Insights into Fungal Mitochondrial Genomes and Inheritance Based on Current Findings from Yeast-like Fungi
Abstract
:1. Introduction
2. Fungal Mitochondrial Genome Structure
3. The Coding Genes of the Fungal Mitochondrial Genome
4. Mobile Elements in the Fungal Mitochondrial Genome
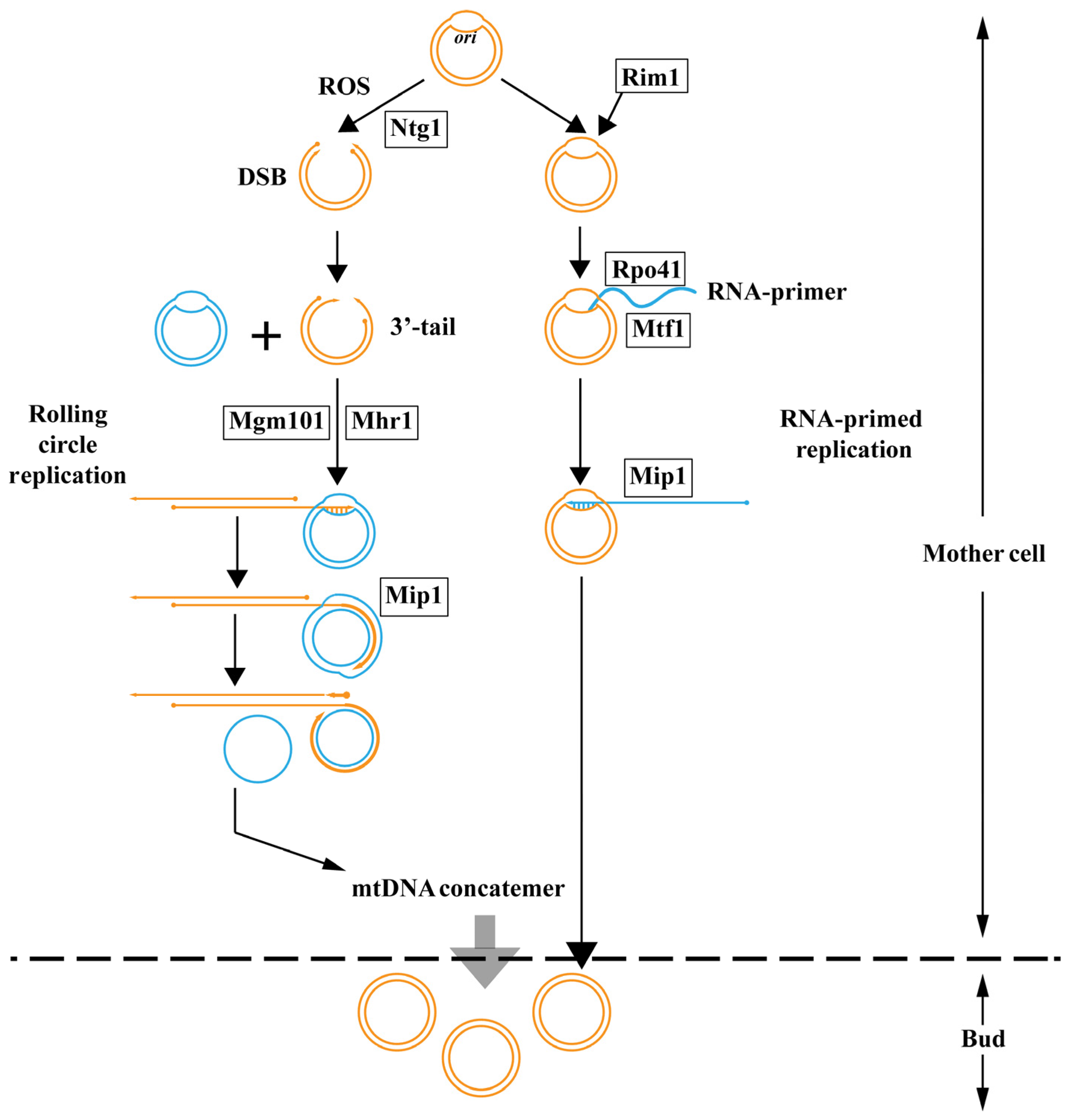
5. The Replication Strategies of Fungal mtDNA
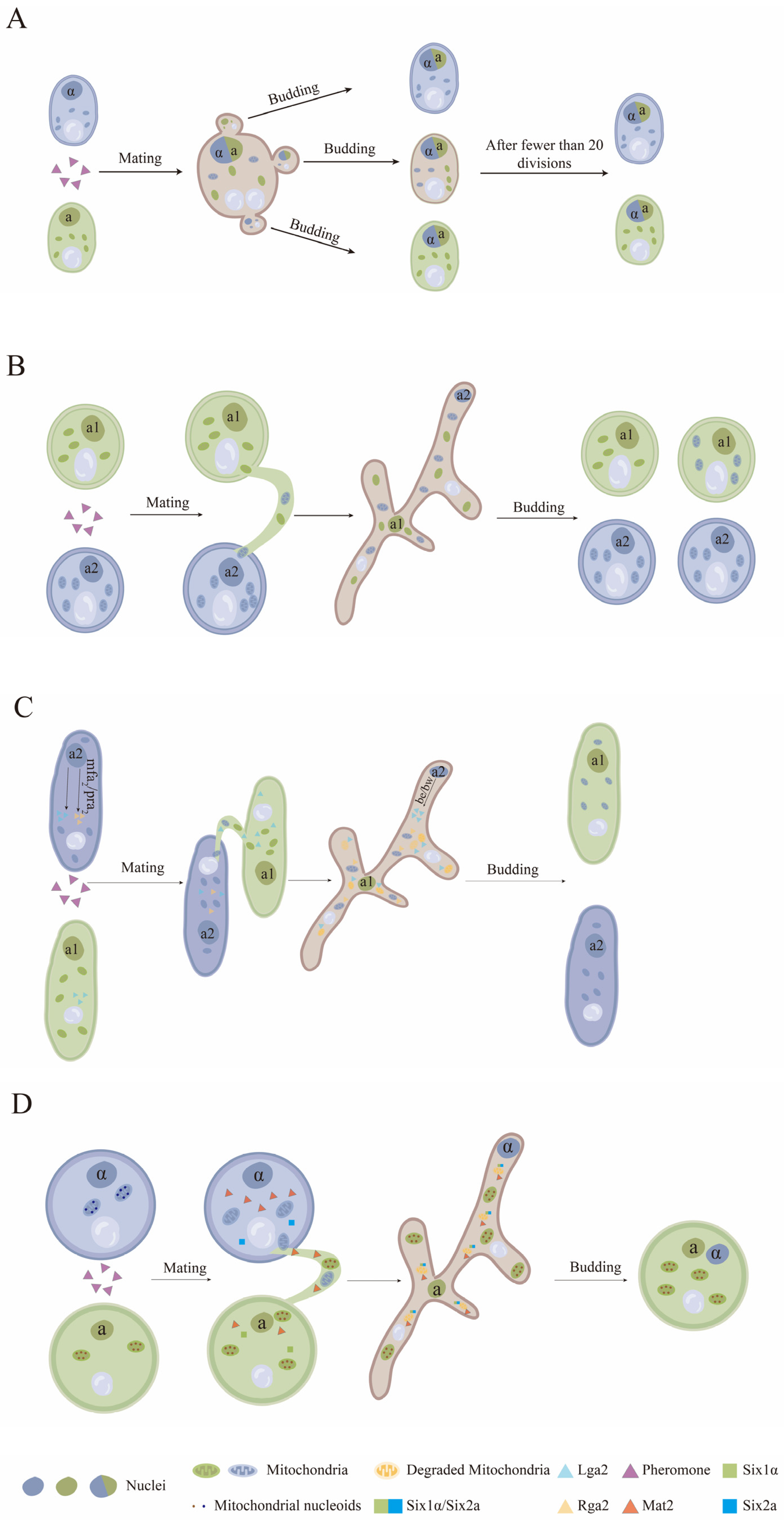
6. The Inheritance of the Fungal Mitochondrial Genome during Bisexual Mating
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284. [Google Scholar] [CrossRef]
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The Origin and Diversification of Mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef]
- Greimann, E.S.; Ward, S.F.; Woodell, J.D.; Hennessey, S.; Kline, M.R.; Moreno, J.A.; Peters, M.; Cruise, J.L.; Montooth, K.L.; Neiman, M.; et al. Phenotypic Variation in Mitochondria-Related Performance Traits Across New Zealand Snail Populations. Integr. Comp. Biol. 2020, 60, 275–287. [Google Scholar] [CrossRef]
- Bonen, L. Mitochondrial genomes: A paradigm of organizational diversity. In Advances in Genome Biology; Verma, R.S., Ed.; Genes and Genomes; JAI: Greenwich, CT, USA, 1998; Volume 5, pp. 415–461. [Google Scholar]
- Kitazaki, K.; Kubo, T. Cost of Having the Largest Mitochondrial Genome: Evolutionary Mechanism of Plant Mitochondrial Genome. J. Bot. 2010, 2010, 620137. [Google Scholar] [CrossRef]
- Nosek, J.; Tomáska, L. Mitochondrial genome diversity: Evolution of the molecular architecture and replication strategy. Curr. Genet. 2003, 44, 73–84. [Google Scholar] [CrossRef]
- Chen, X.J.; Clark-Walker, G.D. Unveiling the mystery of mitochondrial DNA replication in yeasts. Mitochondrion 2018, 38, 17–22. [Google Scholar] [CrossRef]
- Fontana, G.A.; Gahlon, H.L. Mechanisms of replication and repair in mitochondrial DNA deletion formation. Nucleic Acids Res. 2020, 48, 11244–11258. [Google Scholar] [CrossRef]
- Mendoza, H.; Perlin, M.H.; Schirawski, J. Mitochondrial Inheritance in Phytopathogenic Fungi-Everything Is Known, or Is It? Int. J. Mol. Sci. 2020, 21, 3883. [Google Scholar] [CrossRef]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef]
- Hollenberg, C.P.; Borst, P.; van Bruggen, E.F. Mitochondrial DNA. V. A 25 micron closed circular duplex DNA molecule in wild-type yeast mitochondria. Stucture and genetic complexity. Biochim. Biophys. Acta 1970, 209, 1–15. [Google Scholar] [CrossRef]
- Wesolowski, M.; Fukuhara, H. Linear mitochondrial deoxyribonucleic acid from the yeast Hansenula mrakii. Mol. Cell. Biol. 1981, 1, 387–393. [Google Scholar] [CrossRef]
- Rycovska, A.; Valach, M.; Tomaska, L.; Bolotin-Fukuhara, M.; Nosek, J. Linear versus circular mitochondrial genomes: Intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiol. Read. Engl. 2004, 150, 1571–1580. [Google Scholar] [CrossRef]
- Steinert, G.; Firket, H.; Steinert, M. Synthesis of desoxyribonucleic acid in the parabasal body of Trypanosoma mega. Exp. Cell Res. 1958, 15, 632–635. [Google Scholar] [CrossRef]
- Handa, H. Linear plasmids in plant mitochondria: Peaceful coexistences or malicious invasions? Mitochondrion 2008, 8, 15–25. [Google Scholar] [CrossRef]
- O’Hanlon, S.J.; Rieux, A.; Farrer, R.A.; Rosa, G.M.; Waldman, B.; Bataille, A.; Kosch, T.A.; Murray, K.A.; Brankovics, B.; Fumagalli, M.; et al. Recent Asian origin of chytrid fungi causing global amphibian declines. Science 2018, 360, 621–627. [Google Scholar] [CrossRef]
- van de Vossenberg, B.T.L.H.; Brankovics, B.; Nguyen, H.D.T.; van Gent-Pelzer, M.P.E.; Smith, D.; Dadej, K.; Przetakiewicz, J.; Kreuze, J.F.; Boerma, M.; van Leeuwen, G.C.M.; et al. The linear mitochondrial genome of the quarantine chytrid Synchytrium endobioticum; insights into the evolution and recent history of an obligate biotrophic plant pathogen. BMC Evol. Biol. 2018, 18, 136. [Google Scholar] [CrossRef]
- Fonseca, P.L.C.; De-Paula, R.B.; Araújo, D.S.; Tomé, L.M.R.; Mendes-Pereira, T.; Rodrigues, W.F.C.; Del-Bem, L.-E.; Aguiar, E.R.G.R.; Góes-Neto, A. Global Characterization of Fungal Mitogenomes: New Insights on Genomic Diversity and Dynamism of Coding Genes and Accessory Elements. Front. Microbiol. 2021, 12, 787283. [Google Scholar] [CrossRef]
- Kosa, P.; Valach, M.; Tomaska, L.; Wolfe, K.H.; Nosek, J. Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: Insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 2006, 34, 2472–2481. [Google Scholar] [CrossRef]
- Wallen, R.M.; Perlin, M.H. An Overview of the Function and Maintenance of Sexual Reproduction in Dikaryotic Fungi. Front. Microbiol. 2018, 9, 503. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res. 2013, 13, 23–33. [Google Scholar] [CrossRef]
- Franco, M.E.E.; López, S.M.Y.; Medina, R.; Lucentini, C.G.; Troncozo, M.I.; Pastorino, G.N.; Saparrat, M.C.N.; Balatti, P.A. The mitochondrial genome of the plant-pathogenic fungus Stemphylium lycopersici uncovers a dynamic structure due to repetitive and mobile elements. PLoS ONE 2017, 12, e0185545. [Google Scholar] [CrossRef]
- Schikora-Tamarit, M.À.; Marcet-Houben, M.; Nosek, J.; Gabaldón, T. Shared evolutionary footprints suggest mitochondrial oxidative damage underlies multiple complex I losses in fungi. Open Biol. 2021, 11, 200362. [Google Scholar] [CrossRef]
- Lavín, J.L.; Oguiza, J.A.; Ramírez, L.; Pisabarro, A.G. Comparative genomics of the oxidative phosphorylation system in fungi. Fungal Genet. Biol. 2008, 45, 1248–1256. [Google Scholar] [CrossRef]
- Kerscher, S.J. Diversity and origin of alternative NADH:ubiquinone oxidoreductases. Biochim. Biophys. Acta 2000, 1459, 274–283. [Google Scholar] [CrossRef]
- Romero-Aguilar, L.; Vázquez-Meza, H.; Guerra-Sánchez, G.; Luqueño-Bocardo, O.I.; Pardo, J.P. The Mitochondrial Alternative Oxidase in Ustilago maydis Is Not Involved in Response to Oxidative Stress Induced by Paraquat. J. Fungi 2022, 8, 1221. [Google Scholar] [CrossRef]
- Cárdenas-Monroy, C.A.; Pohlmann, T.; Piñón-Zárate, G.; Matus-Ortega, G.; Guerra, G.; Feldbrügge, M.; Pardo, J.P. The mitochondrial alternative oxidase Aox1 is needed to cope with respiratory stress but dispensable for pathogenic development in Ustilago maydis. PLoS ONE 2017, 12, e0173389. [Google Scholar] [CrossRef]
- Mendoza, H.; Culver, C.D.; Lamb, E.A.; Schroeder, L.A.; Khanal, S.; Müller, C.; Schirawski, J.; Perlin, M.H. Identification and Functional Characterization of a Putative Alternative Oxidase (Aox) in Sporisorium reilianum f. sp. zeae. J. Fungi 2022, 8, 148. [Google Scholar] [CrossRef]
- Zaccaron, A.Z.; De Souza, J.T.; Stergiopoulos, I. The mitochondrial genome of the grape powdery mildew pathogen Erysiphe necator is intron rich and exhibits a distinct gene organization. Sci. Rep. 2021, 11, 13924. [Google Scholar] [CrossRef]
- Bietenhader, M.; Martos, A.; Tetaud, E.; Aiyar, R.S.; Sellem, C.H.; Kucharczyk, R.; Clauder-Münster, S.; Giraud, M.-F.; Godard, F.; Salin, B.; et al. Experimental relocation of the mitochondrial ATP9 gene to the nucleus reveals forces underlying mitochondrial genome evolution. PLoS Genet. 2012, 8, e1002876. [Google Scholar] [CrossRef]
- Wai, A.; Hausner, G. The mitochondrial genome of Ophiostoma himal-ulmi and comparison with other fungi causing Dutch elm disease. Can. J. Microbiol. 2021, 67, 584–598. [Google Scholar] [CrossRef]
- Li, W.; Zhang, F.; Gao, L.-Z. SMRT-based mitochondrial genome of the edible mushroom Morchella conica. Mitochondrial DNA Part B Resour. 2020, 5, 3201–3202. [Google Scholar] [CrossRef]
- Paquin, B.; Laforest, M.J.; Forget, L.; Roewer, I.; Wang, Z.; Longcore, J.; Lang, B.F. The fungal mitochondrial genome project: Evolution of fungal mitochondrial genomes and their gene expression. Curr. Genet. 1997, 31, 380–395. [Google Scholar] [CrossRef]
- Turmel, M.; Lemieux, C.; Burger, G.; Lang, B.F.; Otis, C.; Plante, I.; Gray, M.W. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor. Two radically different evolutionary patterns within green algae. Plant Cell 1999, 11, 1717–1730. [Google Scholar] [CrossRef]
- Groot, G.S.; Mason, T.L.; Van Harten-Loosbroek, N. Var1 is associated with the small ribosomal subunit of mitochondrial ribosomes in yeast. Mol. Gen. Genet. MGG 1979, 174, 339–342. [Google Scholar] [CrossRef]
- Burke, J.M.; RajBhandary, U.L. Intron within the large rRNA gene of N. crassa mitochondria: A long open reading frame and a consensus sequence possibly important in splicing. Cell 1982, 31, 509–520. [Google Scholar] [CrossRef]
- Korovesi, A.G.; Ntertilis, M.; Kouvelis, V.N. Mt-rps3 is an ancient gene which provides insight into the evolution of fungal mitochondrial genomes. Mol. Phylogenet. Evol. 2018, 127, 74–86. [Google Scholar] [CrossRef]
- Paquin, B.; Lang, B.F. The mitochondrial DNA of Allomyces macrogynus: The complete genomic sequence from an ancestral fungus. J. Mol. Biol. 1996, 255, 688–701. [Google Scholar] [CrossRef]
- Wai, A.; Shen, C.; Carta, A.; Dansen, A.; Crous, P.W.; Hausner, G. Intron-encoded ribosomal proteins and N-acetyltransferases within the mitochondrial genomes of fungi: Here today, gone tomorrow? Mitochondrial DNA Part DNA Mapp. Seq. Anal. 2019, 30, 573–584. [Google Scholar] [CrossRef]
- Smits, P.; Smeitink, J.A.M.; van den Heuvel, L.P.; Huynen, M.A.; Ettema, T.J.G. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007, 35, 4686–4703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, X.-N.; Zhang, X.-L.; Liu, X.-Z.; Zhang, Y.-J. Complete mitochondrial genome of the endophytic fungus Pestalotiopsis fici: Features and evolution. Appl. Microbiol. Biotechnol. 2017, 101, 1593–1604. [Google Scholar] [CrossRef] [PubMed]
- Aguileta, G.; de Vienne, D.M.; Ross, O.N.; Hood, M.E.; Giraud, T.; Petit, E.; Gabaldón, T. High variability of mitochondrial gene order among fungi. Genome Biol. Evol. 2014, 6, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Hsiang, T.; Li, S.; Lin, L.; Wang, Q.; Chen, Q.; Xie, B.; Ming, R. Comparison of the Mitochondrial Genome Sequences of Six Annulohypoxylon stygium Isolates Suggests Short Fragment Insertions as a Potential Factor Leading to Larger Genomic Size. Front. Microbiol. 2018, 9, 2079. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, X.-Y.; Huang, B. The complete mitochondrial genome of Linnemannia amoeboidea (W. Gams) Vandepol & Bonito (Mortierellales: Mortierellaceae). Mitochondrial DNA Part B Resour. 2022, 7, 374–376. [Google Scholar] [CrossRef]
- Seif, E.R.; Forget, L.; Martin, N.C.; Lang, B.F. Mitochondrial RNase P RNAs in ascomycete fungi: Lineage-specific variations in RNA secondary structure. RNA 2003, 9, 1073–1083. [Google Scholar] [CrossRef]
- Deng, Y.; Zhang, X.; Xie, B.; Lin, L.; Hsiang, T.; Lin, X.; Lin, Y.; Zhang, X.; Ma, Y.; Miao, W.; et al. Intra-specific comparison of mitochondrial genomes reveals host gene fragment exchange via intron mobility in Tremella fuciformis. BMC Genom. 2020, 21, 426. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, P.L.C.; Badotti, F.; De-Paula, R.B.; Araújo, D.S.; Bortolini, D.E.; Del-Bem, L.-E.; Azevedo, V.A.; Brenig, B.; Aguiar, E.R.G.R.; Góes-Neto, A. Exploring the Relationship Among Divergence Time and Coding and Non-coding Elements in the Shaping of Fungal Mitochondrial Genomes. Front. Microbiol. 2020, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Johansen, S.D. Group I introns: Moving in new directions. RNA Biol. 2009, 6, 375–383. [Google Scholar] [CrossRef]
- Stoddard, B.L. Homing endonucleases from mobile group I introns: Discovery to genome engineering. Mob. DNA 2014, 5, 7. [Google Scholar] [CrossRef]
- Wu, B.; Hao, W. Horizontal transfer and gene conversion as an important driving force in shaping the landscape of mitochondrial introns. G3 Genes Genomes Genet. 2014, 4, 605–612. [Google Scholar] [CrossRef]
- Sanchez-Puerta, M.V.; Abbona, C.C.; Zhuo, S.; Tepe, E.J.; Bohs, L.; Olmstead, R.G.; Palmer, J.D. Multiple recent horizontal transfers of the cox1 intron in Solanaceae and extended co-conversion of flanking exons. BMC Evol. Biol. 2011, 11, 277. [Google Scholar] [CrossRef]
- Lang, B.F.; Laforest, M.-J.; Burger, G. Mitochondrial introns: A critical view. Trends Genet. 2007, 23, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, D.; Reeb, V.; Simon, D.M.; Lutzoni, F. Phylogenetic analyses suggest reverse splicing spread of group I introns in fungal ribosomal DNA. BMC Evol. Biol. 2005, 5, 68. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, O.; Zingler, N. Group II introns: Structure, folding and splicing mechanism. Biol. Chem. 2007, 388, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Lambowitz, A.M.; Zimmerly, S. Mobile group II introns. Annu. Rev. Genet. 2004, 38, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, K.; Schmidt, U. Group II introns: Structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 249–303. [Google Scholar] [CrossRef] [PubMed]
- Lambowitz, A.M.; Zimmerly, S. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 2011, 3, a003616. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, A.J.F.; Kraus, S.R.; Barton, R.; Court, D.A.; Myers, C.J.; Bertrand, H. Heterokaryotic transmission of senescence plasmid DNA in Neurospora. Curr. Genet. 1990, 17, 139–145. [Google Scholar] [CrossRef]
- van Diepeningen, A.D.; Debets, A.J.M.; Slakhorst, S.M.; Hoekstra, R.F. Mitochondrial pAL2-1 plasmid homologs are senescence factors in Podospora anserina independent of intrinsic senescence. Biotechnol. J. 2008, 3, 791–802. [Google Scholar] [CrossRef]
- Andrade, B.S.; Góes-Neto, A. Phylogenetic analysis of DNA and RNA polymerases from a Moniliophthora perniciosa mitochondrial plasmid reveals probable lateral gene transfer. Genet. Mol. Res. 2015, 14, 14105–14114. [Google Scholar] [CrossRef]
- Férandon, C.; Xu, J.; Barroso, G. The 135 kbp mitochondrial genome of Agaricus bisporus is the largest known eukaryotic reservoir of group I introns and plasmid-related sequences. Fungal Genet. Biol. 2013, 55, 85–91. [Google Scholar] [CrossRef]
- Nieuwenhuis, M.; Groeneveld, J.; Aanen, D.K. Horizontal transfer of tRNA genes to mitochondrial plasmids facilitates gene loss from fungal mitochondrial DNA. Curr. Genet. 2023, 69, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Maas, M.F.P.M.; de Boer, H.J.; Debets, A.J.M.; Hoekstra, R.F. The mitochondrial plasmid pAL2-1 reduces calorie restriction mediated life span extension in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 2004, 41, 865–871. [Google Scholar] [CrossRef]
- Maas, M.F.P.M.; van Mourik, A.; Hoekstra, R.F.; Debets, A.J.M. Polymorphism for pKALILO based senescence in Hawaiian populations of Neurospora intermedia and Neurospora tetrasperma. Fungal Genet. Biol. 2005, 42, 224–232. [Google Scholar] [CrossRef]
- Lecrenier, N.; Foury, F. New features of mitochondrial DNA replication system in yeast and man. Gene 2000, 246, 37–48. [Google Scholar] [CrossRef]
- Clark-Walker, G.D.; McArthur, C.R.; Sriprakash, K.S. Location of transcriptional control signals and transfer RNA sequences in Torulopsis glabrata mitochondrial DNA. EMBO J. 1985, 4, 465–473. [Google Scholar] [CrossRef]
- Sekito, T.; Okamoto, K.; Kitano, H.; Yoshida, K. The complete mitochondrial DNA sequence of Hansenula wingei reveals new characteristics of yeast mitochondria. Curr. Genet. 1995, 28, 39–53. [Google Scholar] [CrossRef]
- Fangman, W.L.; Henly, J.W.; Brewer, B.J. RPO41-independent maintenance of [rho-] mitochondrial DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990, 10, 10–15. [Google Scholar] [CrossRef]
- Gerhold, J.M.; Aun, A.; Sedman, T.; Jõers, P.; Sedman, J. Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol. Cell 2010, 39, 851–861. [Google Scholar] [CrossRef]
- Gerhold, J.M.; Sedman, T.; Visacka, K.; Slezakova, J.; Tomaska, L.; Nosek, J.; Sedman, J. Replication intermediates of the linear mitochondrial DNA of Candida parapsilosis suggest a common recombination based mechanism for yeast mitochondria. J. Biol. Chem. 2014, 289, 22659–22670. [Google Scholar] [CrossRef]
- Ling, F.; Shibata, T. Recombination-dependent mtDNA partitioning: In vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J. 2002, 21, 4730–4740. [Google Scholar] [CrossRef]
- Ling, F.; Hori, A.; Shibata, T. DNA recombination-initiation plays a role in the extremely biased inheritance of yeast [rho-] mitochondrial DNA that contains the replication origin ori5. Mol. Cell. Biol. 2007, 27, 1133–1145. [Google Scholar] [CrossRef] [PubMed]
- Nardozzi, J.D.; Wang, X.; Mbantenkhu, M.; Wilkens, S.; Chen, X.J. A properly configured ring structure is critical for the function of the mitochondrial DNA recombination protein, Mgm101. J. Biol. Chem. 2012, 287, 37259–37268. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Ling, F. DNA recombination protein-dependent mechanism of homoplasmy and its proposed functions. Mitochondrion 2007, 7, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Kaniak-Golik, A.; Skoneczna, A. Mitochondria-nucleus network for genome stability. Free Radic. Biol. Med. 2015, 82, 73–104. [Google Scholar] [CrossRef] [PubMed]
- Hayles, J.; Nurse, P. Genetics of the fission yeast Schizosaccharomyces pombe. Annu. Rev. Genet. 1992, 26, 373–402. [Google Scholar] [CrossRef] [PubMed]
- Birky, C.W. The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Annu. Rev. Genet. 2001, 35, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Xu, J. Mitochondrial inheritance: Diverse patterns and mechanisms with an emphasis on fungi. Mycology 2012, 3, 158–166. [Google Scholar] [CrossRef]
- Xu, J.; Wang, P. Mitochondrial inheritance in basidiomycete fungi. Fungal Biol. Rev. 2015, 29, 209–219. [Google Scholar] [CrossRef]
- Yan, Z.; Sun, S.; Shahid, M.; Xu, J. Environment factors can influence mitochondrial inheritance in the fungus Cryptococcus neoformans. Fungal Genet. Biol. 2007, 44, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wilson, A.; Xu, J. Mitochondrial DNA inheritance in the human fungal pathogen Cryptococcus gattii. Fungal Genet. Biol. 2015, 75, 1–10. [Google Scholar] [CrossRef]
- Chen, X.J.; Butow, R.A. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005, 6, 815–825. [Google Scholar] [CrossRef]
- Birky, C.W.; Demko, C.A.; Perlman, P.S.; Strausberg, R. Uniparental inheritance of mitochondrial genes in yeast: Dependence on input bias of mitochondrial DNA and preliminary investigations of the mechanism. Genetics 1978, 89, 615–651. [Google Scholar] [CrossRef]
- Yockteng, R.; Marthey, S.; Chiapello, H.; Gendrault, A.; Hood, M.E.; Rodolphe, F.; Devier, B.; Wincker, P.; Dossat, C.; Giraud, T. Expressed sequences tags of the anther smut fungus, Microbotryum violaceum, identify mating and pathogenicity genes. BMC Genom. 2007, 8, 272. [Google Scholar] [CrossRef]
- Wilch, G.; Ward, S.; Castle, A. Transmission of mitochondrial DNA in Ustilago violacea. Curr. Genet. 1992, 22, 135–140. [Google Scholar] [CrossRef]
- Kronstad, J.W.; Leong, S.A. The b mating-type locus of Ustilago maydis contains variable and constant regions. Genes Dev. 1990, 4, 1384–1395. [Google Scholar] [CrossRef]
- Gillissen, B.; Bergemann, J.; Sandmann, C.; Schroeer, B.; Bölker, M.; Kahmann, R. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 1992, 68, 647–657. [Google Scholar] [CrossRef]
- Fedler, M.; Luh, K.-S.; Stelter, K.; Nieto-Jacobo, F.; Basse, C.W. The a2 mating-type locus genes lga2 and rga2 direct uniparental mitochondrial DNA (mtDNA) inheritance and constrain mtDNA recombination during sexual development of Ustilago maydis. Genetics 2009, 181, 847–860. [Google Scholar] [CrossRef]
- Nieto-Jacobo, F.; Pasch, D.; Basse, C.W. The mitochondrial Dnm1-like fission component is required for lga2-induced mitophagy but dispensable for starvation-induced mitophagy in Ustilago maydis. Eukaryot. Cell 2012, 11, 1154–1166. [Google Scholar] [CrossRef]
- Yadav, V.; Sun, S.; Heitman, J. Uniparental nuclear inheritance following bisexual mating in fungi. eLife 2021, 10, e66234. [Google Scholar] [CrossRef]
- Matha, A.R.; Lin, X. Current Perspectives on Uniparental Mitochondrial Inheritance in Cryptococcus neoformans. Pathogens 2020, 9, 743. [Google Scholar] [CrossRef]
- Yan, Z.; Xu, J. Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics 2003, 163, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, R.; Lin, X. Prezygotic and postzygotic control of uniparental mitochondrial DNA inheritance in Cryptococcus neoformans. mBio 2013, 4, e00112–e00113. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Fu, C.; Ianiri, G.; Heitman, J. The Pheromone and Pheromone Receptor Mating-Type Locus Is Involved in Controlling Uniparental Mitochondrial Inheritance in Cryptococcus. Genetics 2020, 214, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Shikanai, T.; Kawamoto, S.; Toh-E, A. Step-wise elimination of α-mitochondrial nucleoids and mitochondrial structure as a basis for the strict uniparental inheritance in Cryptococcus neoformans. Sci. Rep. 2020, 10, 2468. [Google Scholar] [CrossRef] [PubMed]
| Eukaryotes | Mitochondrial Genome Size | Structure | Characterization of Genes in Mitochondrial Genomes |
|---|---|---|---|
| Animals | 10–50 Kb | Circular | Highly conserved set of genes with a few intergenic regions and introns |
| Plants | 60 Kb–12 Mb | Circular (most) or linear (a small amount) | A large number of genes with large intergenic repetitions and a variable number of introns |
| Fungi | 10–350 Kb | Circular or linear | Highly diverse sets of genes with various sequences of intergenic regions and types of introns |
| Phylum | Number of Published Fungal Mitochondrial Genomes | Mitochondrial Genome Size (bp) |
|---|---|---|
| Ascomycota | 693 | 11,198–272,497 |
| Basidiomycota | 403 | 13,032–343,690 |
| Chytridiomycota | 180 | 19,473–225,604 |
| Zygomycota | 30 | 26,612–83,361 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Zhang, L.; Su, J.; Ye, Q.; Li, Y.; Liu, D.; Cui, H.; Zhang, Y.; Ye, Z. Insights into Fungal Mitochondrial Genomes and Inheritance Based on Current Findings from Yeast-like Fungi. J. Fungi 2024, 10, 441. https://doi.org/10.3390/jof10070441
Tang J, Zhang L, Su J, Ye Q, Li Y, Liu D, Cui H, Zhang Y, Ye Z. Insights into Fungal Mitochondrial Genomes and Inheritance Based on Current Findings from Yeast-like Fungi. Journal of Fungi. 2024; 10(7):441. https://doi.org/10.3390/jof10070441
Chicago/Turabian StyleTang, Jintian, Leilei Zhang, Jinghan Su, Qingwen Ye, Yukang Li, Dinghang Liu, Haifeng Cui, Yafen Zhang, and Zihong Ye. 2024. "Insights into Fungal Mitochondrial Genomes and Inheritance Based on Current Findings from Yeast-like Fungi" Journal of Fungi 10, no. 7: 441. https://doi.org/10.3390/jof10070441






