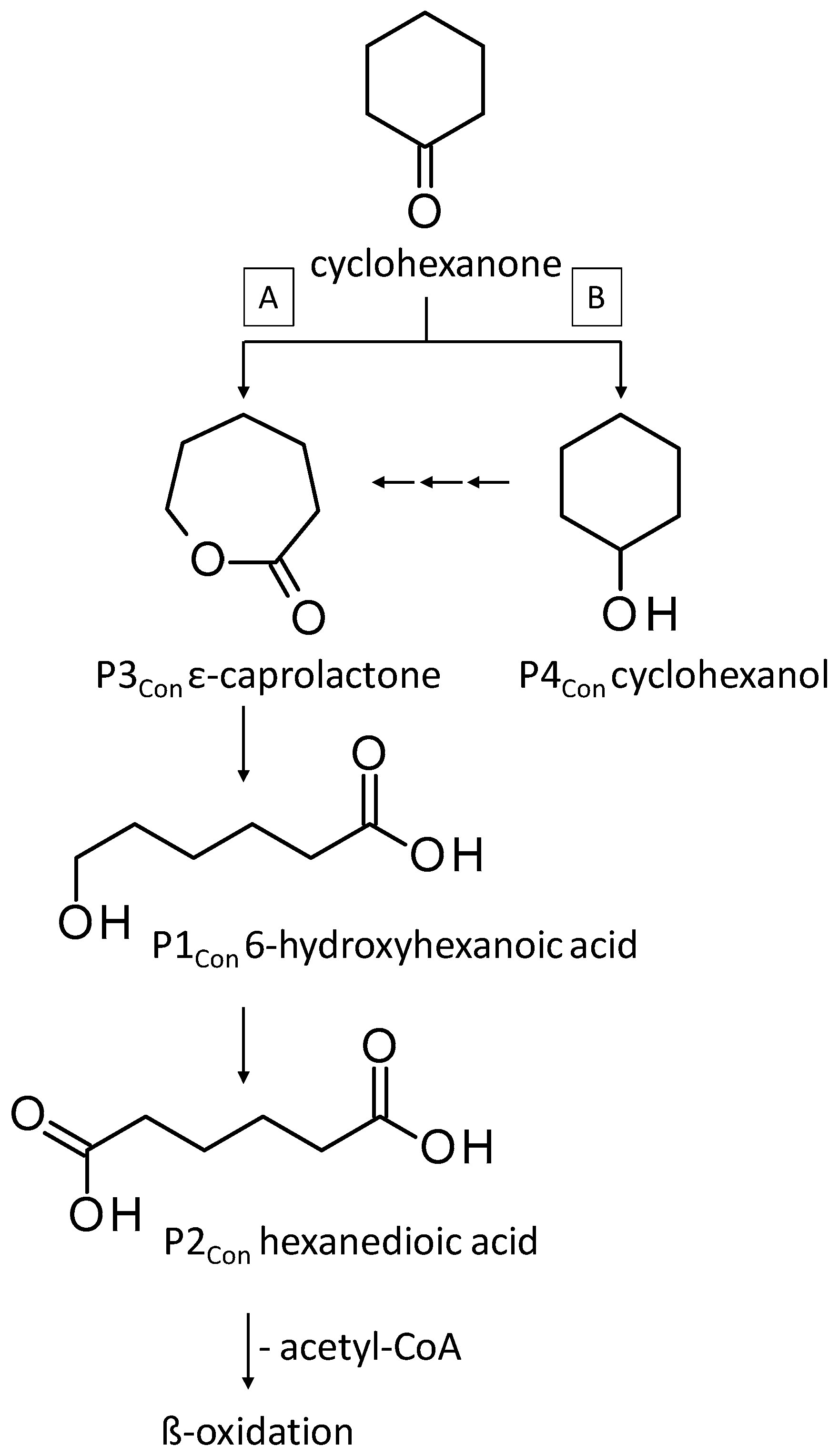
3.3.2. Biodegradation of Cyclohexanone
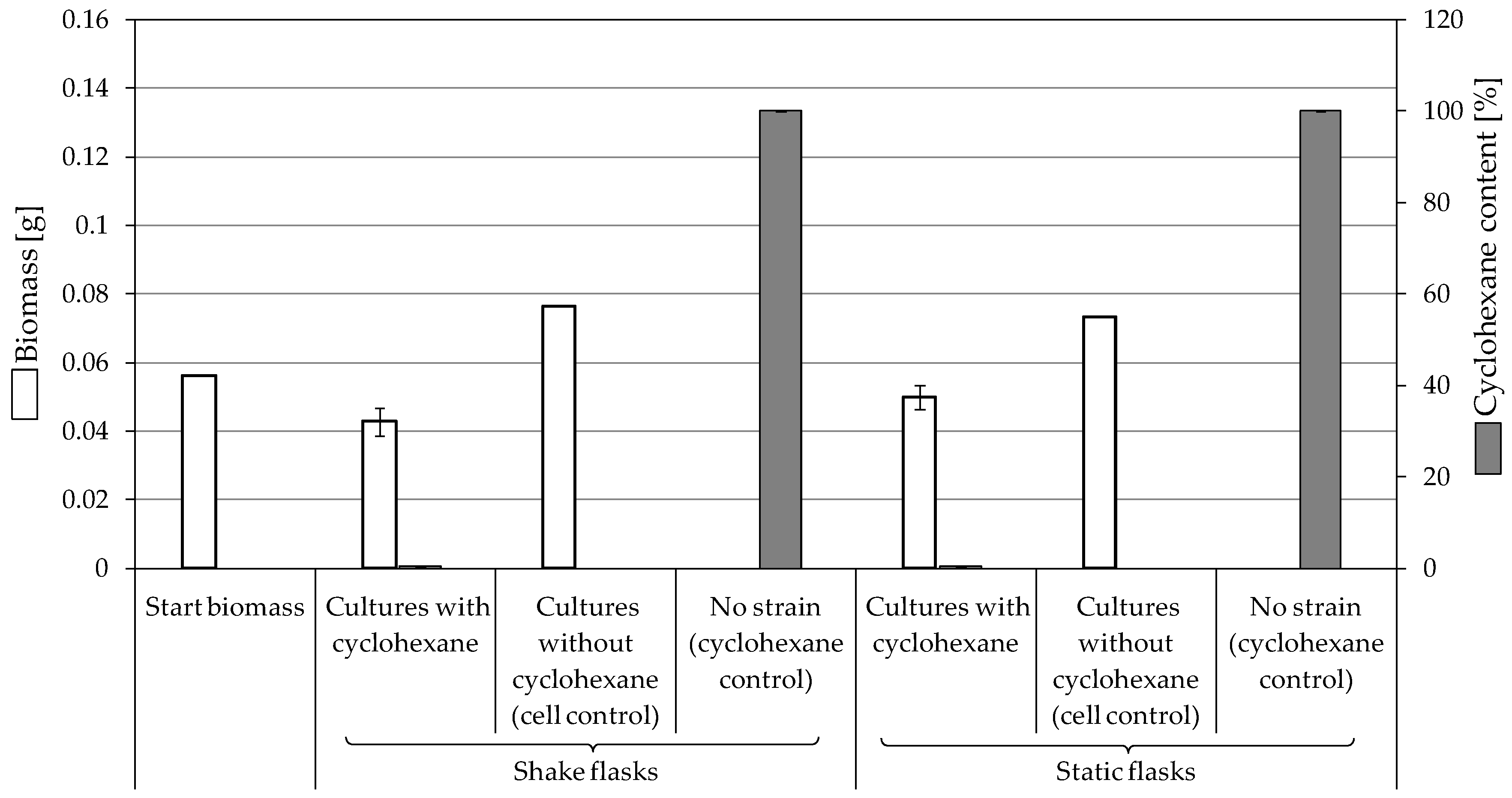
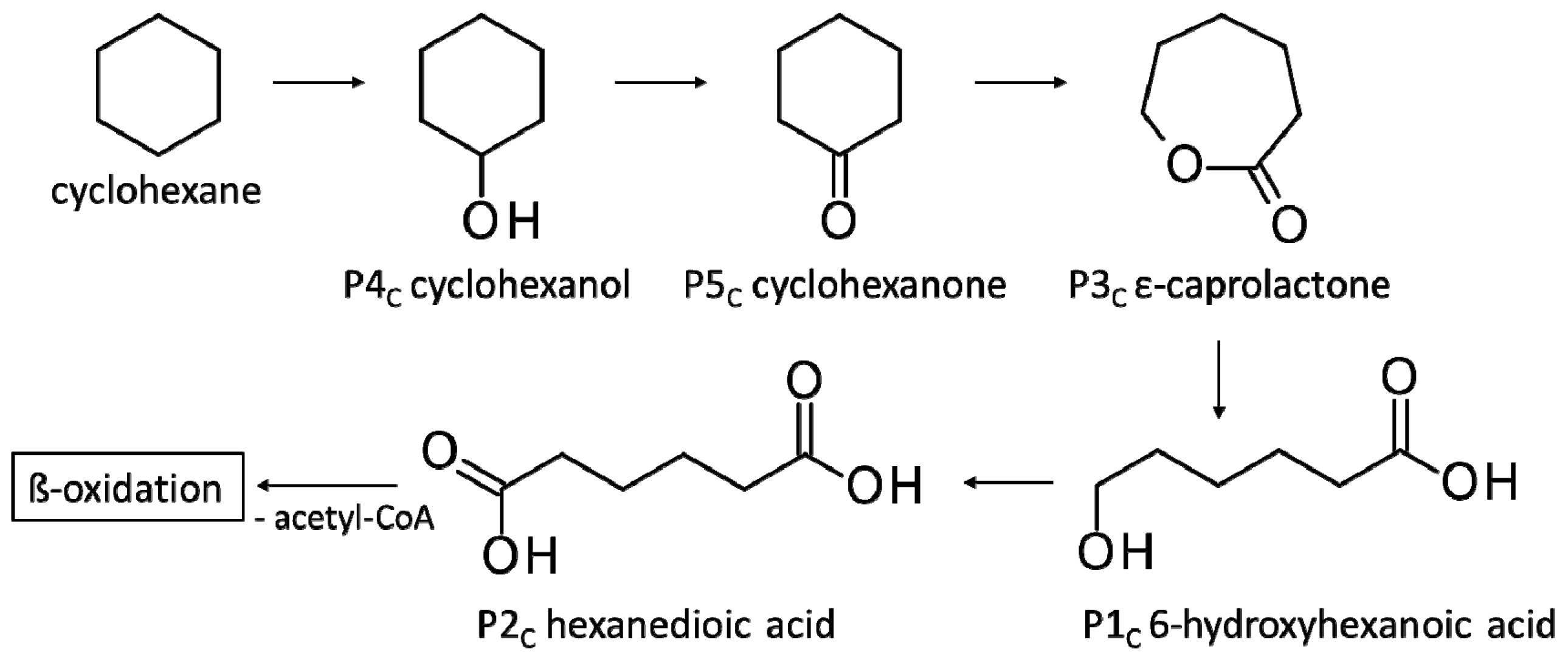
3.3.3. Biodegradation of Cyclohexane
4. Discussion
4.1. Consumption and Degradation of Tetradecane
4.2. Consumption and Degradation of Cyclohexanone and Cyclohexane
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Satubaldina, A. Bringing Kazakhstan to the World: Strategic Partnership, Energy Security: Macron’s Visit to Kazakhstan Redefines Bilateral Ties. The Astana Times. 3 November 2023. Available online: https://astanatimes.com/2023/11/strategic-partnership-energy-security-macrons-visit-to-kazakhstan-redefines-bilateral-ties/ (accessed on 30 November 2023).
- Zabanova, Y.; Williams, S.; Andresh, M.; Melnikov, Y.; Tumysheva, A. EU-Kazakhstan Green Hydrogen Partnership, Mapping Barriers and Establishing a Roadmap. EPICO KlimaInnovation (Energy and Climate Policy and Innovation Council e.V.); ISBN 978-3-98574-192-2. Available online: https://www.kas.de/documents/d/guest/eu-kazakhstan-green-hydrogen-partnership (accessed on 30 November 2023).
- Statista. Primary Energy Consumption Worldwide from 2019 to 2022, by Fuel Type. 2023. Available online: https://www.statista.com/statistics/265619/primary-energy-consumption-worldwide-by-fuel/ (accessed on 30 November 2023).
- Thakur, A.; Koul, B. Chapter 7—Impact of oil exploration and spillage on marine environments. In Advances in Oil-Water Separation; Papita, D., Suvendu, M., Jitendra, K.P., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 115–135. ISBN 978-03-238-997-89. [Google Scholar] [CrossRef]
- Zhuang, J.; Zhang, R.; Zeng, Y.; Dai, T.; Ye, Z.; Gao, Q.; Yang, Y.; Guo, X.; Li, G.; Zhou, J. Petroleum pollution changes microbial diversity and network complexity of soil profile in an oil refinery. Front. Microbiol. 2023, 14, 1193189. [Google Scholar] [CrossRef]
- Reuters. Gulf of Mexico Oil Spill Shuts in around 3% of Daily Output, November 23. Available online: https://www.reuters.com/business/energy/gulf-mexico-oil-spill-shuts-around-3-daily-output-2023-11-22/2023 (accessed on 30 November 2023).
- Liu, J.; Wang, L.; Chen, F.; Hu, W.; Dong, C.; Wang, Y.; Han, Y. Molecular characterization of hydrocarbons in petroleum by ultrahigh-resolution mass spectrometry. Energies 2023, 16, 4296. [Google Scholar] [CrossRef]
- Lestan, D.; Lamar, R.T. Development of fungal inocula for bioaugmentation of contaminated soils. Appl. Environ. Microbiol. 1996, 62, 2045–2052. [Google Scholar] [CrossRef]
- Hina Khatoon, H.; Rai, J.P.N.; Jillani, A. Chapter 7—Role of fungi in bioremediation of contaminated soil. In Fungi Bio-Prospects in Sustainable Agriculture, Environment and Nano-Technology; Vijay, K.S., Maulin, P.S., Shobhika, P., Ajay, K., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 121–156. ISBN 978-01-282-192-56. [Google Scholar] [CrossRef]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques–classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 2016, 32, 180. [Google Scholar] [CrossRef]
- Medaura, M.C.; Guivernau, M.; Moreno-Ventas, X.; Prenafeta-Boldú, F.X.; Viñas, M. Bioaugmentation of native fungi, an efficient strategy for the bioremediation of an aged industrially polluted soil with heavy hydrocarbons. Front. Microbiol. 2021, 12, 626436. [Google Scholar] [CrossRef]
- Gaid, M.; Pöpke, D.; Reinhard, A.; Berzhanova, R.; Mukasheva, T.; Urich, T.; Mikolasch, A. Characterization of the mycoremediation of n-alkanes and branched-chain alkanes by filamentous fungi from oil-polluted soil samples in Kazakhstan. Microorganisms 2023, 11, 2195. [Google Scholar] [CrossRef]
- Kristanti, R.A.; Hadibarata, T.; Toyama, T.; Tanaka, Y.; Mori, K. Bioremediation of crude oil by white rot fungi Polyporus sp. S133. J. Microbiol. Biotechnol. 2011, 21, 995–1000. [Google Scholar] [CrossRef]
- Mikolasch, A.; Donath, M.; Reinhard, A.; Herzer, C.; Zayadan, B.; Urich, T.; Schauer, F. Diversity and degradative capabilities of bacteria and fungi isolated from oil-contaminated and hydrocarbon-polluted soils in Kazakhstan. Appl. Microbiol. Biotechnol. 2019, 103, 7261–7274. [Google Scholar] [CrossRef]
- Al-Dhabaan, F.A. Mycoremediation of crude oil contaminated soil by specific fungi isolated from Dhahran in Saudi Arabia. Saudi J. Biol. Sci. 2021, 28, 73–77. [Google Scholar] [CrossRef]
- Awe, S.; Mikolasch, A.; Hammer, E.; Schauer, F. Degradation of phenylalkanes and characterization of aromatic intermediates acting as growth inhibiting substances in hydrocarbon utilizing yeast Candida maltosa. Int. Biodeterior. Biodegrad. 2008, 62, 408–414. [Google Scholar] [CrossRef]
- Awe, S.; Mikolasch, A.; Schauer, F. Formation of coumarines during the degradation of alkyl substituted aromatic oil components by the yeast Trichosporon asahii. Appl. Microbiol. Biotechnol. 2009, 84, 965–976. [Google Scholar] [CrossRef]
- Prenafeta-Boldú, F.X.; de Hoog, G.S.; Summerbell, R.C. Fungal communities in hydrocarbon degradation. In Microbial Communities Utilizing Hydrocarbons and Lipids: Members, Metagenomics and Ecophysiology; McGenity, T., Ed.; Handbook of Hydrocarbon and Lipid Microbiology; Springer: Cham, Switzerland, 2019; ISBN 978-3-030-14785-3. [Google Scholar] [CrossRef]
- Mikolasch, A.; Berzhanova, R.; Omirbekova, A.; Reinhard, A.; Zühlke, D.; Meister, M.; Mukasheva, T.; Riedel, K.; Urich, T.; Schauer, F. Moniliella spathulata, an oil-degrading yeast, which promotes growth of barley in oil-polluted soil. Appl. Microbiol. Biotechnol. 2021, 105, 401–415. [Google Scholar] [CrossRef]
- Novianty, R.; Saryono, A.; Pratiwi, N.W.; Hidayah, A.; Juliantari, E. The diversity of fungi consortium isolated from polluted soil for degrading petroleum hydrocarbon. Biodiversitas 2021, 22, 5077–5084. [Google Scholar] [CrossRef]
- Horel, A.; Schiewer, S. Microbial degradation of different hydrocarbon fuels with mycoremediation of volatiles. Microorganisms 2020, 8, 163. [Google Scholar] [CrossRef]
- Karatayev, M.; Clarke, M.L. Current energy resources in Kazakhstan and the future potential of renewables: A Review. Energy Procedia 2014, 59, 97–104. [Google Scholar] [CrossRef]
- Issayeva, A.U.; Mametova, A.; Baiduisenova, T.; Kossauova, A.; Zhumakhanova, R.; Zhumadulayeva, A.; Ashirbayeva, S.; Patasheva, A. The Effect of oil pollution of the gray soils on revegetation in the south of Kazakhstan. J. Ecol. Eng. 2023, 24, 28–33. [Google Scholar] [CrossRef]
- Kalia, A.; Sharma, S.; Semor, N.; Babele, P.K.; Sagar, S.; Bhatia, R.K.; Walia, A. Recent advancements in hydrocarbon bioremediation and future challenges: A review. Three Biotech. 2022, 12, 135. [Google Scholar] [CrossRef]
- Mikolasch, A.; Reinhard, A.; Alimbetova, A.; Omirbekova, A.; Pasler, L.; Schumann, P.; Kabisch, J.; Mukasheva, T.; Schauer, F. From oil spills to barley growth—Oil-degrading soil bacteria and their promoting effects. J. Basic. Microbiol. 2016, 56, 1252–1273. [Google Scholar] [CrossRef]
- Mikolasch, A.; Omirbekova, A.; Schumann, P.; Reinhard, A.; Sheikhany, H.; Berzhanova, R.; Mukasheva, T.; Schauer, F. Enrichment of aliphatic, alicyclic and aromatic acids by oil-degrading bacteria isolated from the rhizosphere of plants growing in oil-contaminated soil from Kazakhstan. Appl. Microbiol. Biotechnol. 2015, 99, 4071–4084. [Google Scholar] [CrossRef] [PubMed]
- Dallinger, A.; Duldhardt, I.; Kabisch, J.; Schlüter, R.; Schauer, F. Biotransformation of cyclohexane and related alicyclic hydrocarbons by Candida maltosa and Trichosporon species. Int. Biodeterior. Biodegrad. 2016, 107, 132–139. [Google Scholar] [CrossRef]
- Nhi-Cong, L.T.; Mikolasch, A.; Awe, S.; Sheikhany, H.; Klenk, H.-P.; Schauer, F. Oxidation of aliphatic, branched chain, and aromatic hydrocarbons by Nocardia cyriacigeorgica isolated from oil-polluted sand samples collected in the Saudi Arabian desert. J. Basic Microbiol. 2010, 50, 241–253. [Google Scholar] [CrossRef]
- Stolk; Scott, D.B. Eupenicillium javanicum (J.F.H. Beyma). Verh. K. Akad. Wet. Tweede Sect. 1929, 26, 17. Available online: http://sweetgum.nybg.org/science/vh/specimen_details.php?irn=2998094 (accessed on 4 June 2024).
- Raper, K.B.; Fennell, D.I. New Species of Penicillium. Mycologia 1948, 40, 507–546. [Google Scholar] [CrossRef]
- Liang, Z.-Y.; Shen, N.X.; Zheng, Y.Y.; Wu, J.-T.; Miao, L.; Fu, X.-M.; Chen, M.; Wang, C.-Y. Two new unsaturated fatty acids from the mangrove rhizosphere soil-derived fungus Penicillium javanicum HK1-22. Bioorganic Chem. 2019, 93, 103331. [Google Scholar] [CrossRef]
- Okeke, B.; Seigle-Murandi, F.; Steiman, R.; Benoit-Guyod, J.L.; Kaouadji, M. Identification of mycotoxin-producing fungal strains: A step in the isolation of compounds active against rice fungal diseases J. Agric. Food Chem. 1993, 41, 1731–1735. [Google Scholar] [CrossRef]
- Lai, L.H.; Zong, M.-H.; Huang, Z.; Ni, Z.-F.; Xu, P.; Lou, W.-Y. Purification, structural elucidation and biological activities of exopolysaccharide produced by the endophytic Penicillium javanicum from Millettia speciosa Champ. J. Biotechnol. 2023, 362, 54–62. [Google Scholar] [CrossRef]
- Ikediugwu, F.E.O.; Ejale, A.U. Root-surface mycoflora of cassava (Manihot esculenta) and post harvest rot of the tubers. Mycopathologia 1980, 71, 67–71. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, Z.; Liang, W.; Gao, B.; Wang, Y.; Chang, J.; Zhu, D. Endophytic fungi from Dongxiang wild rice (Oryza rufipogon Griff.) show diverse catalytic potential for converting glycyrrhizin. 3 Biotech 2022, 12, 79. [Google Scholar] [CrossRef]
- Oudot, J.; Dupont, J.; Haloui, S.; Roquebert, M.F. Biodegradation potential of hydrocarbon-assimilating tropical fungi. Soil Biol. Biochem. 1993, 25, 1167–1173. [Google Scholar] [CrossRef]
- Lombard, L.; Sandoval-Denis, M.; Lamprecht, S.C.; Crous, P.W. Epitypification of Fusarium oxysporum—Clearing the taxonomic chaos. Persoonia 2019, 43, 1–47. [Google Scholar] [CrossRef]
- Oudot, J.; Fusey, P.; Abdelouahid, D.E.; Haloui, S.; Roquebert, M.F. Degradation potentials of bacteria and fungi isolated from fuel-contaminated soil. Can. J. Microbiol. 1987, 33, 232–243. [Google Scholar] [CrossRef]
- Pozdnyakova, N.N.; Balandina, S.A.; Turkovskaya, O.V. Degradative activity of fungi towards oil hydrocarbons under high temperature. Theor. Appl. Ecol. 2019, 4, 69–75. [Google Scholar] [CrossRef]
- Pozdnyakova, N.N.; Varese, G.C.; Prigione, V.; Dubrovskaya, E.; Balandina, S.A.; Turkovskaya, O.V. Degradative properties of two newly isolated strains of the ascomycetes Fusarium oxysporum and Lecanicillium aphanocladii. Int. Microbiol. 2019, 22, 103–110. [Google Scholar] [CrossRef]
- Premnath, N.; Mohanrasu, K.; Guru Raj Rao, R.; Dinesh, G.H.; Prakash, G.S.; Ananthi, V.; Ponnuchamy, K.; Muthusamy, G.; Arun, A. A crucial review on polycyclic aromatic hydrocarbons—Environmental occurrence and strategies for microbial degradation. Chemosphere 2021, 280, 130608. [Google Scholar] [CrossRef]
- Silva, I.S.; Grossman, M.; Durranta, L.R. Degradation of polycyclic aromatic hydrocarbons (2–7 rings) under microaerobic and very-low-oxygen conditions by soil fungi. Int. Biodeterior. Biodegrad. 2009, 63, 224–229. [Google Scholar] [CrossRef]
- Jacques, R.J.S.; Okeke, B.C.; Bento, F.M.; Teixeira, A.S.; Peralba, M.C.R.; Camargo, F.A.O. Microbial consortium bioaugmentation of a polycyclic aromatic hydrocarbons contaminated soil. Bioresour. Technol. 2008, 99, 2637–2643. [Google Scholar] [CrossRef]
- Spini, G.; Spina, F.; Poli, A.; Blieux, A.-L.; Regnier, T.; Gramellini, C.; Varese, G.C.; Puglisi, E. Molecular and microbiological insights on the enrichment procedures for the isolation of petroleum degrading bacteria and fungi. Front. Microbiol. 2018, 9, 2543. [Google Scholar] [CrossRef]
- Micheli, P. Nova Plantarum Genera, 1st ed.; Genesis Publications: Frorence, Italy, 1729; ISBN 0904351092. Available online: https://www.genesis-publications.com/book/0904351092/nova-plantarum-genera (accessed on 4 June 2024).
- Thom, C.; Raper, K.B. A Manual of the Aspergilli, 1st ed.; Waverly Press, INC-The Williams & Wilkins Company: Baltimore, MD, USA, 1945; Available online: https://archive.org/download/manualofaspergil00thom/manualofaspergil00thom.pdf (accessed on 5 June 2024).
- Romero, M.S.; Giudicessi, L.S.; Vitale, G.R. Is the fungus Aspergillus a threat to cultural heritage? J. Cult. Herit. 2021, 51, 107–124. [Google Scholar] [CrossRef]
- Raper, K.B.; Fennell, D.I. The Genus Aspergillus; Williams & Wilkins: Baltimore, MD, USA, 1965; ASIN: B01L9WEW4C. [Google Scholar]
- Baker, S.E.; Bennett, J.W. An overview of the genus aspergillus. In The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods; Gustavo, H., Goldman, S.A.O., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 3–13. ISBN 978-0429-129-162. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Fennell, D.I.; Raper, K.B. New species and varieties of Aspergillus. Mycologia 1955, 47, 68–89. [Google Scholar] [CrossRef]
- Kozlowski, M.; Stepien, P.P. Restriction enzyme analysis of mitochondrial DNA of members of the genus Aspergillus as an aid in taxonomy. J. Gen. Microbiol. 1982, 128, 471–476. [Google Scholar] [CrossRef]
- Mageswari, A.; Choi, Y.; Thao, L.D.; Lee, D.; Kim, D.H.; Hong, S.B. Re-identification of Aspergillus subgenus Nidulantes strains and description of three unrecorded species from Korea. Mycobiology 2024, 52, 92–101. [Google Scholar] [CrossRef]
- Visagie, C.M.; Yilmaz, N.; Kocsubé, S.; Frisvad, J.C.; Hubka, V.; Samson, R.A.; Houbraken, J. A review of recently introduced Aspergillus, Penicillium, Talaromyces and other Eurotiales species. Stud. Mycol. 2024, 107, 1–66. [Google Scholar] [CrossRef]
- Volke-Sepúlveda, T.L.; Gutiérrez-Rojas, M.; Favela-Torres, E. Biodegradation of hexadecane in liquid and solid-state fermentations by Aspergillus niger. Bioresour. Technol. 2003, 87, 81–86. [Google Scholar] [CrossRef]
- Volke-Sepúlveda, T.; Gutiérrez-Rojas, M.; Favela-Torres, E. Biodegradation of high concentrations of hexadecane by Aspergillus niger in a solid-state system: Kinetic analysis. Bioresour. Technol. 2006, 97, 1583–1591. [Google Scholar] [CrossRef]
- Al-Hawash, A.B.; Zhang, J.L.; Li, S.; Liu, J.S.; Ghalib, H.B.; Zhang, X.Y.; Ma, F.Y. Biodegradation of n-hexadecane by Aspergillus sp RFC-1 and its mechanism. Ecotoxicol. Environ. Saf. 2018, 164, 398–408. [Google Scholar] [CrossRef]
- Elshafie, A.; AlKindi, A.Y.; Al-Busaidi, S.; Bakheit, C.; Albahry, S.N. Biodegradation of crude oil and n-alkanes by fungi isolated from Oman. Mar. Pollut. Bull. 2007, 54, 1692–1696. [Google Scholar] [CrossRef]
- Barnes, N.M.; Khodse, V.B.; Lotlikar, N.P.; Meena, R.M.; Damare, S.R. Bioremediation potential of hydrocarbon-utilizing fungi from select marine niches of India. 3 Biotech 2018, 8, 21. [Google Scholar] [CrossRef]
- Burghal, A.A.; Abu-Mejdad, N.M.J.A.; Al-Tamimi, W.H. Mycodegradation of crude oil by fungal species isolated from petroleum contaminated soil. Int. J. Innov. Res. Sci. Eng. Technol. 2016, 2, 1517–1524. [Google Scholar]
- Obire, O.; Anyanwu, E.C. Impact of various concentrations of crude oil on fungal populations of soil. Int. J. Environ. Sci. Technol. 2009, 6, 211–218. [Google Scholar] [CrossRef]
- Miles, S.M.; Asiedu, E.; Balaberda, A.-L.; Ulrich, A.C. Oil sands process affected water sourced Trichoderma harzianum demonstrates capacity for mycoremediation of naphthenic acid fraction compounds. Chemosphere 2020, 258, 127281. [Google Scholar] [CrossRef]
- Ferreira, F.V.; Musumeci, M.A. Trichoderma as biological control agent: Scope and prospects to improve efficacy. World J. Microbiol. Biotechnol. 2021, 37, 90. [Google Scholar] [CrossRef]
- Tripathi, P.; Singh, P.C.; Mishra, A.; Chauhan, P.S.; Dwivedi, S.; Bais, R.T.; Tripathi, R.D. Trichoderma: A potential bioremediator for environmental clean up. Clean Technol. Environ. Policy 2013, 15, 541–550. [Google Scholar] [CrossRef]
- Jonathan, S.G.; Asemoloye, M.; Ahmad, R.; Olawuyi, O.J.; Adejoye, D. Response of a Newly Identified Trichoderma harzianum KY488466 to Crude Oil Pollution and Its Expression of Peroxidase Genes. 2017, pp. 1–10. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3089090 (accessed on 12 February 2024).
- Van Beilen, J.B.; Li, Z.; Duetz, W.A.; Smits, T.H.M.; Witholt, B. Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci. Technol. 2003, 58, 427–440. [Google Scholar] [CrossRef]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- Wentzel, A.; Ellingsen, T.E.; Kotlar, H.-K.; Zotchev, S.B.; Throne-Holst, M. Bacterial metabolism of long chain n-alkanes. Appl. Microbiol. Biotechnol. 2007, 76, 1209–1221. [Google Scholar] [CrossRef]
- Palmgren, K.; Ivarssan, M. Mycoremediation of n-alkanes under aerobic conditions—A review. Fungal Interact. 2024, 1, 100001. [Google Scholar] [CrossRef]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [CrossRef]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Interactions of cyclic hydrocarbons with biological membranes. JBC 1994, 269, 8022–8028. [Google Scholar] [CrossRef]
- Amund, O.O.; Ilori, M.O.; Adebusoye, S.A.; Musa, K.I. Utilisation of alicyclic compounds by soil bacteria. Nat. Sci. 2006, 4, 65–68. [Google Scholar]
- Salamanca, D.; Engesser, K.H. Isolation and characterization of two novel strains capable of using cyclohexane as carbon source. Environ. Sci. Pollut. Res. 2014, 21, 12757–12766. [Google Scholar] [CrossRef]
- Schlüter, R.; Dallinger, A.; Kabisch, J.; Duldhardt, I.; Schauer, F. Fungal biotransformation of short-chain n-alkylcycloalkanes. Appl. Microbiol. Biotechnol. 2019, 103, 4137–4151. [Google Scholar] [CrossRef]
- Morgan, P.; Watkinson, R.J. Biodegradation of components of petroleum. In Biochemistry of Microbial Degradation; Ratledge, C., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994; pp. 1–31. [Google Scholar]
- Mandal, D.; Ahmad, A.; Khan, M.I.; Kumar, R. Biocatalytic transformation of cyclohexanone by Fusarium sp. J. Mol. Catal. A Chem. 2002, 181, 237–241. [Google Scholar] [CrossRef]
- Hidayat, A.; Tachibana, S. Crude oil and n-octadecane degradation under saline conditions by Fusarium sp., F092. J. Environ. Sci. Technol. 2013, 6, 29–40. [Google Scholar] [CrossRef][Green Version]


| Isolate * | Scientific Name ** | Abbreviation | Soil Source | Isolation Substrate |
|---|---|---|---|---|
| SBUG-M1743 | Aspergillus sp. | St1 | Park near Almaty train station | Tetradecane |
| SBUG-M1744 | Penicillium javanicum | St2 | Oil deposit “Aktöbe” | |
| SBUG-M1750 | Trichoderma harzianum | St3 | Park near Almaty train station | |
| SBUG-M1746 | Fusarium oxysporum | St4 | Oil depot “Ozen” | Cyclohexanone |
| SBUG-M1748 | Fusarium oxysporum | St5 | Park near Almaty train station | |
| SBUG-M1768 | Fusarium oxysporum | St6 | Oil deposit “Aktöbe” | Crude oil |
| SBUG-M1769 | Fusarium oxysporum | St7 | ||
| SBUG-M1770 | Penicillium javanicum | St8 |
| Products | Retention Time (min) | St1 | St2 | St3 | ||||
|---|---|---|---|---|---|---|---|---|
| Type of Culture | ||||||||
| Shake | Static | Shake | Static | Shake | Static | |||
| P1T | Tetradecanoic acid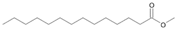 | 32.50 | + | + | + | + | + | + |
| P2T | Dodecanoic acid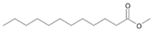 | 26.87 | + | + | + | + | + | + |
| P3T | Decanoic acid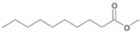 | 20.66 | + | + | - | - | - | + |
| P4T | Hexanedioic acid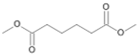 | 18.20 | - | - | - | - | - | - |
| P5T | Octanoic acid | 13.94 | + | + | + | + | - | + |
| P6T | Hexanoic acid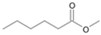 | 7.22 | + | + | - | - | + | - |
| Products | Retention Time (min) | St6 | St7 | ST8 | |
|---|---|---|---|---|---|
| P1T | Tetradecanoic acid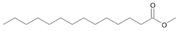 | 32.50 | - | - | - |
| P2T | Dodecanoic acid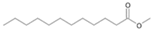 | 26.87 | + | + | + |
| P3T | Decanoic acid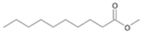 | 20.66 | + | + | + |
| P4T | Hexanedioic acid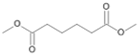 | 18.20 | + | + | - |
| P5T | Octanoic acid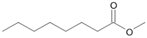 | 13.94 | + | + | + |
| P6T | Hexanoic acid | 7.22 | + | + | - |
| Products | Retention Time (min) | St4 | St5 | |||
|---|---|---|---|---|---|---|
| Type of Culture | ||||||
| Shake | Static | Shake | Static | |||
| P1Con | 6-Hydroxyhexanoic acid *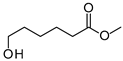 | 22.4 * | + | - | + | - |
| P2Con | Hexanedioic acid *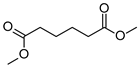 | 18.2 * | + | + | + | + |
| P3Con | ε-Caprolactone **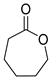 | 8.7 ** | + | + | + | + |
| P4Con | Cyclohexanol ** | 4.4 ** | + | + | + | + |
| Products | Retention Time (min) | Shake | Static | |
|---|---|---|---|---|
| P1C | 6-Hydroxyhexanoic acid *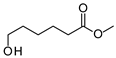 | 22.4 * | + | + |
| P2C | Hexanedioic acid * | 18.2 * | + | + |
| P3C | ε-Caprolactone **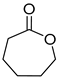 | 8.7 ** | + | + |
| P4C | Cyclohexanol **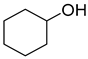 | 4.4 ** | + | + |
| P5C | Cyclohexanone ** | 4.3 ** | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaid, M.; Jentzsch, W.; Beermann, H.; Reinhard, A.; Meister, M.; Berzhanova, R.; Mukasheva, T.; Urich, T.; Mikolasch, A. Comparative Bioremediation of Tetradecane, Cyclohexanone and Cyclohexane by Filamentous Fungi from Polluted Habitats in Kazakhstan. J. Fungi 2024, 10, 436. https://doi.org/10.3390/jof10060436
Gaid M, Jentzsch W, Beermann H, Reinhard A, Meister M, Berzhanova R, Mukasheva T, Urich T, Mikolasch A. Comparative Bioremediation of Tetradecane, Cyclohexanone and Cyclohexane by Filamentous Fungi from Polluted Habitats in Kazakhstan. Journal of Fungi. 2024; 10(6):436. https://doi.org/10.3390/jof10060436
Chicago/Turabian StyleGaid, Mariam, Wiebke Jentzsch, Hannah Beermann, Anne Reinhard, Mareike Meister, Ramza Berzhanova, Togzhan Mukasheva, Tim Urich, and Annett Mikolasch. 2024. "Comparative Bioremediation of Tetradecane, Cyclohexanone and Cyclohexane by Filamentous Fungi from Polluted Habitats in Kazakhstan" Journal of Fungi 10, no. 6: 436. https://doi.org/10.3390/jof10060436
APA StyleGaid, M., Jentzsch, W., Beermann, H., Reinhard, A., Meister, M., Berzhanova, R., Mukasheva, T., Urich, T., & Mikolasch, A. (2024). Comparative Bioremediation of Tetradecane, Cyclohexanone and Cyclohexane by Filamentous Fungi from Polluted Habitats in Kazakhstan. Journal of Fungi, 10(6), 436. https://doi.org/10.3390/jof10060436








